Prognostic Significance of Amino Acid Metabolism-Related Genes in Prostate Cancer Retrieved by Machine Learning
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Preparation and Differential Gene Expression Analysis
2.2. Functional Enrichment Analysis
2.3. Survival Analysis
2.4. Kaplan–Meier Survival Estimate
3. Results
3.1. Prostate Cancer Amino Acid Metabolism-Related Gene Expression Appears to Be Highly Aberrant
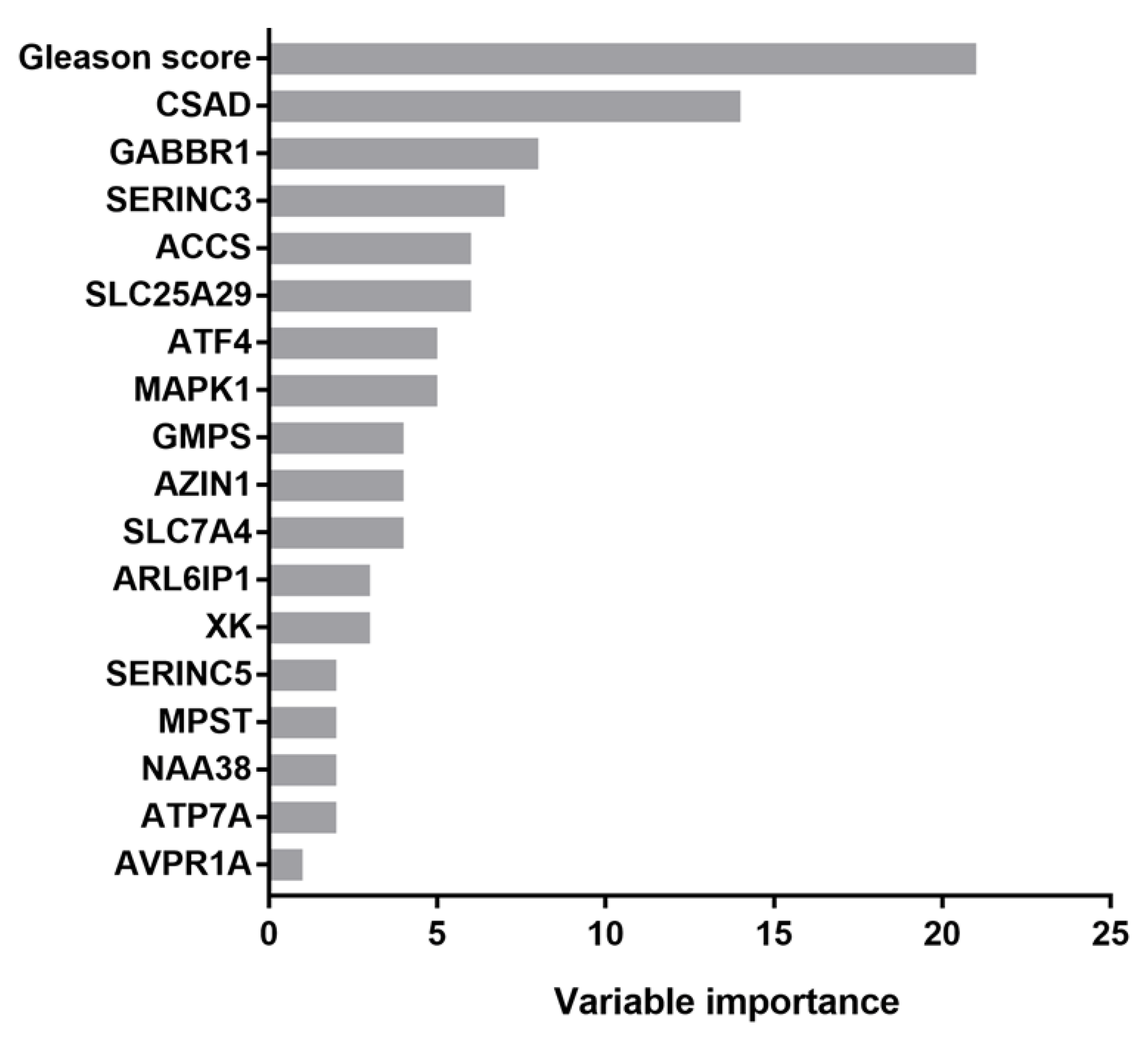
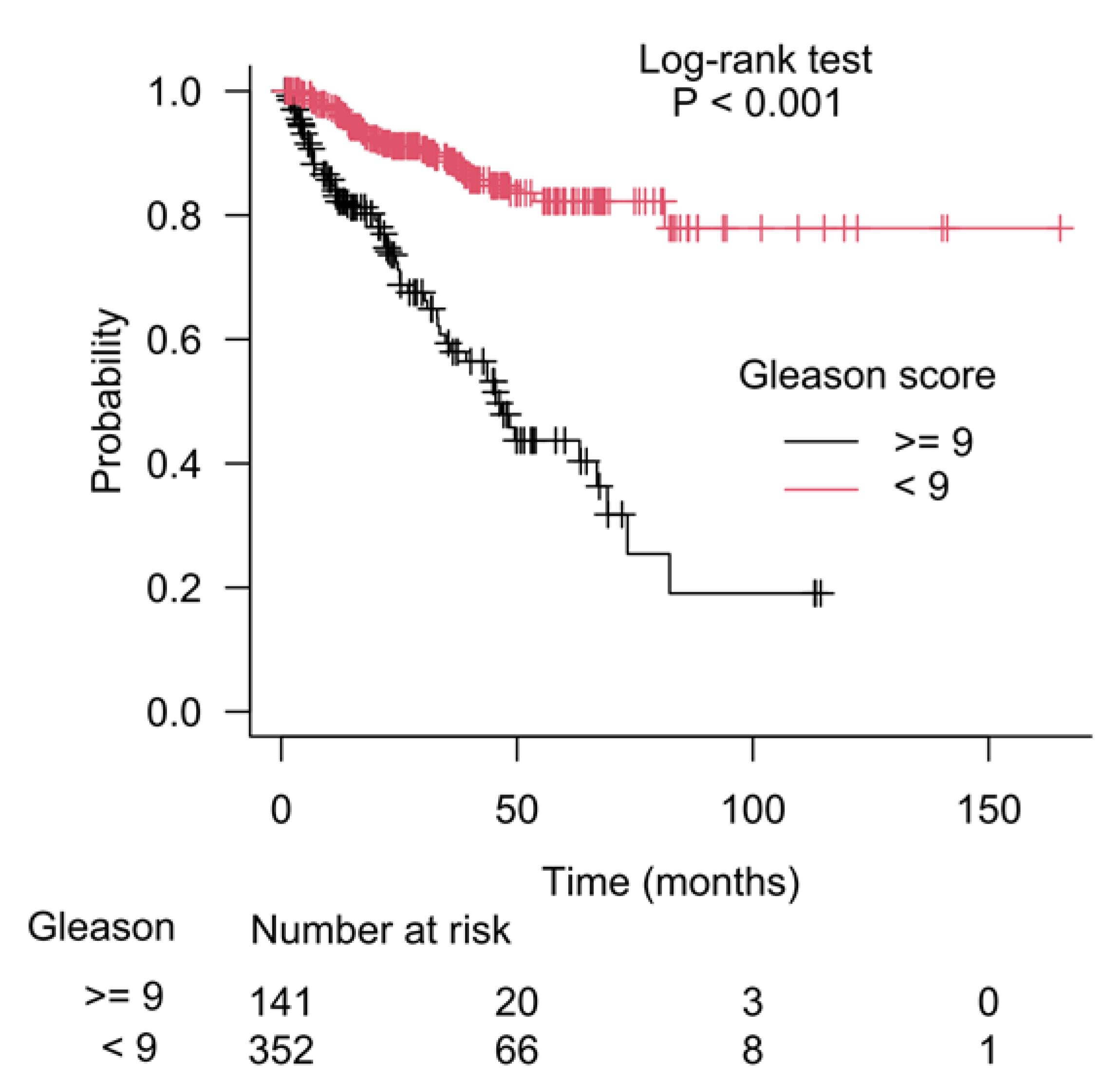
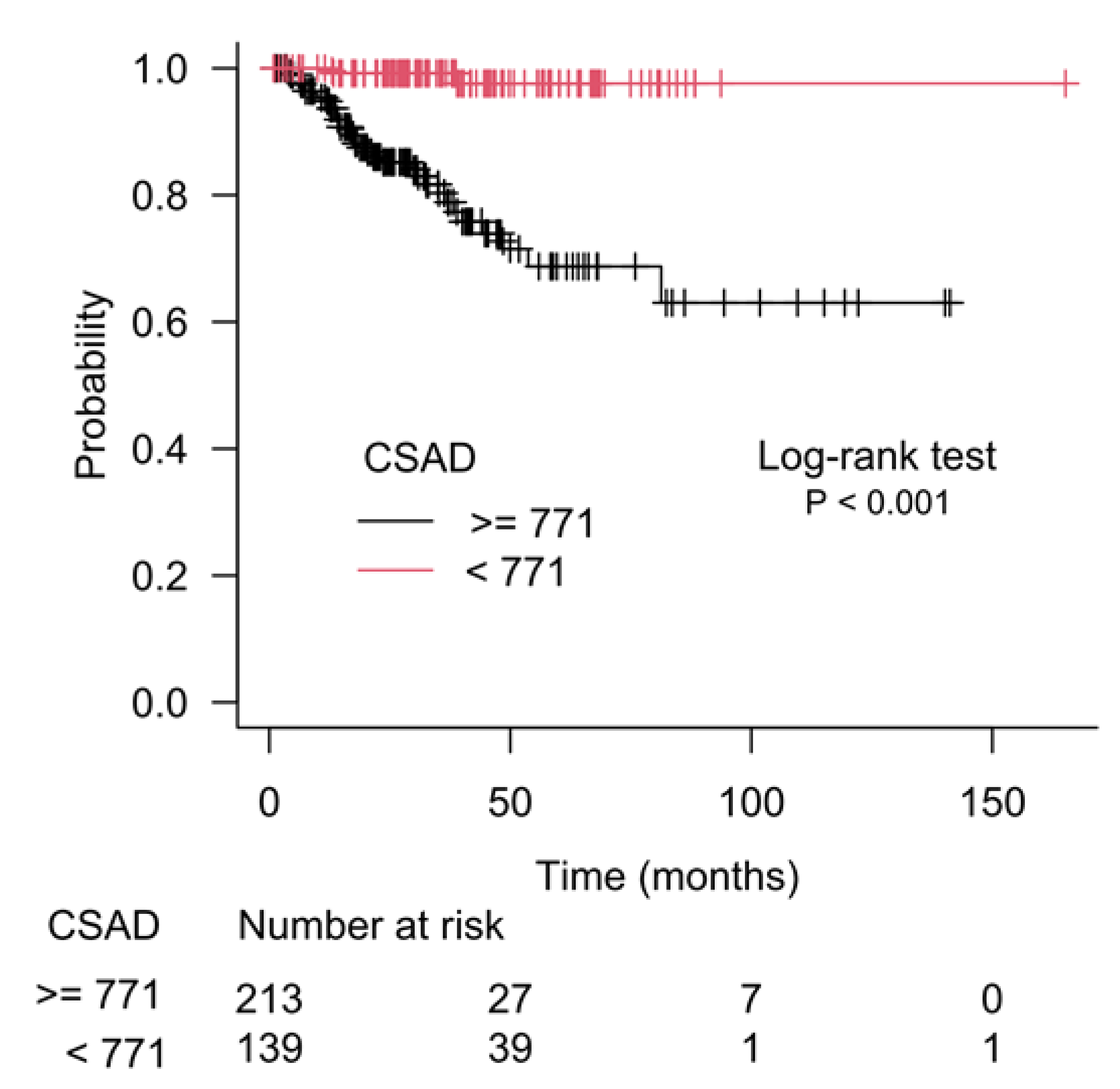
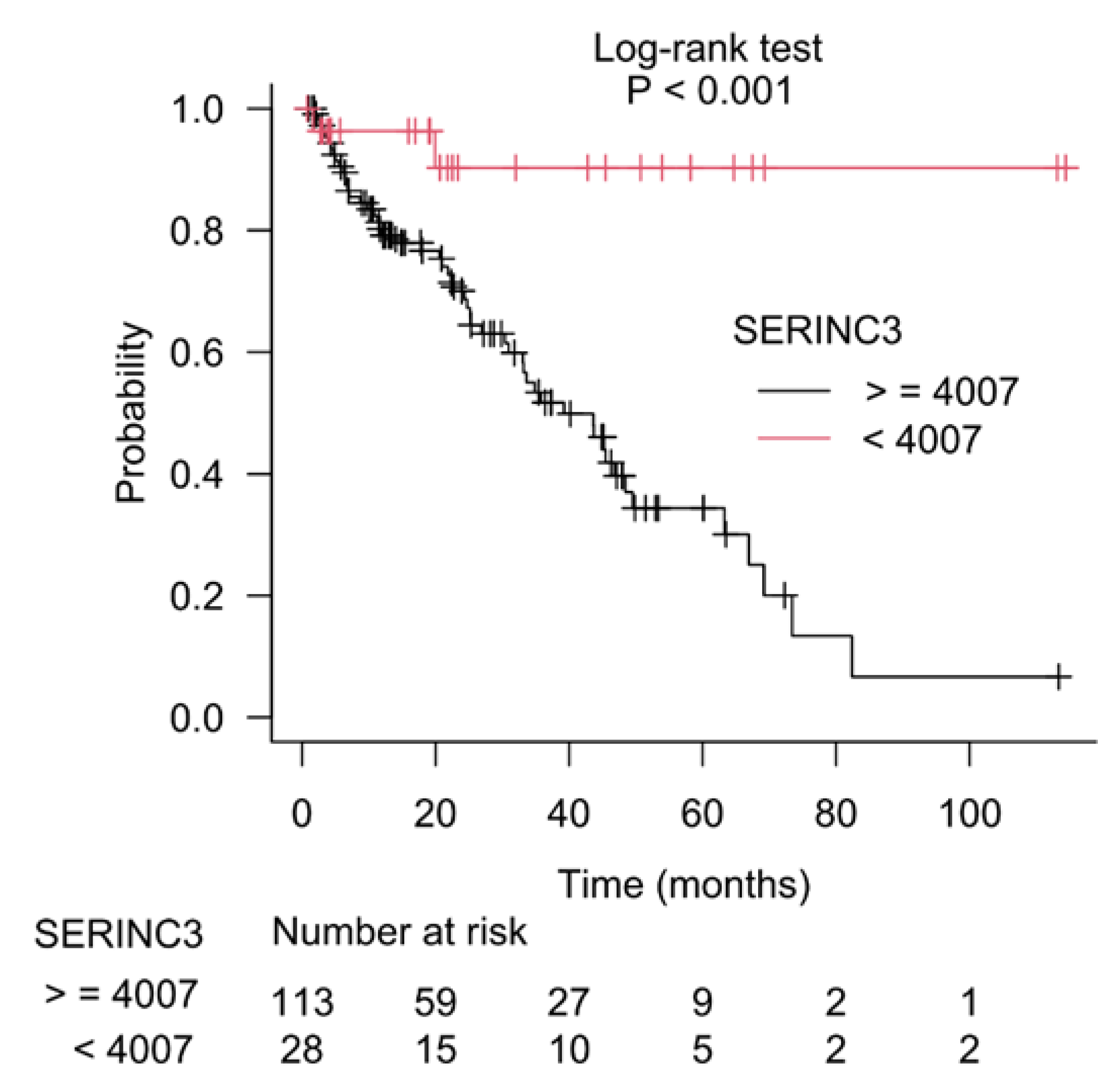
3.2. CSAD and SERINC3 Genes Further Refine the Prognostic Value of the Gleason Score in Prostate Cancer
3.3. Kaplan–Meier Estimate on Prostate Cancer Patients Stratified According to Gleason Score and CSAD and SERINC3 Expression
4. Discussion
4.1. Metabolites and Metabolism-Related Genes in the Prognosis of Prostate Cancer
4.2. Differentially Expressed Amino Acid Metabolism-Related Genes in Prostate Cancer
4.3. Prognostic Value of Amino Acid Metabolism-Related Genes in Prostate Cancer
4.4. Methodological Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Zhao, Y.; Tao, Z.; Li, L.; Zheng, J.; Chen, X. Predicting Biochemical-Recurrence-Free Survival Using a Three-Metabolic-Gene Risk Score Model in Prostate Cancer Patients. BMC Cancer 2022, 22, 239. [Google Scholar] [CrossRef]
- van den Broeck, T.; van den Bergh, R.C.N.; Briers, E.; Cornford, P.; Cumberbatch, M.; Tilki, D.; de Santis, M.; Fanti, S.; Fossati, N.; Gillessen, S.; et al. Biochemical Recurrence in Prostate Cancer: The European Association of Urology Prostate Cancer Guidelines Panel Recommendations. Eur. Urol. Focus 2020, 6, 231–234. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Zadra, G.; Loda, M. Metabolic Vulnerabilities of Prostate Cancer: Diagnostic and Therapeutic Opportunities. Cold Spring Harb Perspect Med 2018, 8, a030569. [Google Scholar] [CrossRef]
- Eidelman, E.; Twum-Ampofo, J.; Ansari, J.; Siddiqui, M.M. The Metabolic Phenotype of Prostate Cancer. Front. Oncol. 2017, 7, 131. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, F.; Cherukuri, M.K.; Choyke, P.L. Metabolic Reprogramming in Prostate Cancer. Br. J. Cancer 2021, 125, 1185–1196. [Google Scholar] [CrossRef]
- Chetta, P.; Zadra, G. Metabolic Reprogramming as an Emerging Mechanism of Resistance to Endocrine Therapies in Prostate Cancer. Cancer Drug Resist. 2021, 4, 143–162. [Google Scholar] [CrossRef]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino Acids in Cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef]
- Yoo, H.C.; Han, J.M. Amino Acid Metabolism in Cancer Drug Resistance. Cells 2022, 11, 140. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2021, 8, xxx. [Google Scholar] [CrossRef]
- Butler, M.; van der Meer, L.T.; van Leeuwen, F.N. Amino Acid Depletion Therapies: Starving Cancer Cells to Death. Trends Endocrinol. Metab. 2021, 32, 367–381. [Google Scholar] [CrossRef]
- Strmiska, V.; Michalek, P.; Eckschlager, T.; Stiborova, M.; Adam, V.; Krizkova, S.; Heger, Z. Prostate Cancer-Specific Hallmarks of Amino Acids Metabolism: Towards a Paradigm of Precision Medicine. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 248–258. [Google Scholar] [CrossRef]
- Schcolnik-Cabrera, A.; Juárez-López, D. Dual Contribution of the MTOR Pathway and of the Metabolism of Amino Acids in Prostate Cancer. Cell Oncol. 2022, 45, 831–859. [Google Scholar] [CrossRef]
- Yang, M.; Vousden, K.H. Serine and One-Carbon Metabolism in Cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef]
- Pan, S.; Fan, M.; Liu, Z.; Li, X.; Wang, H. Serine, Glycine and One-Carbon Metabolism in Cancer (Review). Int. J. Oncol. 2021, 58, 158–170. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Linares, J.F.; Duran, A.; Cordes, T.; L’Hermitte, A.; Badur, M.G.; Bhangoo, M.S.; Thorson, P.K.; Richards, A.; Rooslid, T.; et al. Increased Serine and One-Carbon Pathway Metabolism by PKCλ/ι Deficiency Promotes Neuroendocrine Prostate Cancer. Cancer Cell 2019, 35, 385–400.e9. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Locasale, J.W.; Reid, M.A. Serine and Methionine Metabolism: Vulnerabilities in Lethal Prostate Cancer. Cancer Cell 2019, 35, 339–341. [Google Scholar] [CrossRef] [Green Version]
- Ganini, C.; Amelio, I.; Bertolo, R.; Candi, E.; Cappello, A.; Cipriani, C.; Mauriello, A.; Marani, C.; Melino, G.; Montanaro, M.; et al. Serine and One-Carbon Metabolisms Bring New Therapeutic Venues in Prostate Cancer. Discov. Oncol. 2021, 12, 45. [Google Scholar] [CrossRef]
- Ndaru, E.; Garibsingh, R.A.A.; Shi, Y.Y.; Wallace, E.; Zakrepine, P.; Wang, J.; Schlessinger, A.; Grewer, C. Novel Alanine Serine Cysteine Transporter 2 (ASCT2) Inhibitors Based on Sulfonamide and Sulfonic Acid Ester Scaffolds. J. Gen. Physiol. 2019, 151, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Saruta, M.; Takahara, K.; Yoshizawa, A.; Niimi, A.; Takeuchi, T.; Nukaya, T.; Takenaka, M.; Zennami, K.; Ichino, M.; Sasaki, H.; et al. Alanine-Serine-Cysteine Transporter 2 Inhibition Suppresses Prostate Cancer Cell Growth In Vitro. J. Clin. Med. 2022, 11, 5466. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Console, L.; Losso, M.A.; Indiveri, C. The Human SLC1A5 (ASCT2) Amino Acid Transporter: From Function to Structure and Role in Cell Biology. Front. Cell Dev. Biol. 2018, 6, 96. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Kim, Y.S.; Choi, E.J.; Hwang, Y.J.; Yun, Y.S.; Bae, S.M.; Park, P.J.; Kim, E.K. Taurine Attenuates Epithelial-Mesenchymal Transition-Related Genes in Human Prostate Cancer Cells. Adv. Exp. Med. Biol. 2017, 975, 1203–1212. [Google Scholar] [CrossRef]
- Song, X.; Yuan, B.; Zhao, S.; Zhao, D. Effect of Taurine on the Proliferation, Apoptosis and MST1/Hippo Signaling in Prostate Cancer Cells. Transl. Cancer Res. 2022, 11, 1705–1712. [Google Scholar] [CrossRef]
- Tang, Y.; Choi, E.J.; Cheong, S.H.; Hwang, Y.J.; Arokiyaraj, S.; Park, P.J.; Moon, S.H.; Kim, E.K. Effect of Taurine on Prostate-Specific Antigen Level and Migration in Human Prostate Cancer Cells. Adv. Exp. Med. Biol. 2015, 803, 203–214. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Sander, C.; Stuart, J.M.; Chang, K.; Creighton, C.J.; et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Silva, T.C.; Colaprico, A.; Olsen, C.; D’Angelo, F.; Bontempi, G.; Ceccarelli, M.; Noushmehr, H. TCGA Workflow: Analyze Cancer Genomics and Epigenomics Data Using Bioconductor Packages. F1000 Res. 2016, 5, 1542. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e17. [Google Scholar] [CrossRef]
- Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New Functionalities in the TCGAbiolinks Package for the Study and Integration of Cancer Data from GDC and GTEX. PLoS Comput. Biol. 2019, 15. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Heath, A.P.; Ferretti, V.; Agrawal, S.; An, M.; Angelakos, J.C.; Arya, R.; Bajari, R.; Baqar, B.; Barnowski, J.H.B.; Burt, J.; et al. The NCI Genomic Data Commons. Nat. Genet. 2021, 53, 257–262. [Google Scholar] [CrossRef]
- Narayanachar Tattar, P.; Vaman, H.J. Survival Analysis; CRC Press: Boca Raton, FL, USA, 2023. [Google Scholar]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Iny Stein, T.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 2016. [Google Scholar] [CrossRef]
- R Core Team. R Core Team; R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: http://www.R-project.org/ (accessed on 1 November 2022).
- Atkinson, E.J.; Therneau, T.M. An Introduction to Recursive Partitioning Using the RPART Routines. Mayo Clin. 2000, 61, xxx. [Google Scholar]
- Therneau, T.; Atkinson, B. Rpart: Recursive Partitioning and Regression Trees. R Package Version 4.1-15. 2019. Available online: https://CRAN.R-project.org/package=rpart (accessed on 1 November 2022).
- Ulm, K.; Kriner, M.; Eberle, S.; Reck, M.; Hessler, S. Statistical Methods to Identify Predictive Factors. In Handbook of Statistics in Clinical Oncology; Crowley, J., Ankerst, D., Eds.; Chapman & Hall/CRC: Boca Raton, FL, USA, 2006; pp. 335–345. [Google Scholar]
- Schumacher, M.; Hollander, N.; Schwarzer, G.; Sauerbrei, W. Prognostic Factor Studies. In Handbook of Statistics in Clinical Oncology; Crowley, J., Ankerst, D., Eds.; Chapman & Hall/CRC: Boca Raton, FL, USA, 2006; pp. 289–333. [Google Scholar]
- Aralica, G.; Šarec Ivelj, M.; Pačić, A.; Baković, J.; Milković Periša, M.; Krištić, A.; Konjevoda, P. Prognostic Significance of Lacunarity in Preoperative Biopsy of Colorectal Cancer. Pathol. Oncol. Res. 2020, 26, 2567–2576. [Google Scholar] [CrossRef]
- Rowe, P. Essential Statistics for the Pharmaceutical Sciences; Wiley Online Library: Hoboken, NJ, USA, 2015. [Google Scholar]
- Kanda, Y. Investigation of the Freely Available Easy-to-Use Software “EZR” for Medical Statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Solorzano, S.R.; Imaz-Rosshandler, I.; Camacho-Arroyo, I.; García-Tobilla, P.; Morales-Montor, G.; Salazar, P.; Arena-Ortiz, M.L.; Rodríguez-Dorantes, M. GABA Promotes Gastrin-Releasing Peptide Secretion in NE/NE-like Cells: Contribution to Prostate Cancer Progression. Sci. Rep. 2018, 8, 10272. [Google Scholar] [CrossRef] [Green Version]
- Testa, U.; Castelli, G.; Pelosi, E. Cellular and Molecular Mechanisms Underlying Prostate Cancer Development: Therapeutic Implications. Medicines 2019, 6, 82. [Google Scholar] [CrossRef] [Green Version]
- Samaržija, I. Post-Translational Modifications That Drive Prostate Cancer Progression. Biomolecules 2021, 11, 247. [Google Scholar] [CrossRef]
- Samaržija, I. Site-Specific and Common Prostate Cancer Metastasis Genes as Suggested by Meta-Analysis of Gene Expression Data. Life 2021, 11, 636. [Google Scholar] [CrossRef]
- Samaržija, I. A Need for Stratification of Metastasis Samples According to Secondary Site in Gene Expression Studies. Biocell 2022, 46, 1747–1750. [Google Scholar] [CrossRef]
- Kelly, R.S.; Heiden, M.G.V.; Giovannucci, E.; Mucci, L.A. Metabolomic Biomarkers of Prostate Cancer: Prediction, Diagnosis, Progression, Prognosis, and Recurrence. Cancer Epidemiol. Biomark. Prev. 2016, 25, 887–906. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, R.; Liang, F.; Zhang, L.; Liang, X. Identification of Metabolism-Associated Prostate Cancer Subtypes and Construction of a Prognostic Risk Model. Front Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, X.; Zhang, L.; Wang, D. Metabolic Characterization and Metabolism-Score of Tumor to Predict the Prognosis in Prostate Cancer. Sci. Rep. 2021, 11, 22486. [Google Scholar] [CrossRef]
- Feng, D.; Shi, X.; Zhang, F.; Xiong, Q.; Wei, Q.; Yang, L. Energy Metabolism-Related Gene Prognostic Index Predicts Biochemical Recurrence for Patients With Prostate Cancer Undergoing Radical Prostatectomy. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef]
- Peng, X.; Zheng, T.; Guo, Y.; Zhu, Y. Amino Acid Metabolism Genes Associated with Immunotherapy Responses and Clinical Prognosis of Colorectal Cancer. Front. Mol. Biosci. 2022, 9. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, J.; Wang, S.; Jiang, Q.; Xu, K. Identification and Validation of a Nine-Gene Amino Acid Metabolism-Related Risk Signature in HCC. Front Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Zhang, F.; Lin, J.; Zhu, D.; Tang, Y.; Lu, Y.; Liu, Z.; Wang, X. Identification of an Amino Acid Metabolism-Associated Gene Signature Predicting the Prognosis and Immune Therapy Response of Clear Cell Renal Cell Carcinoma. Front. Oncol. 2022, 12. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Chai, R.C.; Wang, Y.Z.; Wang, Z.; Liu, X.; Wu, F.; Jiang, T. Amino Acid Metabolism-Related Gene Expression-Based Risk Signature Can Better Predict Overall Survival for Glioma. Cancer Sci. 2019, 110, 321–333. [Google Scholar] [CrossRef]
- Li, W.; Zou, Z.; An, N.; Wang, M.; Liu, X.; Mei, Z. A Multifaceted and Feasible Prognostic Model of Amino Acid Metabolism-Related Genes in the Immune Response and Tumor Microenvironment of Head and Neck Squamous Cell Carcinomas. Front. Oncol. 2022, 12. [Google Scholar] [CrossRef]
- Xu, M.; Sakamoto, S.; Matsushima, J.; Kimura, T.; Ueda, T.; Mizokami, A.; Kanai, Y.; Ichikawa, T. Up-Regulation of LAT1 during Antiandrogen Therapy Contributes to Progression in Prostate Cancer Cells. J. Urol. 2016, 195, 1588–1597. [Google Scholar] [CrossRef] [Green Version]
- Cordova, R.A.; Misra, J.; Amin, P.H.; Klunk, A.J.; Damayanti, N.P.; Carlson, K.R.; Elmendorf, A.J.; Kim, H.-G.; Mirek, E.T.; Elzey, B.D.; et al. GCN2 EIF2 Kinase Promotes Prostate Cancer by Maintaining Amino Acid Homeostasis. Elife 2022, 11, e81083. [Google Scholar] [CrossRef]
- Zhu, Q.; Meng, Y.; Li, S.; Xin, J.; Du, M.; Wang, M.; Cheng, G. Association of Genetic Variants in Autophagy-Lysosome Pathway Genes with Susceptibility and Survival to Prostate Cancer. Gene 2022, 808, 145953. [Google Scholar] [CrossRef]
- Rii, J.; Sakamoto, S.; Sugiura, M.; Kanesaka, M.; Fujimoto, A.; Yamada, Y.; Maimaiti, M.; Ando, K.; Wakai, K.; Xu, M.; et al. Functional Analysis of LAT3 in Prostate Cancer: Its Downstream Target and Relationship with Androgen Receptor. Cancer Sci. 2021, 112, 3871–3883. [Google Scholar] [CrossRef]
- Sun, J.; Nagel, R.; Zaal, E.A.; Ugalde, A.P.; Han, R.; Proost, N.; Song, J.; Pataskar, A.; Burylo, A.; Fu, H.; et al. SLC 1A3 Contributes to L-asparaginase Resistance in Solid Tumors. EMBO J. 2019, 38, e102147. [Google Scholar] [CrossRef]
- Verma, S.; Shankar, E.; Chan, E.R.; Gupta, S. Metabolic Reprogramming and Predominance of Solute Carrier Genes during Acquired Enzalutamide Resistance in Prostate Cancer. Cells 2020, 9, 2535. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Wang, X.; Ping, Y.; Wang, D.; Cao, Y.; Dai, Y.; Liu, W.; Tao, Z. Five-Gene Signature Associating with Gleason Score Serve as Novel Biomarkers for Identifying Early Recurring Events and Contributing to Early Diagnosis for Prostate Adenocarcinoma. J. Cancer 2021, 12, 3626–3647. [Google Scholar] [CrossRef]
- Meng, J.; Guan, Y.; Wang, B.; Chen, L.; Chen, J.; Zhang, M.; Liang, C. Risk Subtyping and Prognostic Assessment of Prostate Cancer Based on Consensus Genes. Commun. Biol. 2022, 5, 233. [Google Scholar] [CrossRef]
- Wu, X.; Lv, D.; Lei, M.; Cai, C.; Zhao, Z.; Eftekhar, M.; Gu, D.; Liu, Y. A 10-Gene Signature as a Predictor of Biochemical Recurrence after Radical Prostatectomy in Patients with Prostate Cancer and a Gleason Score ≥ 7. Oncol. Lett. 2020, 20, 2906–2918. [Google Scholar] [CrossRef]
- Glinsky, G.V.; Glinskii, A.B.; Stephenson, A.J.; Hoffman, R.M.; Gerald, W.L. Gene Expression Profiling Predicts Clinical Outcome of Prostate Cancer. J. Clin. Investig. 2004, 113, 913–923. [Google Scholar] [CrossRef]
- Mou, Z.; Spencer, J.; Knight, B.; John, J.; McCullagh, P.; McGrath, J.S.; Harries, L.W. Gene Expression Analysis Reveals a 5-Gene Signature for Progression-Free Survival in Prostate Cancer. Front. Oncol. 2022, 12. [Google Scholar] [CrossRef]
- Zhou, R.; Feng, Y.; Ye, J.; Han, Z.; Liang, Y.; Chen, Q.; Xu, X.; Huang, Y.; Jia, Z.; Zhong, W. Prediction of Biochemical Recurrence-Free Survival of Prostate Cancer Patients Leveraging Multiple Gene Expression Profiles in Tumor Microenvironment. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Lu, G.; Cai, W.; Wang, X.; Huang, B.; Zhao, Y.; Shao, Y.; Wang, D. Identifying Prognostic Signatures in the Microenvironment of Prostate Cancer. Transl. Androl. Urol. 2021, 10. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Z. A Gleason Score-Related Outcome Model for Human Prostate Cancer: A Comprehensive Study Based on Weighted Gene Co-Expression Network Analysis. Cancer Cell Int. 2020, 20, 159. [Google Scholar] [CrossRef]
- Gao, P.; Yang, C.; Nesvick, C.L.; Feldman, M.J.; Sizdahkhani, S.; Liu, H.; Chu, H.; Yang, F.; Tang, L.; Tian, J.; et al. Hypotaurine Evokes a Malignant Phenotype in Glioma through Aberrant Hypoxic Signaling. Oncotarget 2016, 7, 15200–15214. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Sun, L.; Wan, F. Molecular Mechanisms of TUG1 in the Proliferation, Apoptosis, Migration and Invasion of Cancer Cells (Review). Oncol. Lett. 2019, 18, 4393–4402. [Google Scholar] [CrossRef] [Green Version]
- Hao, S.D.; Ma, J.X.; Liu, Y.; Liu, P.J.; Qin, Y. Long Non-Coding TUG1 Accelerates Prostate Cancer Progression through Regulating MiR-128-3p/YES1 Axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 619–632. [Google Scholar] [CrossRef]
- Yang, B.; Tang, X.; Wang, Z.; Sun, D.; Wei, X.; Ding, Y. TUG1 Promotes Prostate Cancer Progression by Acting as a CeRNA of MiR-26a. Biosci. Rep. 2018, 38, BSR20180677. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Yang, J.; Chong, T.; Huang, Y.; Liu, Y.; Li, H. TUG1 Knockdown Inhibits the Tumorigenesis and Progression of Prostate Cancer by Regulating MicroRNA-496/Wnt/β-Catenin Pathway. Anticancer Drugs 2020, 31, 592–600. [Google Scholar] [CrossRef]
- Xiu, D.; Liu, L.; Cheng, M.; Sun, X.; Ma, X. Knockdown of LncRNA TUG1 Enhances Radiosensitivity of Prostate Cancer via the TUG1/MiR-139-5p/SMC1A Axis. Onco. Targets Ther. 2020, 13, 2319–2331. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Wang, Y.; Peng, S. The High Expression of LncRNA TUG1 Correlates with Progressive Tumor Condition and Less Satisfying Survival Profiles in Prostate Cancer Patients. Transl. Cancer Res. 2019, 8. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347. [Google Scholar] [CrossRef]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.K.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [Green Version]
- Tu, M.; Saputo, S. From Beginning to End: Expanding the SERINC3 Interactome Through an in Silico Analysis. Bioinform. Biol. Insights 2022, 16. [Google Scholar] [CrossRef]
- Xu, S.; Zheng, Z.; Pathak, J.L.; Cheng, H.; Zhou, Z.; Chen, Y.; Wu, Q.; Wang, L.; Zeng, M.; Wu, L. The Emerging Role of the Serine Incorporator Protein Family in Regulating Viral Infection. Front. Cell Dev. Biol. 2022, 10, 856468. [Google Scholar] [CrossRef]
- Birge, R.B.; Boeltz, S.; Kumar, S.; Carlson, J.; Wanderley, J.; Calianese, D.; Barcinski, M.; Brekken, R.A.; Huang, X.; Hutchins, J.T.; et al. Phosphatidylserine Is a Global Immunosuppressive Signal in Efferocytosis, Infectious Disease, and Cancer. Cell Death Differ. 2016, 23, 962–978. [Google Scholar] [CrossRef] [Green Version]
- Voelkel-Johnson, C.; Norris, J.S.; White-Gilbertson, S. Interdiction of Sphingolipid Metabolism Revisited: Focus on Prostate Cancer. Adv. Cancer Res. 2018, 140, 265–293. [Google Scholar] [CrossRef]

| No Progression | Progression | ||
|---|---|---|---|
| N, total | 400 | 93 | |
| Age, years | <60 | 166 (41.5%) | 34 (36.6%) |
| ≥60 | 234 (58.5%) | 59 (63.4%) | |
| Gleason score | 6 | 44 (11%) | 1 (1.1%) |
| 7 | 221 (55.3%) | 24 (25.8%) | |
| 8 | 49 (12.3) | 13 (14%) | |
| 9 | 84 (21%) | 53 (57%) | |
| 10 | 2 (0.5%) | 2 (2.2%) | |
| Clinical T stage | cT1 | 158 (39.5%) | 17 (18.3%) |
| cT2 | 137 (34.3%) | 35 (37.6%) | |
| cT3 | 28 (7%) | 24 (25.8%) | |
| cT4 | 1 (0.3%) | 1 (1.1%) | |
| NA | 76 (19%) | 16 (17.2%) | |
| Clinical M stage | cM0 | 362 (90.5%) | 89 (95.7%) |
| cM1 | 2 (0.5%) | 1 (1.1%) | |
| NA | 36 (9%) | 3 (3.2%) | |
| Pathologic T stage | pT2 | 172 (43%) | 14 (15.1%) |
| pT3 | 215 (53.8%) | 75 (80.7%) | |
| pT4 | 7 (1.8%) | 3 (3.2%) | |
| NA | 6 (1.5%) | 1 (1.1%) | |
| Pathologic N stage | pN0 | 280 (70%) | 62 (66.7%) |
| pN1 | 56 (14%) | 22 (23.7%) | |
| NA | 64 (16%) | 9 (9.7%) | |
| Residual tumor | R0 | 266 (66.5%) | 46 (49.5%) |
| R1 | 102 (25.5%) | 44 (47.3%) | |
| R2 | 5 (1.3%) | 0 | |
| RX | 13 (3.3%) | 2 (2.2%) | |
| NA | 14 (3.5%) | 1 (1.1%) | |
| Radiation therapy | Yes | 48 (12%) | 46 (49.5%) |
| No | 313 (78.3%) | 43 (46.2%) | |
| NA | 39 (9.8%) | 4 (4.3%) | |
| GO Molecular Function (First 10 Terms) and Biological Process (Last 10 Terms) Categories | Overlap | p-Value | Adj. p-Value | Genes |
|---|---|---|---|---|
| Amino acid transmembrane transporter activity (GO:0015171) | 16/49 | 3.26 × 10−24 | 6.98 × 10−22 | SLC36A1; SLC6A19; SLC38A1; SLC47A1; SLC43A1; SLC3A1; SLC38A11; SLC7A11; SLC6A1; SLC7A1; SLC7A4; SLC7A5; SLC6A6; PDPN; SLC16A2; SLC38A5 |
| L-amino acid transmembrane transporter activity (GO:0015179) | 13/53 | 5.15 × 10−18 | 5.51 × 10−16 | SLC36A1; SLC38A1; SLC47A1; SLC43A1; SLC1A3; SLC3A1; SLC7A11; SLC7A1; SLC7A5; SLC25A15; SLC25A12; SLC25A22; SLC38A5 |
| Organic anion transmembrane transporter activity (GO:0008514) | 17/144 | 1.67 × 10−17 | 1.19 × 10−15 | SLC36A1; SLC38A1; SLC1A3; SLC3A1; SLC6A1; SLC7A1; SLC6A6; SLC25A15; SLC7A5; GJA1; PDPN; SFXN3; SLC25A21; SFXN2; SLC25A12; SLC25A22; SLC38A5 |
| Carboxylic acid transmembrane transporter activity (GO:0046943) | 12/57 | 7.73 × 10−16 | 4.14 × 10−14 | SLC36A1; SLC7A4; SLC7A5; SLC6A6; SLC38A1; PDPN; SLC3A1; SLC6A11; SLC38A11; SLC16A2; SLC7A1; SLC38A5 |
| Neutral amino acid transmembrane transporter activity (GO:0015175) | 9/32 | 2.00 × 10−13 | 8.57 × 10−12 | SLC36A1; SLC6A6; SLC7A5; SLC6A19; SLC38A1; SLC43A1; SFXN3; SFXN2; SLC38A5 |
| Cation transmembrane transporter activity (GO:0008324) | 9/48 | 1.10 × 10−11 | 3.94 × 10−10 | SLC36A1; SLC6A6; SLC7A5; SLC25A15; SLC38A1; SFXN3; SFXN2; SLC7A1; SLC38A5 |
| Pyridoxal phosphate binding (GO:0030170) | 6/21 | 2.18 × 10−9 | 6.66 × 10−8 | SDS; OAT; SHMT2; CBS; PSAT1; ACCS |
| Amino acid: sodium symporter activity (GO:0005283) | 5/12 | 3.35 × 10−9 | 7.96 × 10−8 | SLC38A1; SLC6A15; SLC1A3; SLC6A11; SLC6A1 |
| Transaminase activity (GO:0008483) | 5/12 | 3.35 × 10−9 | 7.96 × 10−8 | OAT; AADAT; PSAT1; BCAT1; BCAT2 |
| Amino acid binding (GO:0016597) | 6/32 | 3.45 × 10−8 | 7.38 × 10−7 | GRM7; SHMT2; NOS1; NAGS; ASS1; GNMT |
| Cellular amino acid catabolic process (GO:0009063) | 25/90 | 2.11 × 10−35 | 2.40 × 10−32 | SHMT2; HAAO; SDSL; DDO; GCSH; IL4I1; TDO2; CBS; SLC25A21; NOS1; PRODH; GLUL; HMGCLL1; ACAD8; MCCC2; SDS; AADAT; GAD1; AMT; PIPOX; GSTZ1; BCAT1; ASPA; IDO1; BCAT2 |
| Alpha-amino acid metabolic process (GO:1901605) | 16/46 | 9.80 × 10−25 | 5.57 × 10−22 | OAT; AADAT; FOLH1B; ASNS; PYCR1; ASS1; GNMT; FOLH1; CPS1; CBS; NOX4; DPEP1; RIMKLA; SLC25A12; GLUL; ASPA |
| Amino acid transport (GO:0006865) | 16/50 | 4.77 × 10−24 | 1.81 × 10−21 | SLC36A1; SLC6A19; SLC38A1; SLC6A17; SLC6A15; SLC43A1; SLC3A1; SLC38A11; SLC16A10; SLC7A11; SLC7A1; SLC7A4; SLC7A5; SLC6A6; PDPN; SLC38A5 |
| Amino acid transmembrane transport (GO:0003333) | 14/45 | 5.77 × 10−21 | 1.64 × 10−18 | SLC36A1; SLC38A1; SLC47A1; SLC38A11; SLC7A11; SLC7A1; SLC6A6; SLC7A5; SFXN3; SFXN2; SLC16A2; SLC25A12; SLC25A22; SLC38A5 |
| Amino acid import (GO:0043090) | 10/22 | 2.73 × 10−17 | 6.21 × 10−15 | SLC36A1; SLC6A6; SLC7A5; SLC47A1; SFXN3; SLC1A3; SFXN2; SLC6A1; SLC16A2; SLC7A1 |
| Glutamine family amino acid metabolic process (GO:0009064) | 11/37 | 1.87 × 10−16 | 3.54 × 10−14 | OAT; GLYATL1; CPS1; AADAT; PYCR1; NAGS; PRODH; RIMKLA; GLUL; NIT2; ART4 |
| Nitrogen compound transport (GO:0071705) | 15/143 | 8.73 × 10−15 | 1.42 × 10−12 | SLC36A1; SLC6A19; SLC38A1; SLC11A1; SLC6A15; SLC43A1; SLC3A1; SLC16A10; SLC7A11; SLC7A1; SLC7A4; SLC6A6; SLC7A5; PDPN; SLC38A5 |
| Organic acid transport (GO:0015849) | 13/100 | 3.43 × 10−14 | 4.88 × 10−12 | SLC36A1; SLC6A19; SLC38A1; SLC6A15; SLC43A1; SLC3A1; SLC16A10; SLC7A11; SLC7A1; SLC7A4; SLC6A6; PDPN; SLC38A5 |
| Import into cell (GO:0098657) | 10/41 | 4.3 × 10−14 | 5.44 × 10−12 | SLC36A1; SLC6A6; SLC7A5; SLC38A1; SLC47A1; SLC1A3; ATP1A2; SLC16A2; SLC7A1; GLUL |
| Aspartate family amino acid metabolic process (GO:0009066) | 9/30 | 1.01 × 10−13 | 1.07 × 10−11 | FOLH1; FOLH1B; SMS; ASNS; SLC25A12; ASPA; NIT2; ASS1 |
| Gene | Function | FC (T/N) | FDR |
|---|---|---|---|
| SLC3A1 | Transports neutral and basic amino acids in the renal tubule and intestinal tract. | 2.72 | 2.93 × 10−5 |
| SLC6A11 | Sodium-dependent transporter that uptakes gamma-aminobutyric acid (GABA), an inhibitory neurotransmitter, which ends the GABA neurotransmission. | 3.72 | 7.57 × 10−13 |
| SLC6A15 | Encodes a member of the solute carrier family 6 protein family, which transports neutral amino acids. | 2.15 | 0.003273 |
| SLC6A17 | Responsible for the presynaptic uptake of neurotransmitters. The encoded vesicular transporter is selective for proline, glycine, leucine and alanine. | 3.61 | 8.27 × 10−10 |
| SLC6A19 | Encodes a system B(0) transmembrane protein that actively transports most neutral amino acids across the apical membrane of epithelial cells. | 6.40 | 0.000127 |
| SLC7A1 | Enables L-arginine transmembrane transporter activity and L-histidine transmembrane transporter activity. | 1.52 | 9.93 × 10−8 |
| SLC7A11 | Encodes a member of a heteromeric, sodium-independent, anionic amino acid transport system that is highly specific for cysteine and glutamate. | 3.67 | 6.95 × 10−22 |
| SLC11A1 | Member of the proton-coupled divalent metal ion transporters family; encodes a multi-pass membrane protein that functions as a divalent transition metal (iron and manganese) transporter involved in iron metabolism. | 1.79 | 2.91 × 10−11 |
| SLC16A10 | Member of a family of plasma membrane amino acid transporters that mediate the Na(+)-independent transport of aromatic amino acids across the plasma membrane. | 1.56 | 0.000213 |
| SLC25A15 | Member of the mitochondrial carrier family. The encoded protein transports ornithine across the inner mitochondrial membrane from the cytosol to the mitochondrial matrix. The protein is an essential component of the urea cycle and functions in ammonium detoxification and biosynthesis of the amino acid arginine. | 1.74 | 3.49 × 10−14 |
| SLC25A21 | Mitochondrial carrier that transports C5-C7 oxodicarboxylates across inner mitochondrial membranes. | 1.98 | 5.81 × 10−12 |
| SLC25A22 | Encodes a mitochondrial glutamate carrier. | 1.88 | 1.43 × 10−24 |
| SLC36A1 | The encoded protein functions as a proton-dependent, small amino acid transporter. | 1.80 | 3.82 × 10−6 |
| SLC38A11 | Predicted to enable amino acid transmembrane transporter activity. | 2.45 | 1.02 × 10−6 |
| SLC43A1 | Belongs to the system L family of plasma membrane carrier proteins that transports large neutral amino acids. | 2.72 | 2.92 × 10−17 |
| SLC1A3 | Member of a high affinity glutamate transporter family. | 0.55 | 1.63 × 10−12 |
| SLC6A1 | The protein encoded by this gene is a gamma-aminobutyric acid (GABA) transporter that localizes to the plasma membrane. | 0.65 | 3.31 × 10−5 |
| SLC6A6 | This gene encodes a multi-pass membrane protein that is a member of a family of sodium and chloride-ion-dependent transporters. The encoded protein transports taurine and beta-alanine. | 0.64 | 1.32 × 10−9 |
| SLC7A4 | Predicted to enable amino acid transmembrane transporter activity. Predicted to be involved in amino acid transport. | 0.53 | 0.001077 |
| SLC7A5 | Enables L-leucine transmembrane transporter activity, L-tryptophan transmembrane transporter activity and thyroid hormone transmembrane transporter activity. | 0.31 | 2.00 × 10−26 |
| SLC16A2 | Encodes an integral membrane protein that functions as a transporter of thyroid hormone. | 0.58 | 1.00 × 10−17 |
| SLC25A12 | Encodes a calcium-binding mitochondrial carrier protein. The encoded protein localizes to the mitochondria and is involved in the exchange of aspartate for glutamate across the inner mitochondrial membrane. | 0.64 | 3.29 × 10−24 |
| SLC38A1 | An important transporter of glutamine, an intermediate in the detoxification of ammonia and the production of urea. | 0.64 | 9.92 × 10−14 |
| SLC38A5 | The encoded protein transports glutamine, asparagine, histidine, serine, alanine and glycine across the cell membrane, but does not transport charged amino acids, imino acids, or N-alkylated amino acids. | 0.45 | 3.30 × 10−16 |
| SLC47A1 | Among its related pathways are the transport of inorganic cations/anions and amino acids/oligopeptides. | 0.39 | 6.09 × 10−29 |
| Gene | Function | FC (T/N) | FDR |
|---|---|---|---|
| AADAT | Aminoadipate aminotransferase. Highly similar to mouse and rat kynurenine aminotransferase II. The rat protein is a homodimer with two transaminase activities. One activity is the transamination of alpha-aminoadipic acid, a final step in the saccaropine pathway, which is the major pathway for L-lysine catabolism. The other activity involves the transamination of kynurenine to produce kynurenine acid, the precursor of kynurenic acid. | 2.01 | 6.04 × 10−12 |
| ACAD8 | Acyl-CoA dehydrogenase family member 8. This gene encodes a member of the acyl-CoA dehydrogenase family of enzymes that catalyzes the dehydrogenation of acyl-CoA derivatives in the metabolism of fatty acids or branch-chained amino acids. The encoded protein is a mitochondrial enzyme that functions in catabolism of the branched-chain amino acid valine. | 1.67 | 2.96 × 10−5 |
| BCAT1 | Branched chain amino acid transaminase 1. This gene encodes the cytosolic form of the enzyme branched-chain amino acid transaminase. This enzyme catalyzes the reversible transamination of branched-chain alpha-keto acids to branched-chain L-amino acids essential for cell growth. | 1.71 | 0.00045 |
| BCAT2 | Branched chain amino acid transaminase 2. This gene encodes a branched-chain aminotransferase found in mitochondria. The encoded protein forms a dimer that catalyzes the first step in the production of the branched-chain amino acids leucine, isoleucine and valine. | 1.51 | 3.01 × 10−12 |
| CBS | Cystathionine beta-synthase. The protein encoded by this gene acts as a homotetramer to catalyze the conversion of homocysteine to cystathionine, the first step in the transsulfuration pathway. | 2.23 | 2.09 × 10−14 |
| GAD1 | Glutamate decarboxylase 1. This gene encodes one of several forms of glutamic acid decarboxylase, identified as a major autoantigen in insulin-dependent diabetes. The enzyme encoded is responsible for catalyzing the production of gamma-aminobutyric acid from L-glutamic acid. | 3.07 | 4.07 × 10−13 |
| GCSH | Glycine cleavage system protein H. The degradation of glycine is brought about by the glycine cleavage system, which is composed of four mitochondrial protein components: P protein (a pyridoxal phosphate-dependent glycine decarboxylase), H protein (a lipoic acid-containing protein), T protein (a tetrahydrofolate-requiring enzyme), and L protein (a lipoamide dehydrogenase). The protein encoded by this gene is the H protein, which transfers the methylamine group of glycine from the P protein to the T protein. | 1.62 | 1.98 × 10−5 |
| GSTZ1 | Glutathione S-transferase zeta 1. This gene is a member of the glutathione S-transferase (GST) super-family that encodes multifunctional enzymes important in the detoxification of electrophilic molecules, including carcinogens, mutagens and several therapeutic drugs, via conjugation with glutathione. This enzyme catalyzes the conversion of maleylacetoacetate to fumarylacetoacatate, which is one of the steps in the phenylalanine/tyrosine degradation pathway. | 1.51 | 1.98 × 10−9 |
| IDO1 | Indoleamine 2,3-dioxygenase 1. This gene encodes indoleamine 2,3-dioxygenase (IDO)—a heme enzyme that catalyzes the first and rate-limiting step in tryptophan catabolism to N-formyl-kynurenine. This enzyme acts on multiple tryptophan substrates, including D-tryptophan, L-tryptophan, 5-hydroxy-tryptophan, tryptamine, and serotonin. | 1.51 | 0.009556 |
| IL4I1 | Interleukin 4 induced 1. This gene encodes a secreted L-amino acid oxidase protein, which primarily catabolizes L-phenylalanine and, to a lesser extent, L-arginine. | 1.81 | 5.50 × 10−10 |
| MCCC2 | Methylcrotonyl-CoA carboxylase subunit 2. This gene encodes the small subunit of 3-methylcrotonyl-CoA carboxylase. This enzyme functions as a heterodimer and catalyzes the carboxylation of 3-methylcrotonyl-CoA to form 3-methylglutaconyl-CoA. | 2.45 | 5.63 × 10−12 |
| SDS | Serine dehydratase. This gene encodes one of three enzymes that are involved in metabolizing serine and glycine. L-serine dehydratase converts L-serine to pyruvate and ammonia and requires pyridoxal phosphate as a cofactor. The encoded protein can also metabolize threonine to NH4+ and 2-ketobutyrate. | 3.32 | 1.36 × 10−14 |
| SDSL | Serine dehydratase like. Predicted to be involved in the isoleucine biosynthetic process and threonine catabolic process. | 1.52 | 3.11 × 10−10 |
| SHMT2 | Serine hydroxymethyltransferase 2. This gene encodes the mitochondrial form of a pyridoxal phosphate-dependent enzyme that catalyzes the reversible reaction of serine and tetrahydrofolate to glycine and 5,10-methylene tetrahydrofolate. The encoded product is primarily responsible for glycine synthesis. The activity of the encoded protein has been suggested to be the primary source of intracellular glycine. | 1.69 | 7.99 × 10−17 |
| SLC25A21 | Solute carrier family 25 member 21. Homolog of the S. cerevisiae ODC proteins, mitochondrial carriers that transport C5-C7 oxodicarboxylates across inner mitochondrial membranes. One of the species transported by ODC is 2-oxoadipate, a common intermediate in the catabolism of lysine, tryptophan and hydroxylysine in mammals. | 1.98 | 5.81 × 10−12 |
| TDO2 | Tryptophan 2,3-dioxygenase. This gene encodes a heme enzyme that plays a critical role in tryptophan metabolism by catalyzing the first and rate-limiting step of the kynurenine pathway. | 3.45 | 0.0044 |
| AMT | Aminomethyltransferase. This gene encodes one of four critical components of the glycine cleavage system. | 0.52 | 7.11 × 10−13 |
| ASPA | Aspartoacylase. This gene encodes an enzyme that catalyzes the conversion of N-acetyl-L-aspartic acid (NAA) to aspartate and acetate. | 0.24 | 6.87 × 10−31 |
| DDO | D-aspartate oxidase. The protein encoded by this gene is a peroxisomal flavoprotein that catalyzes the oxidative deamination of D-aspartate and N-methyl D-aspartate. | 0.50 | 1.01 × 10−15 |
| GLUL | Glutamate-ammonia ligase. The protein encoded by this gene belongs to the glutamine synthetase family. It catalyzes the synthesis of glutamine from glutamate and ammonia in an ATP-dependent reaction. | 0.64 | 7.49 × 10−16 |
| HAAO | 3-Hydroxyanthranilate 3,4-dioxygenase is a monomeric cytosolic protein belonging to the family of intramolecular dioxygenases containing nonheme ferrous iron. HAAO catalyzes the synthesis of quinolinic acid (QUIN) from 3-hydroxyanthranilic acid. | 0.45 | 2.36 × 10−19 |
| HMGCLL1 | 3-Hydroxymethyl-3-methylglutaryl-CoA lyase like 1. Non-mitochondrial 3-hydroxymethyl-3-methylglutaryl-CoA lyase that catalyzes the cation-dependent cleavage of (S)-3-hydroxy-3-methylglutaryl-CoA into acetyl-CoA and acetoacetate, a key step in ketogenesis. | 0.32 | 5.18 × 10−15 |
| NOS1 | Nitric oxide synthase 1. The protein encoded by this gene belongs to the family of nitric oxide synthases, which synthesize nitric oxide from L-arginine. | 0.28 | 2.12 × 10−14 |
| PIPOX | Pipecolic acid and sarcosine oxidase. Enables L-pipecolate oxidase activity and sarcosine oxidase activity. Involved in L-lysine catabolic process to acetyl-CoA via L-pipecolate. | 0.39 | 3.84 × 10−24 |
| PRODH | Proline dehydrogenase 1. This gene encodes a mitochondrial protein that catalyzes the first step in proline degradation. | 0.35 | 1.70 × 10−24 |
| Risk Subgroup | Hazard Ratio | Rule |
|---|---|---|
| Very low risk | 0.088 | Gleason score < 9 AND CSAD < 771 |
| Low risk | 0.480 | Gleason score ≥ 9 AND SERINC3 < 4007 |
| Medium risk | 0.974 | Gleason score < 9 AND CSAD ≥ 771 |
| High risk | 2.923 | Gleason score ≥ 9 AND SERINC3 ≥ 4007 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samaržija, I.; Trošelj, K.G.; Konjevoda, P. Prognostic Significance of Amino Acid Metabolism-Related Genes in Prostate Cancer Retrieved by Machine Learning. Cancers 2023, 15, 1309. https://doi.org/10.3390/cancers15041309
Samaržija I, Trošelj KG, Konjevoda P. Prognostic Significance of Amino Acid Metabolism-Related Genes in Prostate Cancer Retrieved by Machine Learning. Cancers. 2023; 15(4):1309. https://doi.org/10.3390/cancers15041309
Chicago/Turabian StyleSamaržija, Ivana, Koraljka Gall Trošelj, and Paško Konjevoda. 2023. "Prognostic Significance of Amino Acid Metabolism-Related Genes in Prostate Cancer Retrieved by Machine Learning" Cancers 15, no. 4: 1309. https://doi.org/10.3390/cancers15041309
APA StyleSamaržija, I., Trošelj, K. G., & Konjevoda, P. (2023). Prognostic Significance of Amino Acid Metabolism-Related Genes in Prostate Cancer Retrieved by Machine Learning. Cancers, 15(4), 1309. https://doi.org/10.3390/cancers15041309






