Simple Summary
Although alterations of intestinal bacterial microbiota have been admitted as playing one of the most important roles in colorectal carcinogenesis, the links between microbiota compositional changes and premalignant colorectal polyps have still not been fully examined. Furthermore, there is a lack of knowledge in terms of defining the precise differences and the correct interpretation of tissue-derived and stool-based bacterial dysbiosis in patients with precancerous colorectal lesions. Thus, this systematic review aims, firstly, to assess whether and how the tissue-derived intestinal microbiota structure differs from the bacterial dysbiosis in fecal samples of patients with simple, advanced colorectal adenoma and carcinoma in situ, and, secondly, to propose the correct selection of each matrix in order to increase sampling accuracy and applicability in future microbiota studies and clinical practice.
Abstract
Alterations in gut microbiota play a pivotal role in the adenoma-carcinoma sequence. However, there is still a notable lack of the correct implementation of tissue and fecal sampling in the setting of human gut microbiota examination. This study aimed to review the literature and to consolidate the current evidence on the use of mucosa and a stool-based matrix investigating human gut microbiota changes in precancerous colorectal lesions. A systematic review of papers from 2012 until November 2022 published on the PubMed and Web of Science databases was conducted. The majority of the included studies have significantly associated gut microbial dysbiosis with premalignant polyps in the colorectum. Although methodological differences hampered the precise fecal and tissue-derived dysbiosis comparison, the analysis revealed several common characteristics in stool-based and fecal-derived gut microbiota structures in patients with colorectal polyps: simple or advanced adenomas, serrated lesions, and carcinomas in situ. The mucosal samples considered were more relevant for the evaluation of microbiota’s pathophysiological involvement in CR carcinogenesis, while non-invasive stool sampling could be beneficial for early CRC detection strategies in the future. Further studies are required to identify and validate mucosa-associated and luminal colorectal microbial patterns and their role in CRC carcinogenesis, as well as in the clinical setting of human microbiota studies.
1. Introduction
With more than 1.9 million new cases and 935,000 deaths, colorectal cancer (CRC) is the third most diagnosed and the second leading cause of death among cancers worldwide [1]. Several risk factors are associated with the development of CRC through a conventional adenoma-carcinoma sequence and serrated pathways. Such factors include genetical mutations, environmental, lifestyle, and dietary habits. Nevertheless, compositional changes in gut microbiota and a shift in the diversity and distribution of bacterial communities determine increased mucosal permeability, bacterial translocation, and the activation of the immune system, causing chronic inflammation and CRC [2,3,4,5].
The collection of bacteria, archaea and eukarya colonizing the human GI tract is termed the human gut microbiota, while the entire habitat (intestines), including the microorganisms, their genes, and the surrounding environmental conditions, is commonly called the gut microbiome. As for gut bacterial dysbiosis, it describes the altered composition and reduced diversity of core bacterial communities living in the gut [2,4].
Recently, several clinical trials investigating the role of intestinal bacterial dysbiosis in the stages of colorectal carcinogenesis have been published [4,5,6,7]. Multiple studies have shown a strong link between alterations in the human intestinal microbiota and the presence of carcinoma lesions in the colorectum [5,6,7,8,9]. Furthermore, some of the microbes, such as Fusobacterium nucleatum (F. nucleatum), Streptococcus gallolyticus (S. bovis), enterotoxigenic Bacteroides fragilis (ETBF), pks (polyketide synthase) + Escherichia coli, Enterococcus faecalis, Peptostreptococcus anaerobius and Parvimonas micra, etc., were accepted as CRC-associated bacteria [10,11,12,13,14,15]. Colorectal adenoma (CRA) is a precancerous lesion of CRC, and recent research also finds it to be associated with an altered gut bacterial community structure, a lower richness, and a higher abundance of proinflammatory bacteria [15,16,17,18,19].
While emerging evidence suggests a link between the gut microbiota and CRC, it is hard to say that certain bacteria play an exceptional causal role in CRC, where secondary alterations in the local gut microbiota due to chronic inflammation dominate. In contrast, CR adenoma as a premalignant state does not induce severe local inflammation and consequent changes in the gut microbiota. Therefore, the indisputable detection of a statistically significant correlation between adenomatous colorectal (CR) polyps, as an early stage of the adenoma-carcinoma cascade, and intestinal microbial dysbiosis, would potentially imply primary microbiota’s role in CR tumorigenesis [20].
Sample collection is another challenging step in human microbiome studies. With the expansion of research in the field, advanced invasive and non-invasive examination models have been engaged in the detection of CRC-associated bacteria and the overall bacterial composition in human samples. Since the 1990s, molecular tools targeting the bacterial 16S ribosomal RNA (rRNA) gene have been applied for the explicit evaluation of the gut microbiota from both feces and tissues. Although both tissue and fecal specimens provide useful information about the composition of gut bacterial communities, most of the studies on the gut microbiome, including those related to CRC and CR adenoma, are still based only on fecal samples, as an easy and non-invasive procedure [21,22]. On the contrary, other clinical trials find no statistically significant association between premalignant CR polypoid lesions and an increase in CRC-associated bacteria in stool samples compared with gut mucosal biopsies, where an increased F. nucleatum abundance has been recognized [23]. Similarly, while some researchers state that bacterial community compositions in feces and mucosa differ completely [23,24], others believe that similar variations in CRC bacterial species can be identified between stool samples and gut mucosal biopsies [25,26]. Moreover, several studies suggest that tissue samples are more relevant for the evaluation of microbiota’s pathophysiological involvement in CR carcinogenesis, while stool samples are more powerful for identifying noninvasive diagnostic or prognostic markers of CRC [23,24,27].
Thus, a discussion arises from the detection of different intestinal microbiota shifts in mucosal and fecal samples of patients with CR neoplasia. Moreover, the proper employment of a sampling matrix and its accurate analysis for the examination of intestinal microbiota’s structural and functional composition is still lacking.
Therefore, there is a need for a profound systematic literature review, firstly, to assess the difference between mucosa-associated (tissue) and luminal (fecal) intestinal microbiota alterations in patients with precancerous colorectal lesions (simple and advanced conventional adenoma, serrated adenoma) and preinvasive cancer (carcinoma in situ (Ca in situ)), compared with healthy control and/or self-control groups, and, secondly, to suggest the potentially correct implementation and assessment of tissue and stool samples in human gut microbiota studies and in the clinical field. The following could induce a new research era based on a comprehensive methodology and the accurate use of a selected type of matrix for the precise analysis of CR carcinogenesis. This would also contribute to the validation of preventive measures for the early detection of colorectal neoplasms, as well as the management of the affected gut microbiota in premalignant mucosal changes prior to the development of CRC.
2. Materials and Methods
The present systematic review was performed according to the Cochrane collaboration-specific protocol [28] and was reported following the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) checklist [29]. The PRISMA checklist was completed according to the mentioned recommendations (Table S1). The present systematic review was prospectively registered in PROSPERO (ID No.: CRD42022376106).
2.1. Eligibility Criteria
Studies that examined the link between CRA or Ca in situ, as precancerous colorectal lesions, and the intestinal tissue- and/or fecal-derived intestinal microbiota composition were eligible for inclusion. The search was restricted to human studies published in the English language from 2012 until November 2022. These studies included adult patients (≥18 years) with a diagnosed CR advanced/non-advanced adenoma, serrated polyps, or Ca in situ undergoing a complete examination of the tissue and/or stool-based bacterial microbiota community structure. Advanced adenomas were defined as those with high-grade dysplasia, villous or tubulovillous histology, or a diameter ≥1 cm, while serrated polyps included sessile serrated (SSA) and traditional serrated adenomas (TSA). The analysis included only studies with a complete bacterial community assessment and healthy or the same patients’ paired normal samples as controls.
2.2. Information Sources
A literature search was performed in the PubMed and Web of Science online databases to identify original comparative studies analyzing the colorectal mucosa-associated and/or luminal microbiota composition in patients with premalignant (adenoma) and preinvasive (Ca in situ) colorectal neoplasia. The most recent search was performed in November 2022.
2.3. Literature Search Strategy
We used the following combination of Medical Subject Headings (MeSH) and keywords with the employment of “AND” or “OR” Boolean operators: “Colorectal adenoma” OR “Colorectal polyp” OR “Colorectal polypoid lesion” OR “Colorectal precancerous lesion” OR “Colorectal neoplasms” OR “Colorectal neoplasia” OR “Colonic neoplasia” AND “Serrated adenoma” OR “Serrated polyp” AND “Colorectal carcinogenesis” OR “Colorectal tumorigenesis” OR “Adenoma-carcinoma sequence” AND “Gut microbiome” OR “Gut microbiota” OR “Intestinal microbiome” OR “Intestinal microbiota” OR “Gut dysbiosis” OR “Intestinal dysbiosis” AND “Mucosa-adherent” OR “Tissue-derived” OR “Mucosa-associated” OR “Stool-based” OR “Luminal” OR “Fecal-derived”.
2.4. Study Selection
All titles and abstracts identified in the electronic databases were screened by two experienced reviewers independently of one another using a piloted electronic database (Microsoft Excel). Following the identification of relevant abstracts, full-text articles were retrieved and re-reviewed. Comments on articles, short notes, letters, conference abstracts, systematic reviews, meta-analyses, review articles, preclinical studies, and duplicates were manually excluded. A manual search was performed to identify additional primary studies and minimize search bias. The literature review was completed with an extensive search using the “related articles” function in PubMed. Studies which did not analyze tissue- and/or stool-based bacterial gut microbiota’s structure in patients with colorectal premalignant or preinvasive neoplasms in comparison with healthy or self-controls and/or CRC were excluded. The endpoint measures of the current review consisted of tissue and fecal-derived microbiota’s bacterial compositional diversity in CRA and Ca in situ.
2.5. Data Extraction
The following data were extracted from each study: first, the author’s name, the date of publication, the sample size, including the number of cases and controls, the microbiota examination method and the matrix type, the abundance and/or prevalence of CRC-associated and other bacteria, α- and β-diversity, and the other main findings of the study. The term α-diversity was described as the variation of microbes in a single sample and expressed by richness (that is, the number of taxa present in a sample) and evenness (that is, how evenly distributed the taxa are within a sample). Contrarily, β-diversity was determined by the variation in microbial communities between the samples in terms of ecological distance, likely reflecting the presence and absence of some rare species. The extracted data was only evaluated at the end of the reviewing process to reduce the selection bias.
2.6. Study Quality Assessment and Risk of Bias
The methodological quality of the selected trials was assessed using the Cochrane Handbook method [28]. For evaluating the quality of non-randomized trials, items of risk in patient selection, baseline comparability, and outcomes/exposure selection and measurement were judged using the Newcastle–Ottawa scale (NOS) [30]. We rated the quality of the studies by awarding stars in each domain as follows: a “good” quality score required 3 or 4 stars in selection, 1 or 2 stars in comparability, and 2 or 3 stars in outcomes; a “fair” quality score required 2 stars in selection, 1 or 2 stars in comparability, and 2 or 3 stars in outcomes; a “poor” quality score was reflected by 0 or 1 star(s) in selection, or 0 stars in comparability, or 0 or 1 star(s) in outcomes. Only good and fair quality studies were included in the further analysis. A summary of the quality evaluation process has been visualized in Table 2 and Table S2 (extended version).
3. Results
Search Results and Study Characteristics
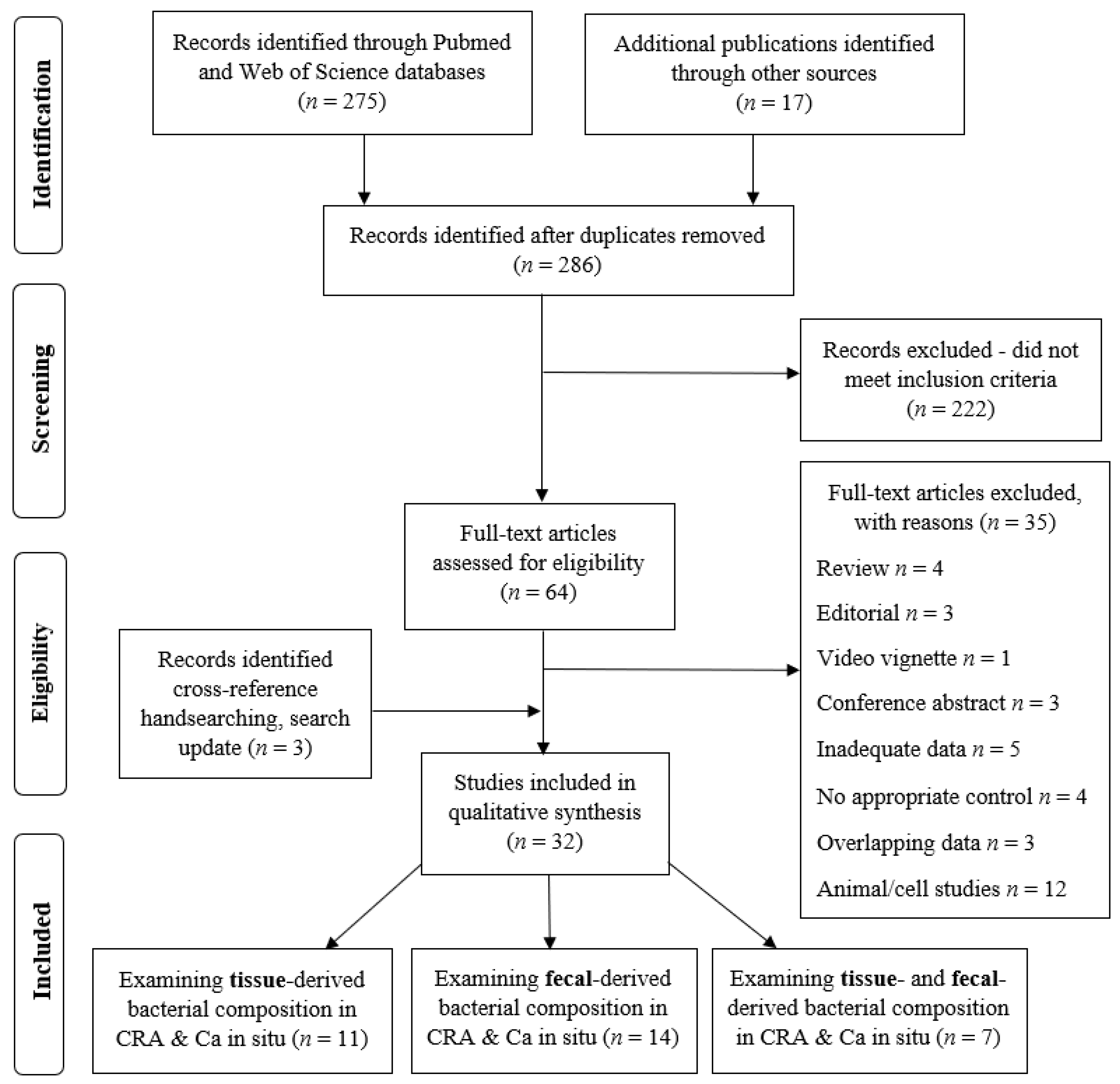
The initial search yielded 292 results; after removing duplicates, 286 articles were screened for eligibility based on the title and abstract, and 64 articles were retrieved for a full-text evaluation. These were assessed for eligibility. A total of 35 were excluded as ineligible for inclusion: 4—review articles, 3—editorials, 1—video vignette, 3—conference abstracts, 5—inadequate data, 3—overlapping data, 4—no appropriate control, and 12—animal/cells study. Three studies were included additionally after the search update. All the included studies were observational: cohort, cross sectional, and case-control studies. No randomized control trials were identified. A total of 32 studies fulfilled the inclusion criteria and were finally selected for a qualitative analysis (Figure 1).
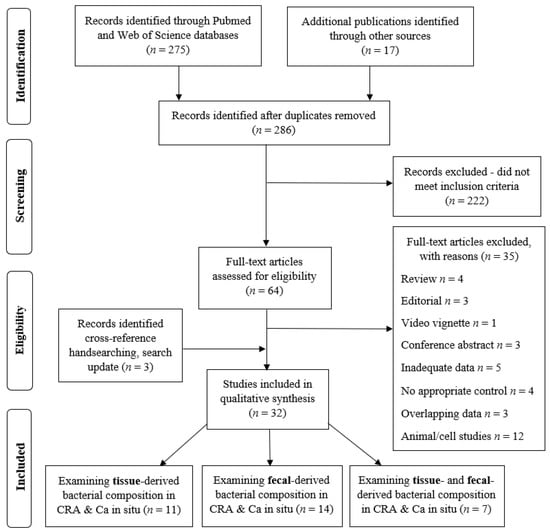
Figure 1.
PRISMA flow diagram indicating the selection of studies for the systematic review.
The included studies were grouped according to the utilized samples and study goals, as follows: (a) studies investigating the composition of gut bacterial tissue-derived microbiota (n = 11), (b) studies examining the structure of gut bacterial stool-based microbiota (n = 14), and (c) those investigating the composition of both tissue- and/vs. fecal-derived gut bacterial microbiota (n = 7) in CRA and Ca in situ.
Most of the studies included used the same ‘human gut microbiota’ term for the evaluation of bacterial communities prevailing in the gut, and the term ‘bacterial dysbiosis’ for the examination of gut bacterial composition and diversity changes. Very few studies referred to the ‘microbiome’ [27,31,32,33,34] and ‘metabolome’ [32,35,36] as study outcomes, and these generally served as a data supplementing factor for the review.
Of the studies included, the comparison of the microbiota was often between conventional adenoma without further specification of type [18,23,24,25,27,30,33,36,37,38,39,40,41,42,43,44,45,46,47,48] or adenoma classified as advanced and non-advanced [32,34,35,49,50,51,52,53], and healthy controls. However, there were few studies that investigated the microbiota’s composition in sessile serrated polyps (SSPs) and traditional serrated adenomas (TSAs) in the serrated pathway specifically [31,51,54]. Only one study aimed to compare microbiota between patients with Ca in situ vs. healthy controls [55].
Eleven studies used DNA analysis with 16S rRNA gene sequencing [27,33,35,37,38,40,45,48,49,51,52] and one was a metagenome-wide association study (MGWAS) [50]. One study used shotgun metagenomic sequencing [39], one used qPCR with liquid (LC−TOFMS) and gas (GC−TOFMS) chromatography time-of-flight mass spectrometry [36], and one used terminal restriction fragment length polymorphism (T-RFLP) with next-generation sequencing (NGS) [55]. The remaining study used ENTERO-test 24 plus MALDI-TOF mass spectrometry [46]. Five other studies also used 454-pyrosequencing [18,25,41,43,44], while five used qPCR [23,24,25,26,47], one used high-throughput sequencing (HTS) and fluorescence in situ hybridization (FISH) [42], one used PCR and FISH [54], and one used only HTS [53]. One study used high-performance liquid chromatography (HPLC) [34], one used ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) metabolomics [32], one used internal transcribed spacer (ITS) ribosomal RNA sequencing and whole-genome shotgun sequencing (WGS) [31], and one used metagenomic sequencing [30] in addition to 16S rRNA gene sequencing.
All the included studies investigated the association between gut bacterial dysbiosis in fecal and/or intestinal tissue samples and precancerous colorectal lesions (and/or CRC). Two parts of the included studies additionally examined the following: the relationship between metabolites and the metagenome-metabolome [32,34,35,36]; genetic mutations [48]; the presence of mucosal biofilm [42]; diagnostic biomarkers [26,27,33,39,40,52,53]; enterotypes and clusters [24,38]; the intracellular microbiota structure [46]; location-specific microbiota [24,54]; “on” and “off” tumor bacterial differences [24]; and the fungal community composition [31] in patients with colorectal polyps.
Twelve studies were evaluated as representative with an estimate of more than 100 subjects per case group.
Most of the studies used tissue specimens directly from the lesion [23,26,42,43,44,45,46,47,48,53,54], while others preferred non-tumor colon or rectal mucosa sampling [25,36,41] for the case group analysis. In several studies both “on” and “off” tumor sampling was planned [24,27,30,31]. Seventy-eight percent of the included studies used stool samples and/or intact colorectal mucosa specimens from healthy patients for the control group [18,23,24,25,26,27,30,31,32,33,34,35,36,37,38,39,40,41,43,45,49,50,51,52,54,55]. In other studies, paired adjacent normal mucosa samples were employed for self-control group formation [47,48]. The remaining studies used both controls [42,44,46,53]. One trial also included adenoma up to 1 cm in addition to neoplasia-free colon tissue sampling for the control group [30].
The characteristics and outcomes of the studies are displayed in Table 1. Additionally, the extended version of the table explicitly describing the type of matrix, the gut microbiota’s structure, and its compositional shifts in the fecal and tissue samples of patients with colorectal adenoma, and/or colorectal cancer vs. healthy controls is provided in a supplementary material file (Table S3).

Table 1.
Summary of human studies investigating precancerous colorectal lesions and healthy control stool and tissue specimens addressing microbial compositional shifts.
Based on the NOS assessment [56], 16 studies had a score of 5/9, 9 studies scored 6/9, and 7 studies scored 7/9 (Table 2). Overall, a high heterogeneity was observed in the study designs, study populations, and the examination methods of the gut microbiota’s composition.

Table 2.
Quality assessment of the selected studies according to the star score of the Newcastle–Ottawa scale (NOS), based on which * are assigned to three criteria, i.e., selection (with a maximum of 4 stars [****]), comparability between case and controls (with a maximum of 2 stars [**]), and ascertainment of effects of microbiota—outcome/exposure (with a maximum of 3 stars [***]) for a potential score ranging from 0 to 9 points. Higher scores indicate a lower risk of bias.
4. Discussion
4.1. Structural Gut Microbiota Profile in Patients with Precancerous and Preinvasive Colorectal Lesions vs. HC
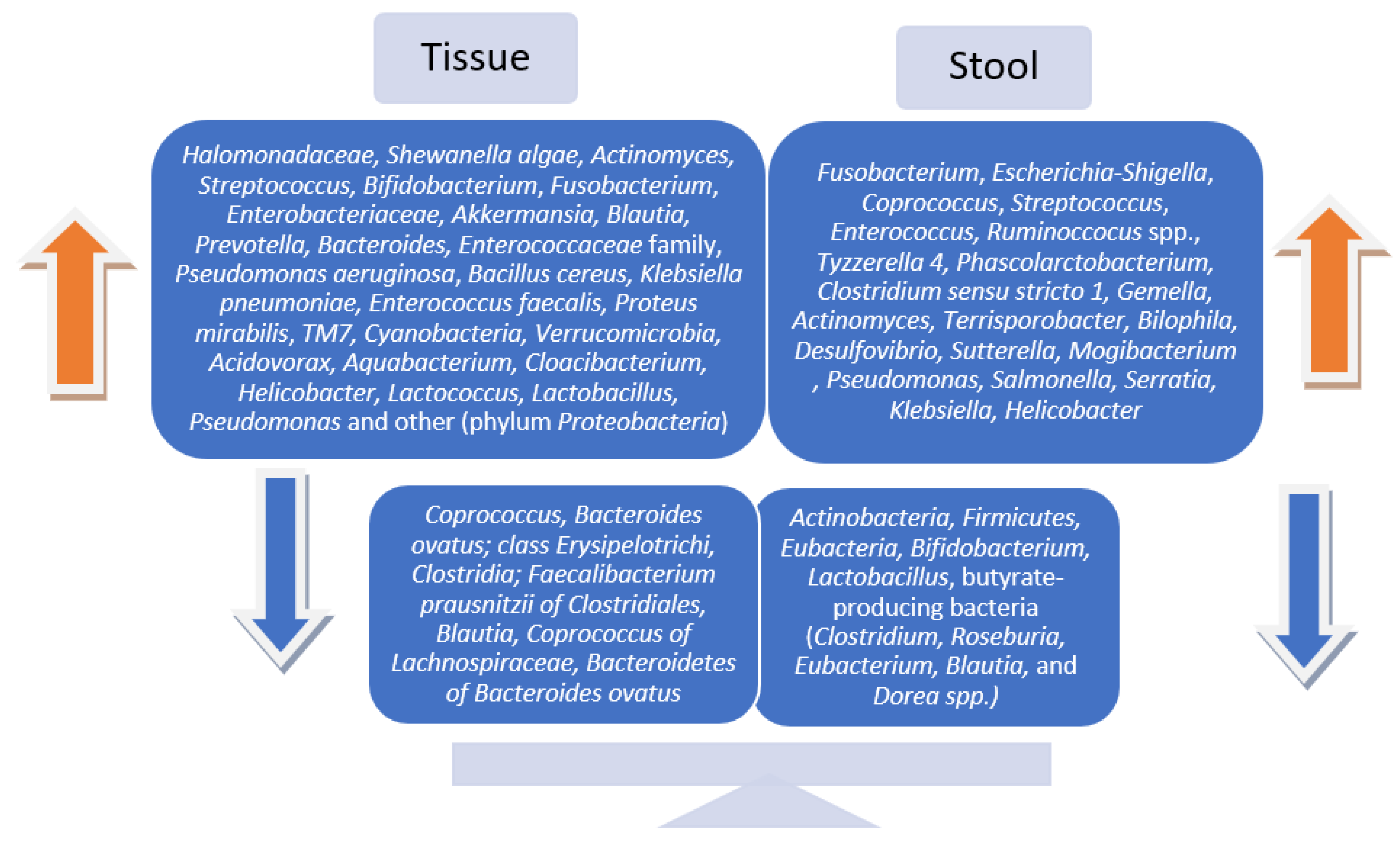
This systematic review revealed certain differences in the gut microbiota diversity and abundance of bacteria in patients with colorectal adenomas and Ca in situ compared to healthy adults. This is supported by most of the included studies. However, several studies reported no significant difference in microbiota diversity [45,48,52], while others did not report any difference in the microbiota’s bacterial composition between subjects with precancerous colorectal lesions and healthy controls [32,55]. Among microbiota’s compositional alterations, the most common were an increased abundance of Fusobacterium, Escherichia-Shigella, Coprococcus, Streptococcus, Enterococcus, and/or Ruminoccocus spp. and a reduction in Actinobacteria, Firmicutes, Eubacteria, Bifidobacterium, Lactobacillus, and butyrate-producing bacteria (Clostridium, Roseburia, Eubacterium, Blautia, and Dorea spp). These were evident in both mucosal and fecal samples in colorectal adenoma vs. healthy controls [23,25,27,37,49,51,55]. No consensus in the α-diversity and β-diversity was evident between the patterns of tissue- and fecal-derived microbiota in preneoplastic colorectal lesions. Overall, a reduction in the diversity/richness of bacterial species in the intestinal microbial community was detected in both tissue and stool samples.
While comparing stool-based microbiota composition between the case and control, eight bacterial species (Actinomyces odontolyticus, Bacteroides fragilis, Clostridium nexile, Fusobacterium varium, Heamophilus parainfluenzae, Prevotella stercorea, Streptococcus gordonii, and Veillonella dispar) and four bacterial genera (Actinomyces, Atopobium, Fusobacterium, and Heamophilus) were significantly associated with the Ca in situ group [55]. Here, the control group significantly differed with the predominant genus being Slackia and sp. Eubacterium coprostanoligens, which is a cholesterol-reducing bacteria and potentially acts as an inhibitor of CRC. However, this observational study was the only one included in the review that examined microbiota changes in patients with colorectal Ca in situ. In addition, there were only six patients forming the case group, leading to some debate on its scientific weight.
Laterally spreading tumors (LSTs) as primary precursors of CRC, due to its special morphology and growth pattern, are extremely difficult to identify during a simple colonoscopy. Thus, there is a need for new sensitive early detection methods, e.g., fecal biomarkers. Interestingly, LSTs are rarely investigated in the light of microbiota signatures. For instance, Shen et al. have demonstrated an increased fecal abundance of the three bacteria ETBF, Peptostreptococcus stomatis (P. stomatis), and Parvimonas micra (P. micra) with considerably high sensitivity and specificity in detecting LST, while tissue-derived microbiota’s composition was shown to be associated with an increase in genus Lactobacillus-Streptococcus and the spp. ETBF–P. stomatis–P. micra [26]. These oral bacteria are defined as early noninvasive biomarkers of LSTs and potentially could also predict the adenoma recurrence risk after resections.
It is worth noting that the number of included studies that differentiated non-advanced adenomas (NAA) from advanced (AA) [32,34,35,49,50,51,52,53], and conventional adenomas from sessile serrated polyps (SSPs) and traditional serrated adenomas (TSAs) was rather low [31,51,54]. Different quantities of bacterial abundance were present at AA in comparison to NAA and HC. Several of the included studies demonstrated a statistically significant decrease in the butyrate producing bacteria, Roseburia, Eubacteria, and Clostridia [49]. Others found a considerable increase in Fusobacterium, Enteroccocus, and Bacteroidetes [50], while Firmicutes phylum and the Firmicutes: Bacteroidetes ratio were depleted in fecal samples of AA, though with no significant difference among the three groups (AA, CRC, and HC) [32].
Hale et al., with an estimated 780 patients included in their trial, reported a statistically significant increase in four genera: Bilophila, Desulfovibrio, Sutterella, and Mogibacterium in the stool samples of patients with diagnosed AA compared to healthy individuals. Bilophilia and Desulfovibrio are known to produce H2S and secondary bile acids, which act as a catalyzer in the A development of CRC [35]. A consistent increase in the genera Fusobacterium, Tyzzerella 4, Phascolarctobacterium, Clostridium sensu stricto 1; Streptococcus, Gemella, Actinomyces, and Terrisporobacter was observed in the fecal microbiota signatures of AA patients vs. healthy controls in a large sample size (n = 758) study. These microbial patterns could potentially supplement fecal immunohistochemical tests for the early non-invasive detection of CRA [52].
The most prominent change in colon tissue specimens of AA vs. healthy controls revealed increased Halomonadaceae and Shewanella algae and depleted Coprococcus and Bacteroides ovatus [53]. Peters et al. divided lesions into proximal and distal, AA and NAA, conventional adenomas (CA) and serrated polyps (SP). Their results showed a lower richness in CA, and especially in AA, and an enrichment of the genera Actinomyces and Streptococcus and a decrease of Erysipelotrichi and Clostridia in SSA compared to healthy controls. Colorectal serrated lesions were linked to the proximal colon location and microbiota dysbiosis was directly dependent on the severity of the lesion along the adenoma-carcinoma sequence and serrated pathway [51]. An increase in the abundance of the genus Fusobacterium was observed in people with serrated colorectal lesions in the reviewed studies [51,54], which was consistent with the literature on F. nucleatum’s primary role in the serrated neoplasia pathway [57].
Moreover, the analysis revealed well-known variations in the CRC-related bacteria found in tissue and stool samples. Lactobacillales were enriched in tumor tissue, Fusobacterium, Porphyromonas, Peptostreptococcus, Gemella, Mogibacterium, and Klebsiella were present in mucosa-adherent flora, while Erysipelotrichaceae, Prevotellaceae, and Coriobacteriaceae were highly abundant in the gut lumen of CRC patients. These prevailing bacterial communities may be related to secondary alterations in the microenvironment of CRC rather than playing a primary role in colorectal carcinogenesis. In contrast, precancerous colorectal lesions, having fewer genetic mutations and only subtle biochemical mucosal changes, have more potential to relate to the discovery of dysbiosis with initiation and acceleration processes in CRC development. Either way, these microbial signatures may resemble those presumably less severe microbiota compositional changes in precancerous colorectal lesions, adjacent tissue, and colon lumen [49,58].
The recent meta-analysis from Mo et al. concluded that the dysbiosis of the off-site (adjacent) tissue in CRC is distinctive and predictive. Tumor-adjacent tissue should not be regarded as healthy tissue and should not be used for self-controls, especially without a healthy control group [59]. In our systematic review, only a few studies employed paired adjacent tissue as self-control samples [47,48], while others used self-controls in addition to healthy patient control groups [42,44,46,53]. Several trials used normal rectal or colon mucosa samples for the case group formation [25,36,41].
Another controversial issue is the formation of biofilm in the colorectum. Biofilm is known as aggregations of microbial communities in a polymeric matrix that adhere to either biological or nonbiological surfaces, especially the colonic mucus layer coming into close contact with the mucosal epithelium itself. This contact eventually leads to altered epithelial functions and procarcinogenic tissue inflammation. One study included in the systematic review revealed a clear association between the presence of biofilm and diminished colonic epithelial cell E-cadherin, enhanced epithelial cell IL-6, and Stat3 activation. Moreover, biofilms were detected not only in tumors, both CRA and CRC, but also on tumor-free mucosa far distant from the tumors. Biofilm detection correlated with bacterial tissue invasion and changes in tissue biology with activated cellular proliferation and microbial dysbiosis [42].
4.2. Gut Microbiota Compositional Patterns in Mucosa-Associated (Tissue) vs. Luminal (Fecal) Samples of Patients with Premalignant Colorectal Polyps
Though the findings were inconsistent, ultimately, the majority of the studies reported statistically significant changes in microbial communities in patients with preneoplastic lesions after examining both tissue and stool samples. Regarding gut microbiota patterns and diversity in fecal vs. tissue samples in people with premalignant colorectal lesions compared to healthy controls, the results remain inconclusive. Several of the included studies declared similar variations in the microbial communities [25,26], while others reported fecal and mucosa-associated microbiotas to be completely distinct and different in composition and diversity [23,24,27]. The most common microbial signatures are displayed in Figure 2.
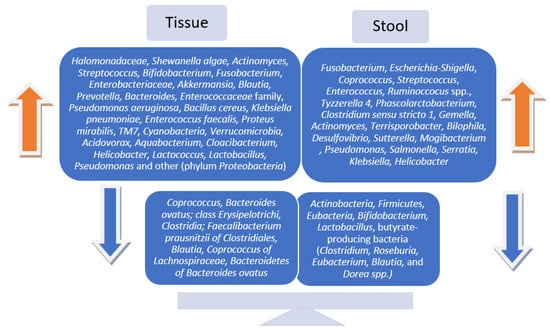
Figure 2.
Bacterial abundances in tissue vs. stool samples in patients with precancerous colorectal lesions.
4.3. Intestinal Microbiota Studies in Patients with Precancerous CR Lesions: Tissue or Stool?
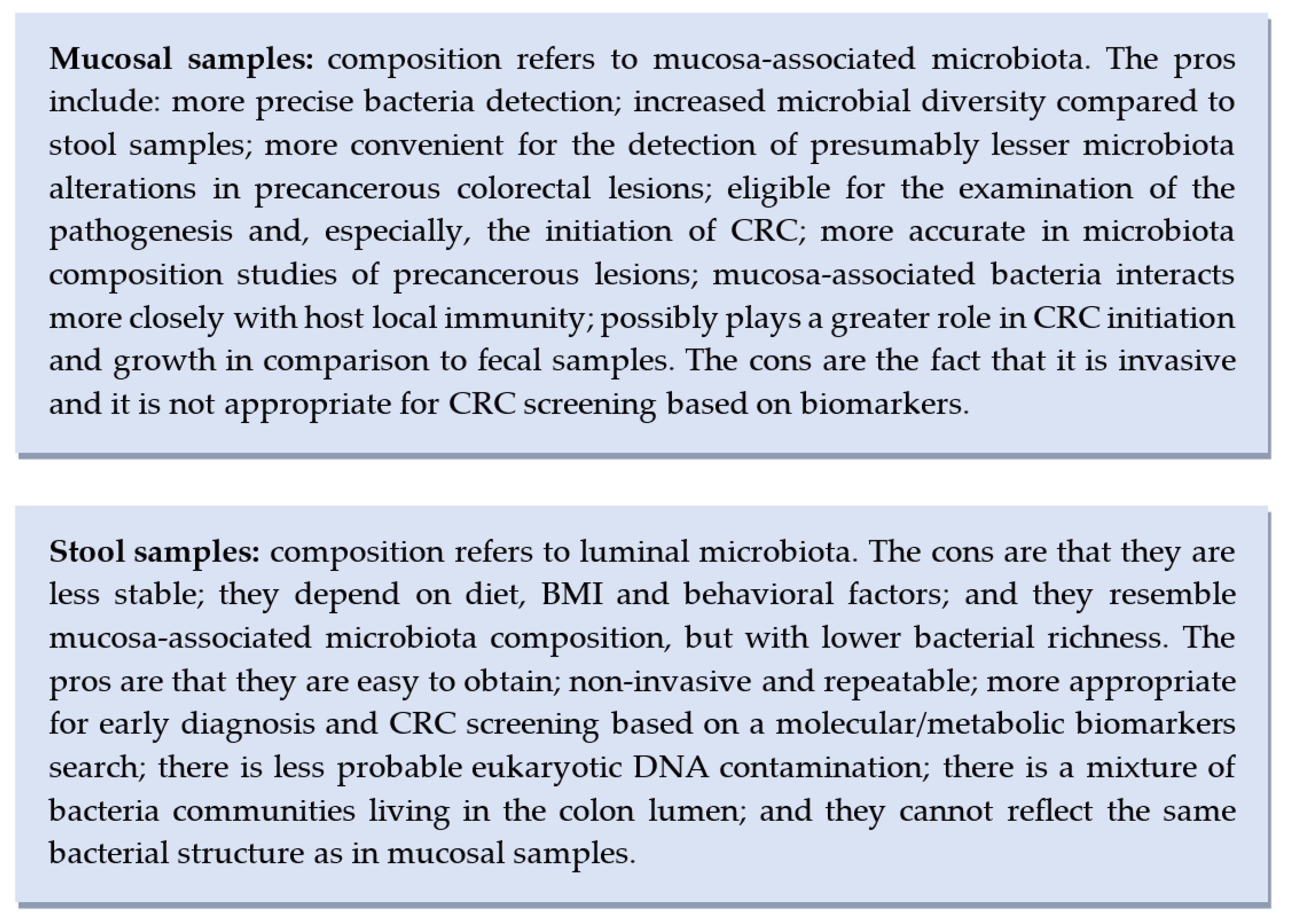
The current literature links microbial dysbiosis with CRC and colorectal precursor lesions. Through exploring the gut microbiota’s structural composition, interactions with the genome, immunome and metabolome, the main goal is to enable the creation of novel and tailored prevention, screening, and therapeutic interventions [59]. According to the included studies’ aims, objectives and outcomes, there is a certain methodology and distinct recommendations for the right selection of matrix. Here we list the main pros and cons for each type of the aforementioned specimens used in gut microbiota studies (Figure 3).

Figure 3.
Advantages and disadvantages of mucosal vs. stool samples usage in gut microbiota studies.
4.4. Limitations of the Review
All the studies included were observational. We did not identify any randomized controlled trials which would meet the eligibility criteria and would be positively quality evaluated for inclusion. The efforts to avoid bias could have been hindered by the fact that non-English trials were not included in the review. Moreover, due to the low number of studies examining gut microbiota composition in both, tissue and/or/vs. fecal samples, we additionally included trials investigating mucosa-associated microbiota alone and those with the aim of examining luminal microbiota (marked accordingly, see Table 1). Similarly, considering the small number of studies looking only at precancerous lesions, we included those looking at premalignant lesions, preinvasive cancer, and CRC along the adenoma-carcinoma sequence. In addition, the results could be hampered by the different study sample sizes, different study populations (according to age, gender, diet, BMI, geographic location, and behavioral factors such as smoking, alcohol consumption, physical activity), different controls (healthy and/or paired normal tissue as self-controls), and different methodologies for the examination of the microbiota composition. The outcomes between the trials, including microbial diversity as well as the abundance of bacteria at phyla, family, and genus taxonomic levels in patients with precancerous colorectal lesions, were inconsistent and at some points, incomparable. Therefore, large sample-size studies examining the composition of gut microbiota in tissue and/or/vs. fecal samples and sharing their metadata are necessary in the future.
4.5. Recommendations and Future Prospects
Considering the large amount of upcoming clinical trials in the field, it is time to rethink our methods and the standardization of specific research practices. There is a significant need for overall recommendations for metagenomic studies which could ensure conceptual results, better comparability, the re-use of metadata, and thus more valuable research input.
The main suggestions are as follows: seek larger sample sizes; use both stool and tissue samples; examine all stages of CRC carcinogenesis; think of both conventional and serrated pathways; continue studies in comprehensive methodology; keep the important data on the type of lesions and the site of sampling performed; consider examination which provides researchers with metabolic data (shotgun sequencing) as well; keep metadata open for the availability of the research community [19,59,60].
Complete network studies investigating the interactions among gut microbiota, diet, the metabolome, genetical alterations, and local immunity responses are paramount for better CRC diagnosis and prevention strategies [60,61,62]. Likewise, understanding the tissue- and fecal-pattern of gut microbiota structure may contribute to novel strategies, such as the early noninvasive stool-based detection of colorectal adenomas and appropriate additional treatment with pre/probiotics, or immunotherapy in people with colorectal neoplasms [63,64].
5. Conclusions
Emerging evidence suggests that gut dysbiosis is one of the major players in the initiation and development of CRC. Ultimately, the findings of this systematic review demonstrate that precancerous colorectal lesions are associated with alterations in gut microbiota composition in both mucosal and fecal samples, in comparison to healthy and self-controls. The majority of studies examining the tissue-associated and/vs. fecal-based structure of microbiota declare a higher presence of Fusobacterium, Escherichia-Shigella, Coprococcus, Streptococcus, Enterococcus, and/or Ruminoccocus spp. and a lower abundance of Actinobacteria, Firmicutes, Eubacteria, Bifidobacterium, Lactobacillus, and butyrate-producing bacteria (Clostridium, Roseburia, Eubacterium, Blautia, and Dorea spp.) in both fecal and tissue specimens, and Faecalibacterium in stool samples from patients with precancerous colorectal lesions compared to healthy controls. Mucosa samples are becoming more relevant in the evaluation of microbiota’s pathophysiological involvement in CR carcinogenesis, while stool samples are more powerful for identifying noninvasive diagnostic or prognostic markers in CRC. Due to the high heterogeneity in terms of methodology and sample size among the included studies, the results are inconclusive. Therefore, further studies with a larger sample size, comprehensive study design, and precise sampling selection are paramount to identify and validate tissue- and fecal-derived colorectal microbial patterns and their role in CRC carcinogenesis. Understanding both the mucosa-associated and luminal pattern of gut microbiota composition could also contribute to CRC diagnosis, prevention, and treatment.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15051602/s1, Table S1: PRISMA checklist; Table S2: Detailed Newcastle–Ottawa scale; Table S3: Extended version of the Table 1 with main characteristics of included studies and microbiota composition analysis.
Author Contributions
Conceptualization, J.V. and T.P.; methodology, J.V.; software, J.V.; validation, J.V., T.P., and K.S.; formal analysis, J.V.; investigation, J.V. and T.P.; resources, J.V.; data curation, J.V.; writing—original draft preparation, J.V. and T.P.; writing—review and editing, J.V. and T.P.; visualization, J.V.; supervision, T.P.; project administration, J.V. and T.P.; funding acquisition, T.P. and K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant (No. S-MIP-22-38) from the Research Council of Lithuania (LMTLT).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Garcia-Etxebarria, K.; Clos-Garcia, M.; Telleria, O.; Nafría, B.; Alonso, C.; Iruarrizaga-Lejarreta, M.; Franke, A.; Crespo, A.; Iglesias, A.; Cubiella, J.; et al. Interplay between Genome, Metabolome and Microbiome in Colorectal Cancer. Cancers 2021, 13, 6216. [Google Scholar] [CrossRef]
- Rasmusson, A.; Zilenaite, D.; Nestarenkaite, A.; Augulis, R.; Laurinaviciene, A.; Ostapenko, V.; Poskus, T.; Laurinavicius, A. Immunogradient Indicators for Antitumor Response Assessment by Automated Tumor-Stroma Interface Zone Detection. Am. J. Pathol. 2020, 190, 1309–1322. [Google Scholar] [CrossRef]
- Oliva, M.; Mulet-Margalef, N.; Ochoa-De-Olza, M.; Napoli, S.; Mas, J.; Laquente, B.; Alemany, L.; Duell, E.J.; Nuciforo, P.; Moreno, V. Tumor-Associated Microbiome: Where Do We Stand? Int. J. Mol. Sci. 2021, 22, 1446. [Google Scholar] [CrossRef]
- Vogtmann, E.; Hua, X.; Zeller, G.; Sunagawa, S.; Voigt, A.Y.; Hercog, R.; Goedert, J.J.; Shi, J.; Bork, P.; Sinha, R. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS ONE 2016, 11, e0155362. [Google Scholar] [CrossRef]
- Yu, L.C.; Wei, S.C.; Ni, Y.H. Impact of microbiota in colorectal carcinogenesis: Lessons from experimental models. Intest. Res. 2018, 16, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Wang, Z.; Li, H.; Cao, Z.; Gao, Z.; Chen, H.; Zhang, X.; Pan, D.; Yang, R.; Zhong, H.; et al. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. J. Gastroenterol. Hepatol. 2020, 35, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Ohigashi, S.; Sudo, K.; Kobayashi, D.; Takahashi, O.; Takahashi, T.; Asahara, T.; Nomoto, K.; Onodera, H. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig Dis. Sci. 2013, 58, 1717–1726. [Google Scholar] [CrossRef]
- Datorre, J.G.; de Carvalho, A.C.; Guimarães, D.P.; Reis, R.M. The Role of Fusobacterium nucleatum in Colorectal Carcinogenesis. Pathobiology 2021, 88, 127–140. [Google Scholar] [CrossRef]
- Viljoen, K.S.; Dakshinamurthy, A.; Goldberg, P.; Blackburn, J.M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS ONE 2015, 10, e0119462. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, L.; Zhao, R.; Long, X.; Coker, O.O.; Sung, J.J.Y. The role of Parvimonas micra in intestinal tumorigenesis in germ-free and conventional APCmin/+ mice. J. Clin. Oncol. 2019, 37, 531. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.A.; Neoh, H.M.; Ab Mutalib, N.S.; Chin, S.F.; Mazlan, L.; Raja Ali, R.A.; Zakaria, A.D.; Ngiu, C.S.; Jamal, R. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci. Rep. 2021, 11, 2925. [Google Scholar] [CrossRef]
- McCoy, A.N.; Araújo-Pérez, F.; Azcárate-Peril, A.; Yeh, J.J.; Sandler, R.S.; Keku, T.O. Fusobacterium is associated with colorectal adenomas. PLoS ONE 2013, 8, e53653. [Google Scholar] [CrossRef]
- Dulal, S.; Keku, T.O. Gut microbiome and colorectal adenomas. Cancer J. 2014, 20, 225–231. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Asadzadeh Aghdaei, H.; Dabiri, H.; Akhavan Sepahi, A.; Modarressi, M.H.; Nazemalhosseini Mojarad, E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 2018, 124, 244–249. [Google Scholar] [CrossRef]
- Shen, X.J.; Rawls, J.F.; Randall, T.; Burcal, L.; Mpande, C.N.; Jenkins, N.; Jovov, B.; Abdo, Z.; Sandler, R.S.; Keku, T.O. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010, 1, 138–147. [Google Scholar] [CrossRef]
- Brim, H.; Yooseph, S.; Zoetendal, E.G.; Lee, E.; Torralbo, M.; Laiyemo, A.O.; Shokrani, B.; Nelson, K.; Ashktorab, H. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS ONE 2013, 8, e81352. [Google Scholar] [CrossRef]
- DeDecker, L.; Coppedge, B.; Avelar-Barragan, J.; Karnes, W.; Whiteson, K. Microbiome distinctions between the CRC carcinogenic pathways. Gut Microbes. 2021, 13, 1854641. [Google Scholar] [CrossRef]
- Olovo, C.V.; Huang, X.; Zheng, X.; Xu, M. Faecal microbial biomarkers in early diagnosis of colorectal cancer. J. Cell Mol. Med. 2021, 25, 10783–10797. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Chiu, J.; Chen, Y.; Huang, Y.; Higashimori, A.; Fang, J.; Brim, H.; Ashktorab, H.; Ng, S.C.; Ng, S.S.M.; et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2017, 23, 2061–2070. [Google Scholar] [CrossRef]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef] [PubMed]
- Mira-Pascual, L.; Cabrera-Rubio, R.; Ocon, S.; Costales, P.; Parra, A.; Suarez, A.; Moris, F.; Rodrigo, L.; Mira, A.; Collado, M.C. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2015, 50, 167–179. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Yu, Y.N.; Yu, T.C.; Zhao, H.J.; Sun, T.T.; Chen, H.M.; Chen, H.Y.; An, H.F.; Weng, Y.R.; Yu, J.; Li, M.; et al. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget 2015, 6, 32013–32026. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Li, J.; Li, J.; Zhang, Y.; Li, X.; Cui, Y.; Gao, Q.; Chen, X.; Chen, Y.; Fang, J.Y. Fecal Enterotoxigenic Bacteroides fragilis-Peptostreptococcus stomatis-Parvimonas micra Biomarker for Noninvasive Diagnosis and Prognosis of Colorectal Laterally Spreading Tumor. Front. Oncol. 2021, 11, 661048. [Google Scholar] [CrossRef]
- Watson, K.M.; Gardner, I.H.; Anand, S.; Siemens, K.N.; Sharpton, T.J.; Kasschau, K.D.; Dewey, E.N.; Martindale, R.; Gaulke, C.A.; Tsikitis, V.L. Colonic Microbial Abundances Predict Adenoma Formers. Ann Surg. 2021, 10, 22–28. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.A. (Eds.) . Cochrane Handbook for Systematic Reviews of Interventions Version 6.3 (Updated February 2022); John Wiley & Sons: Hoboken, NJ, USA, 2022. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Avelar-Barragan, J.; DeDecker, L.; Lu, Z.N.; Coppedge, B.; Karnes, W.E.; Whiteson, K.L. Distinct colon mucosa microbiomes associated with tubular adenomas and serrated polyps. NPJ Biofilms Microbiomes 2022, 8, 69. [Google Scholar] [CrossRef]
- Clos-Garcia, M.; Garcia, K.; Alonso, C.; Iruarrizaga-Lejarreta, M.; D’Amato, M.; Crespo, A.; Iglesias, A.; Cubiella, J.; Bujanda, L.; Falcón-Pérez, J.M. Integrative Analysis of Fecal Metagenomics and Metabolomics in Colorectal Cancer. Cancers 2020, 12, 1142. [Google Scholar] [CrossRef]
- Wei, P.L.; Hung, C.S.; Kao, Y.W.; Lin, Y.C.; Lee, C.Y.; Chang, T.H.; Shia, B.C.; Lin, J.C. Classification of Changes in the Fecal Microbiota Associated with Colonic Adenomatous Polyps Using a Long-Read Sequencing Platform. Genes 2020, 11, 1374. [Google Scholar] [CrossRef]
- Bosch, S.; Acharjee, A.; Quraishi, M.N.; Rojas, P.; Bakkali, A.; Jansen, E.E.; Brizzio Brentar, M.; Kuijvenhoven, J.; Stokkers, P.; Struys, E.; et al. The potential of fecal microbiota and amino acids to detect and monitor patients with adenoma. Gut Microbes. 2022, 14, 2038863. [Google Scholar] [CrossRef]
- Hale, V.L.; Chen, J.; Johnson, S.; Harrington, S.C.; Yab, T.C.; Smyrk, T.C.; Nelson, H.; Boardman, L.A.; Druliner, B.R.; Levin, T.R.; et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol Biomark. Prev. 2017, 26, 85–94. [Google Scholar] [CrossRef]
- Nugent, J.L.; McCoy, A.N.; Addamo, C.J.; Jia, W.; Sandler, R.S.; Keku, T.O. Altered tissue metabolites correlate with microbial dysbiosis in colorectal adenomas. J. Proteome Res. 2014, 13, 1921–1929. [Google Scholar] [CrossRef]
- Goedert, J.J.; Gong, Y.; Hua, X.; Zhong, H.; He, Y.; Peng, P.; Yu, G.; Wang, W.; Ravel, J.; Shi, J.; et al. Fecal Microbiota Characteristics of Patients with Colorectal Adenoma Detected by Screening: A Population-based Study. EBioMedicine 2015, 2, 597–603. [Google Scholar] [CrossRef]
- Yang, T.W.; Lee, W.H.; Tu, S.J.; Huang, W.C.; Chen, H.M.; Sun, T.H.; Lin, C.C. Enterotype-based Analysis of Gut Microbiota along the Conventional Adenoma-Carcinoma Colorectal Cancer Pathway. Sci. Rep. 2019, 9, 10923. [Google Scholar] [CrossRef]
- Zhang, J.; He, Y.; Xia, L.; Yi, J.; Wang, Z.; Zhao, Y.; Song, X.; Li, J.; Liu, H.; Liang, X.; et al. Expansion of Colorectal Cancer Biomarkers Based on Gut Bacteria and Viruses. Cancers 2022, 4, 4662. [Google Scholar] [CrossRef] [PubMed]
- Hua, H.; Sun, Y.; He, X.; Chen, Y.; Teng, L.; Lu, C. Intestinal Microbiota in Colorectal Adenoma-Carcinoma Sequence. Front. Med. 2022, 9, 888340. [Google Scholar] [CrossRef]
- Sanapareddy, N.; Legge, R.M.; Jovov, B.; McCoy, A.; Burcal, L.; Araujo-Perez, F.; Randall, T.A.; Galanko, J.; Benson, A.; Sandler, R.S.; et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012, 6, 1858–1868. [Google Scholar] [CrossRef]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef]
- Geng, J.; Song, Q.; Tang, X.; Liang, X.; Fan, H.; Peng, H.; Guo, Q.; Zhang, Z. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014, 6, 26. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Zheng, J.; Hu, G.; Wang, J.; Huang, C.; Lou, L.; Wang, X.; Zeng, Y. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci. Rep. 2016, 6, 26337. [Google Scholar] [CrossRef]
- Xu, K.; Jiang, B. Analysis of Mucosa-Associated Microbiota in Colorectal Cancer. Med. Sci. Monit. 2017, 23, 4422–4430. [Google Scholar] [CrossRef] [PubMed]
- Wachsmannova, L.; Majek, J.; Zajac, V.; Stevurkova, V.; Ciernikova, S. The study of bacteria in biopsies from Slovak colorectal adenoma and carcinoma patients. Neoplasma 2018, 65, 644–648. [Google Scholar] [CrossRef]
- Bundgaard-Nielsen, C.; Baandrup, U.T.; Nielsen, L.P.; Sørensen, S. The presence of bacteria varies between colorectal adenocarcinomas, precursor lesions and non-malignant tissue. BMC Cancer 2019, 19, 399. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, X.; Xu, H.; Li, S.; Lau, H.C.; Chen, Q.; Zhang, B.; Zhao, L.; Chen, H.; Sung, J.J.; et al. Microbial Community Heterogeneity Within Colorectal Neoplasia and its Correlation With Colorectal Carcinogenesis. Gastroenterology 2021, 160, 2395–2408. [Google Scholar] [CrossRef]
- Chen, H.M.; Yu, Y.N.; Wang, J.L.; Lin, Y.W.; Kong, X.; Yang, C.Q.; Yang, L.; Liu, Z.J.; Yuan, Y.Z.; Liu, F.; et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013, 97, 1044–1052. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Peters, B.A.; Dominianni, C.; Shapiro, J.A.; Church, T.R.; Wu, J.; Miller, G.; Yuen, E.; Freiman, H.; Lustbader, I.; Salik, J.; et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 2016, 4, 69. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, M.; Lu, B.; Liu, C.; Ma, Y.; Liu, L.; Miao, X.; Qin, J.; Chen, H.; Dai, M. Leveraging Fecal Microbial Markers to Improve the Diagnostic Accuracy of the Fecal Immunochemical Test for Advanced Colorectal Adenoma. Clin. Transl. Gastroenterol. 2021, 12, e00389. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Zhou, Y.L.; He, J.; Feng, Z.Q.; Zhang, L.; Lai, X.B.; Zhou, J.X.; Wang, H. Characterizing the composition of intestinal microflora by 16S rRNA gene sequencing. World J. Gastroenterol. 2020, 26, 614–626. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, Y.; Fu, X.; Zhou, X.; Peng, Y.; Shi, L.; Chen, T.; Wu, Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int. J. Cancer. 2016, 139, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol. Rep. 2016, 35, 325–333. [Google Scholar] [CrossRef]
- Stang, A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010, 25, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Han, D.S.; Oh, Y.H.; Lee, A.R.; Lee, Y.R.; Eun, C.S. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci. Rep. 2016, 6, 25271. [Google Scholar] [CrossRef]
- Stary, L.; Mezerova, K.; Skalicky, P.; Zboril, P.; Raclavsky, V. Are we any closer to screening for colorectal cancer using microbial markers? A critical review. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc. Czech Repub. 2017, 161, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Huang, P.; Yang, C.; Xiao, S.; Zhang, G.; Ling, F.; Li, L. Meta-analysis of 16S rRNA Microbial Data Identified Distinctive and Predictive Microbiota Dysbiosis in Colorectal Carcinoma Adjacent Tissue. mSystems 2020, 5, e00138-20. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Dabiri, H.; Asadzadeh-Aghdaei, H.; Sepahi, A.A.; Modarressi, M.H.; Nazemalhosseini-Mojarad, E. The gut microflora assay in patients with colorectal cancer: In feces or tissue samples? Iran J. Microbiol. 2019, 11, 1–6. [Google Scholar] [CrossRef]
- Zackular, J.P.; Rogers, M.A.; Ruffin MT 4th Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef] [PubMed]
- Vacante, M.; Ciuni, R.; Basile, F.; Biondi, A. Gut Microbiota and Colorectal Cancer Development: A Closer Look to the Adenoma-Carcinoma Sequence. Biomedicines 2020, 8, 489. [Google Scholar] [CrossRef] [PubMed]
- Aprile, F.; Bruno, G.; Palma, R.; Mascellino, M.T.; Panetta, C.; Scalese, G.; Oliva, A.; Severi, C.; Pontone, S. Microbiota Alterations in Precancerous Colon Lesions: A Systematic Review. Cancers 2021, 13, 3061. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).