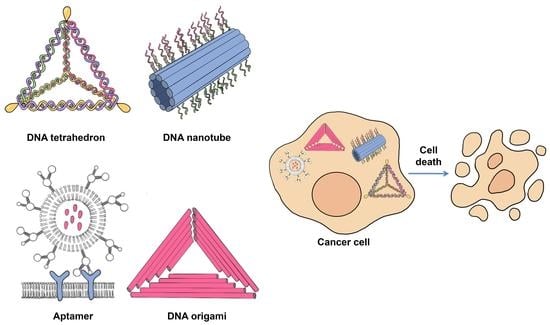
DNA-Based Nanomaterials as Drug Delivery Platforms for Increasing the Effect of Drugs in Tumors
Abstract
:Simple Summary
Abstract
1. Introduction
2. DNA-Based Materials
2.1. DNA Tetrahedron
2.2. DNA Origami
2.3. DNA Nanotube
2.4. Aptamers
2.4.1. AS1411 Aptamer
2.4.2. MUC-1 Aptamer
2.4.3. PSMA Aptamer
2.4.4. CD28 Aptamer
2.4.5. CD44 Aptamer
2.4.6. CD71 Aptamer
2.4.7. CD117 Aptamer
2.4.8. CD133 Aptamer
2.4.9. EGFR Aptamer
2.4.10. HER2 Aptamer
2.4.11. VEGF Aptamer
2.4.12. EpCAM Aptamer
2.4.13. Spiegelmers
3. Delivery of DNA-Based Nanomaterials
3.1. Biodistribution and Biosafety of DNA-Based Nanomaterials
3.2. Cellular Uptake of DNA Nanostructures
3.2.1. Delivery Mechanism of DNA Tetrahedrons
3.2.2. Delivery Mechanism of DNA Origami
3.2.3. Delivery Mechanism of DNA Nanotubes
3.2.4. Aptamer Delivery Mechanism
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, T.; Tian, T.; Zhou, R.; Li, S.; Ma, W.; Zhang, Y.; Liu, N.; Shi, S.; Li, Q.; Xie, X.; et al. Design, Fabrication and Applications of Tetrahedral DNA Nanostructure-Based Multifunctional Complexes in Drug Delivery and Biomedical Treatment. Nat. Protoc. 2020, 15, 2728–2757. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tu, J.; Wang, D.; Zhu, H.; Maity, S.K.; Qu, X.; Bogaert, B.; Pei, H.; Zhang, H. Programmable and Multifunctional DNA-Based Materials for Biomedical Applications. Adv. Mater. 2018, 30, 1703658. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Kong, H.; Cui, Y.; Ren, N.; Li, Q.; Ma, J.; Cui, R.; Zhang, Y.; Shi, J.; Li, Q.; et al. Systematic Study in Mammalian Cells Showing No Adverse Response to Tetrahedral DNA Nanostructure. ACS Appl. Mater. Interfaces 2018, 10, 15442–15448. [Google Scholar] [CrossRef]
- Pei, H.; Zuo, X.; Zhu, D.; Huang, Q.; Fan, C. Functional DNA Nanostructures for Theranostic Applications. Acc. Chem. Res. 2014, 47, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Li, J.; Li, Q.; Huang, Q.; Shi, J.; Yan, H.; Fan, C. Single-Particle Tracking and Modulation of Cell Entry Pathways of a Tetrahedral DNA Nanostructure in Live Cells. Angew. Chem. Int. Ed. 2014, 53, 7745–7750. [Google Scholar] [CrossRef]
- Bagalkot, V.; Farokhzad, O.C.; Langer, R.; Jon, S. An Aptamer–Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform. Angew. Chem. Int. Ed. 2006, 45, 8149–8152. [Google Scholar] [CrossRef] [PubMed]
- Erben, C.M.; Goodman, R.P.; Turberfield, A.J. Single-Molecule Protein Encapsulation in a Rigid DNA Cage. Angew. Chem. Int. Ed. 2006, 45, 7414–7417. [Google Scholar] [CrossRef]
- Ma, W.; Zhan, Y.; Zhang, Y.; Mao, C.; Xie, X.; Lin, Y. The Biological Applications of DNA Nanomaterials: Current Challenges and Future Directions. Signal Transduct. Target. Ther. 2021, 6, 351. [Google Scholar] [CrossRef]
- Belfiore, L.; Saunders, D.N.; Ranson, M.; Thurecht, K.J.; Storm, G.; Vine, K.L. Towards Clinical Translation of Ligand-Functionalized Liposomes in Targeted Cancer Therapy: Challenges and Opportunities. J. Control. Release 2018, 277, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kou, L.; Yao, Q.; Zhang, H.; Chu, M.; Bhutia, Y.D.; Chen, R.; Ganapathy, V. Transporter-Targeted Nano-Sized Vehicles for Enhanced and Site-Specific Drug Delivery. Cancers 2020, 12, 2837. [Google Scholar] [CrossRef]
- Kou, L.; Bhutia, Y.D.; Yao, Q.; He, Z.; Sun, J.; Ganapathy, V. Transporter-Guided Delivery of Nanoparticles to Improve Drug Permeation across Cellular Barriers and Drug Exposure to Selective Cell Types. Front. Pharmacol. 2018, 9, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, W.; Zhao, Y.; Wu, X.; Sun, D.; Yao, Y.; Wang, F.; Zhang, M.; Li, C.; Qin, J.; Zheng, C. Nucleoside Transporter-Guided Cytarabine-Conjugated Liposomes for Intracellular Methotrexate Delivery and Cooperative Choriocarcinoma Therapy. J. Nanobiotechnol. 2021, 19, 184. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506. [Google Scholar] [CrossRef]
- Duangrat, R.; Udomprasert, A.; Kangsamaksin, T. Tetrahedral DNA Nanostructures as Drug Delivery and Bioimaging Platforms in Cancer Therapy. Cancer Sci. 2020, 111, 3164–3173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, Z.; Yang, J. DNA Tetrahedron Delivery Enhances Doxorubicin-Induced Apoptosis of HT-29 Colon Cancer Cells. Nanoscale Res. Lett. 2017, 12, 495. [Google Scholar] [CrossRef] [PubMed]
- Wiraja, C.; Zhu, Y.; Lio, D.C.S.; Yeo, D.C.; Xie, M.; Fang, W.; Li, Q.; Zheng, M.; Van Steensel, M.; Wang, L.; et al. Framework Nucleic Acids as Programmable Carrier for Transdermal Drug Delivery. Nat. Commun. 2019, 10, 1147. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Zhan, X.; Zhang, Z.; Chen, K.; Wang, M.; Sun, Y.; He, B.; Liang, Y. Tetrahedral DNA Nanostructures for Effective Treatment of Cancer: Advances and Prospects. J. Nanobiotechnology 2021, 19, 412. [Google Scholar] [CrossRef]
- Xie, X.; Shao, X.; Ma, W.; Zhao, D.; Shi, S.; Li, Q.; Lin, Y. Overcoming Drug-Resistant Lung Cancer by Paclitaxel Loaded Tetrahedral DNA Nanostructures. Nanoscale 2018, 10, 5457–5465. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-R.; Kim, D.-R.; Lee, T.; Yhee, J.Y.; Kim, B.-S.; Kwon, I.C.; Ahn, D.-R. Drug Delivery by a Self-Assembled DNA Tetrahedron for Overcoming Drug Resistance in Breast Cancer Cells. Chem. Commun. 2013, 49, 2010. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Lee, H.-B.-R.; Johnson, R.W.; Tanskanen, J.T.; Liu, N.; Kim, M.-G.; Pang, C.; Ahn, C.; Bent, S.F.; Bao, Z. Selective Metal Deposition at Graphene Line Defects by Atomic Layer Deposition. Nat. Commun. 2014, 5, 4781. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Xu, H.; Kopelman, R.; Philbert, M.A. Photodynamic Characterization and In Vitro Application of Methylene Blue-Containing Nanoparticle Platforms¶. Photochem. Photobiol. 2007, 81, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Yousuf, S.; Enoch, I.V.M.V. Binding Interactions of Naringenin and Naringin with Calf Thymus DNA and the Role of β-Cyclodextrin in the Binding. AAPS PharmSciTech 2013, 14, 770–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.-R.; Bang, D.; Ahn, D.-R. Nano-Formulation of a Photosensitizer Using a DNA Tetrahedron and Its Potential for in Vivo Photodynamic Therapy. Biomater. Sci. 2016, 4, 605–609. [Google Scholar] [CrossRef]
- Kim, K.-R.; Hwang, D.; Kim, J.; Lee, C.-Y.; Lee, W.; Yoon, D.S.; Shin, D.; Min, S.-J.; Kwon, I.C.; Chung, H.S.; et al. Streptavidin-Mirror DNA Tetrahedron Hybrid as a Platform for Intracellular and Tumor Delivery of Enzymes. J. Control. Release 2018, 280, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Huang, Y.; Gao, P.; Chen, T. Nucleus-Targeted DNA Tetrahedron as a Nanocarrier of Metal Complexes for Enhanced Glioma Therapy. Chem. Commun. 2018, 54, 9394–9397. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jiang, Q.; He, L.; Zhan, P.; Liu, Q.; Liu, S.; Fu, M.; Liu, J.; Li, C.; Ding, B. Self-Assembled Double-Bundle DNA Tetrahedron for Efficient Antisense Delivery. ACS Appl. Mater. Interfaces 2018, 10, 23693–23699. [Google Scholar] [CrossRef]
- Zhang, C.; Han, M.; Zhang, F.; Yang, X.; Du, J.; Zhang, H.; Li, W.; Chen, S. Enhancing Antitumor Efficacy of Nucleoside Analog 5-Fluorodeoxyuridine on HER2-Overexpressing Breast Cancer by Affibody-Engineered DNA Nanoparticle. Int. J. Nanomed. 2020, 15, 885–900. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Strauss, M.T.; Schueder, F.; Haas, D.; Nickels, P.C.; Jungmann, R. Quantifying Absolute Addressability in DNA Origami with Molecular Resolution. Nat. Commun. 2018, 9, 1600. [Google Scholar] [CrossRef]
- Kearney, C.J.; Lucas, C.R.; O’Brien, F.J.; Castro, C.E. DNA Origami: Folded DNA-Nanodevices That Can Direct and Interpret Cell Behavior. Adv. Mater. 2016, 28, 5509–5524. [Google Scholar] [CrossRef] [Green Version]
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA Origami. Nat. Rev. Methods Prim. 2021, 1, 13. [Google Scholar] [CrossRef]
- Babatunde, B.; Arias, D.S.; Cagan, J.; Taylor, R.E. Generating DNA Origami Nanostructures through Shape Annealing. Appl. Sci. 2021, 11, 2950. [Google Scholar] [CrossRef]
- Chen, T.; Ren, L.; Liu, X.; Zhou, M.; Li, L.; Xu, J.; Zhu, X. DNA Nanotechnology for Cancer Diagnosis and Therapy. Int. J. Mol. Sci. 2018, 19, 1671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiden, J.; Bastings, M.M.C. DNA Origami Nanostructures for Controlled Therapeutic Drug Delivery. Curr. Opin. Colloid Interface Sci. 2021, 52, 101411. [Google Scholar] [CrossRef]
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.; Lin, L.; Qiu, D.; Wang, Z.-G.; Zou, G.; Liang, X.; Yan, H.; et al. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403. [Google Scholar] [CrossRef]
- Majikes, J.M.; Nash, J.A.; LaBean, T.H. Search for Effective Chemical Quenching to Arrest Molecular Assembly and Directly Monitor DNA Nanostructure Formation. Nanoscale 2017, 9, 1637–1644. [Google Scholar] [CrossRef]
- Halley, P.D.; Lucas, C.R.; McWilliams, E.M.; Webber, M.J.; Patton, R.A.; Kural, C.; Lucas, D.M.; Byrd, J.C.; Castro, C.E. Daunorubicin-Loaded DNA Origami Nanostructures Circumvent Drug-Resistance Mechanisms in a Leukemia Model. Small 2016, 12, 308–320. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.; Ma, X.; Xue, X.; Jiang, Q.; Song, L.; Dai, L.; Zhang, C.; Jin, S.; Yang, K.; Ding, B.; et al. A Photosensitizer-Loaded DNA Origami Nanosystem for Photodynamic Therapy. ACS Nano 2016, 10, 3486–3495. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, A.R.; Anderson, N.; Kizer, M.; Halvorsen, K.; Wang, X. Beyond the Fold: Emerging Biological Applications of DNA Origami. ChemBioChem 2016, 17, 1081–1089. [Google Scholar] [CrossRef]
- O’Neill, P.; Rothemund, P.W.K.; Kumar, A.; Fygenson, D.K. Sturdier DNA Nanotubes via Ligation. Nano Lett. 2006, 6, 1379–1383. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Yu, L.; Shen, W. DNA Nanotechnology and Its Applications in Biomedical Research. J. Biomed. Nanotechnol. 2014, 10, 2350–2370. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, Y.; Liu, P.; Wang, L.; Lin, J.; Fan, C. Biomimetic DNA Nanotubes: Nanoscale Channel Design and Applications. Angew. Chem. Int. Ed. 2019, 58, 8996–9011. [Google Scholar] [CrossRef]
- Aldaye, F.A.; Lo, P.K.; Karam, P.; McLaughlin, C.K.; Cosa, G.; Sleiman, H.F. Modular Construction of DNA Nanotubes of Tunable Geometry and Single- or Double-Stranded Character. Nat. Nanotechnol. 2009, 4, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R. Introducing Bacteria and Synthetic Biomolecules along Engineered DNA Fibers. Small 2021, 17, 2100136. [Google Scholar] [CrossRef]
- Liang, L.; Shen, J.-W.; Wang, Q. Molecular Dynamics Study on DNA Nanotubes as Drug Delivery Vehicle for Anticancer Drugs. Colloids Surf. B Biointerfaces 2017, 153, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.J.W.; Stephanopoulos, N. Functionalizing DNA Nanostructures for Therapeutic Applications. WIREs Nanomed. Nanobiotechnol. 2021, 13, e1729. [Google Scholar] [CrossRef]
- Ko, S.; Liu, H.; Chen, Y.; Mao, C. DNA Nanotubes as Combinatorial Vehicles for Cellular Delivery. Biomacromolecules 2008, 9, 3039–3043. [Google Scholar] [CrossRef] [Green Version]
- Kocabey, S.; Ekim Kocabey, A.; Schneiter, R.; Rüegg, C. Membrane-Interacting DNA Nanotubes Induce Cancer Cell Death. Nanomaterials 2021, 11, 2003. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Lv, W.Y.; Yan, Y.; Yang, F.F.; Zhen, S.J.; Huang, C.Z. Nucleolin-Targeted DNA Nanotube for Precise Cancer Therapy through Förster Resonance Energy Transfer-Indicated Telomerase Responsiveness. Anal. Chem. 2021, 93, 3526–3534. [Google Scholar] [CrossRef]
- Kong, H.Y.; Byun, J. Nucleic Acid Aptamers: New Methods for Selection, Stabilization, and Application in Biomedical Science. Biomol. Ther. 2013, 21, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Dutta, B.; Barick, K.C.; Hassan, P.A. Recent Advances in Active Targeting of Nanomaterials for Anticancer Drug Delivery. Adv. Colloid Interface Sci. 2021, 296, 102509. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santosh, B.; Yadava, P.K. Nucleic Acid Aptamers: Research Tools in Disease Diagnostics and Therapeutics. Biomed Res. Int. 2014, 2014, 540451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef]
- Hernández-Jiménez, M.; Martín-Vílchez, S.; Ochoa, D.; Mejía-Abril, G.; Román, M.; Camargo-Mamani, P.; Luquero-Bueno, S.; Jilma, B.; Moro, M.A.; Fernández, G.; et al. First-in-Human Phase I Clinical Trial of a TLR4-Binding DNA Aptamer, ApTOLL: Safety and Pharmacokinetics in Healthy Volunteers. Mol. Ther.-Nucleic Acids 2022, 28, 124–135. [Google Scholar] [CrossRef]
- Liu, S.; Xu, Y.; Jiang, X.; Tan, H.; Ying, B. Translation of Aptamers toward Clinical Diagnosis and Commercialization. Biosens. Bioelectron. 2022, 208, 114168. [Google Scholar] [CrossRef]
- Yuan, W.; Wan, L.; Peng, H.; Zhong, Y.; Cai, W.; Zhang, Y.; Ai, W.; Wu, J. The Influencing Factors and Functions of DNA G-quadruplexes. Cell Biochem. Funct. 2020, 38, 524–532. [Google Scholar] [CrossRef]
- Jia, W.; Yao, Z.; Zhao, J.; Guan, Q.; Gao, L. New Perspectives of Physiological and Pathological Functions of Nucleolin (NCL). Life Sci. 2017, 186, 1–10. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic Applications of AS1411 Aptamer, an Update Review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef]
- Hashemi, M.; Shamshiri, A.; Saeedi, M.; Tayebi, L.; Yazdian-Robati, R. Aptamer-Conjugated PLGA Nanoparticles for Delivery and Imaging of Cancer Therapeutic Drugs. Arch. Biochem. Biophys. 2020, 691, 108485. [Google Scholar] [CrossRef]
- Zhao, P.; Tang, Z.-W.; Lin, H.-C.; Djuanda, D.; Zhu, Z.; Niu, Q.; Zhao, L.-M.; Qian, Y.-N.; Cao, G.; Shen, J.-L.; et al. VEGF Aptamer/i-Motif-Based Drug Co-Delivery System for Combined Chemotherapy and Photodynamic Therapy. Photodiagnosis Photodyn. Ther. 2021, 36, 102547. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A Phase II Trial of AS1411 (a Novel Nucleolin-Targeted DNA Aptamer) in Metastatic Renal Cell Carcinoma. Investig. New Drugs. 2014, 32, 178–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-Quadruplex Oligonucleotide AS1411 as a Cancer-Targeting Agent: Uses and Mechanisms. Biochim. Biophys. Acta-Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef]
- Tong, X.; Ga, L.; Ai, J.; Wang, Y. Progress in Cancer Drug Delivery Based on AS1411 Oriented Nanomaterials. J. Nanobiotechnol. 2022, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Nabavinia, M.M.S.; Gholoobi, A.; Charbgoo, F.; Nabavinia, M.M.S.; Ramezani, M.; Abnous, K. Anti-MUC1 Aptamer: A Potential Opportunity for Cancer Treatment. Med. Res. Rev. 2017, 37, 1518–1539. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. Functional Targeting of the MUC1 Oncogene in Human Cancers. Cancer Biol. Ther. 2009, 8, 1197–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousefi, M.; Dehghani, S.; Nosrati, R.; Zare, H.; Evazalipour, M.; Mosafer, J.; Tehrani, B.S.; Pasdar, A.; Mokhtarzadeh, A.; Ramezani, M. Aptasensors as a New Sensing Technology Developed for the Detection of MUC1 Mucin: A Review. Biosens. Bioelectron. 2019, 130, 1–19. [Google Scholar] [CrossRef]
- Esfandyari-Manesh, M.; Mohammadi, A.; Atyabi, F.; Nabavi, S.M.; Ebrahimi, S.M.; Shahmoradi, E.; Varnamkhasti, B.S.; Ghahremani, M.H.; Dinarvand, R. Specific Targeting Delivery to MUC1 Overexpressing Tumors by Albumin-Chitosan Nanoparticles Conjugated to DNA Aptamer. Int. J. Pharm. 2016, 515, 607–615. [Google Scholar] [CrossRef]
- Ma, D.; Hopf, C.E.; Malewicz, A.D.; Donovan, G.P.; Senter, P.D.; Goeckeler, W.F.; Maddon, P.J.; Olson, W.C. Potent Antitumor Activity of an Auristatin-Conjugated, Fully Human Monoclonal Antibody to Prostate-Specific Membrane Antigen. Clin. Cancer Res. 2006, 12, 2591–2596. [Google Scholar] [CrossRef] [Green Version]
- Lupold, S.E. Aptamers and Apple Pies: A Mini-Review of PSMA Aptamers and Lessons from Donald S. Coffey. Am. J. Clin. Exp. Urol. 2018, 6, 78–86. [Google Scholar] [PubMed]
- Lupold, S.E.; Hicke, B.J.; Lin, Y.; Coffey, D.S. Identification and Characterization of Nuclease-Stabilized RNA Molecules That Bind Human Prostate Cancer Cells via the Prostate-Specific Membrane Antigen. Cancer Res. 2002, 62, 4029–4033. [Google Scholar]
- Salaheldin, T.A.; Bharali, D.J.; Mousa, S.A. Functionalized Nano-Targeted Moieties in Management of Prostate Cancer. Futur. Oncol. 2020, 16, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-Aptamer Bioconjugates. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef] [Green Version]
- Dhar, S.; Gu, F.X.; Langer, R.; Farokhzad, O.C.; Lippard, S.J. Targeted Delivery of Cisplatin to Prostate Cancer Cells by Aptamer Functionalized Pt(IV) Prodrug-PLGA–PEG Nanoparticles. Proc. Natl. Acad. Sci. USA 2008, 105, 17356–17361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Teply, B.; Sherifi, I.; Sung, J.; Luther, G.; Gu, F.; Levynissenbaum, E.; Radovicmoreno, A.; Langer, R.; Farokhzad, O. Formulation of Functionalized PLGA–PEG Nanoparticles for in Vivo Targeted Drug Delivery. Biomaterials 2007, 28, 869–876. [Google Scholar] [CrossRef] [Green Version]
- McNamara, J.O.; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell Type–Specific Delivery of SiRNAs with Aptamer-SiRNA Chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef]
- Farokhzad, O.C.; Cheng, J.; Teply, B.A.; Sherifi, I.; Jon, S.; Kantoff, P.W.; Richie, J.P.; Langer, R. Targeted Nanoparticle-Aptamer Bioconjugates for Cancer Chemotherapy in Vivo. Proc. Natl. Acad. Sci. USA 2006, 103, 6315–6320. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Tai, Z.; Gu, F.; Hu, C.; Zhu, Q.; Gao, S. Aptamer-Mediated Delivery of Docetaxel to Prostate Cancer through Polymeric Nanoparticles for Enhancement of Antitumor Efficacy. Eur. J. Pharm. Biopharm. 2016, 107, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.E.; Lee, K.H.; Park, Y.S.; Oh, D.-K.; Oh, S.; Kim, K.-S.; Kim, D.-E. RNA Aptamer-Conjugated Liposome as an Efficient Anticancer Drug Delivery Vehicle Targeting Cancer Cells in Vivo. J. Control. Release 2014, 196, 234–242. [Google Scholar] [CrossRef]
- Xu, W.; Siddiqui, I.A.; Nihal, M.; Pilla, S.; Rosenthal, K.; Mukhtar, H.; Gong, S. Aptamer-Conjugated and Doxorubicin-Loaded Unimolecular Micelles for Targeted Therapy of Prostate Cancer. Biomaterials 2013, 34, 5244–5253. [Google Scholar] [CrossRef] [Green Version]
- Bandekar, A.; Zhu, C.; Jindal, R.; Bruchertseifer, F.; Morgenstern, A.; Sofou, S. Anti–Prostate-Specific Membrane Antigen Liposomes Loaded with 225 Ac for Potential Targeted Antivascular α-Particle Therapy of Cancer. J. Nucl. Med. 2014, 55, 107–114. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Jeong, Y.Y.; Jon, S. A Drug-Loaded Aptamer-Gold Nanoparticle Bioconjugate for Combined CT Imaging and Therapy of Prostate Cancer. ACS Nano 2010, 4, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Hernández, C.D.; Rodríguez-Martínez, G.; Cortés-Ramírez, S.A.; Morales-Pacheco, M.; Cruz-Burgos, M.; Losada-García, A.; Reyes-Grajeda, J.P.; González-Ramírez, I.; González-Covarrubias, V.; Camacho-Arroyo, I.; et al. Aptamers as Theragnostic Tools in Prostate Cancer. Biomolecules 2022, 12, 1056. [Google Scholar] [CrossRef] [PubMed]
- Nozari, A.; Berezovski, M.V. Aptamers for CD Antigens: From Cell Profiling to Activity Modulation. Mol. Ther.-Nucleic Acids 2017, 6, 29–44. [Google Scholar] [CrossRef] [Green Version]
- Pastor, F.; Soldevilla, M.M.; Villanueva, H.; Kolonias, D.; Inoges, S.; de Cerio, A.L.; Kandzia, R.; Klimyuk, V.; Gleba, Y.; Gilboa, E.; et al. CD28 Aptamers as Powerful Immune Response Modulators. Mol. Ther.-Nucleic Acids 2013, 2, e98. [Google Scholar] [CrossRef]
- Soldevilla, M.M.; Villanueva, H.; Casares, N.; Lasarte, J.J.; Bendandi, M.; Inoges, S.; de Cerio, A.L.-D.; Pastor, F. MRP1-CD28 Bi-Specific Oligonucleotide Aptamers: Target Costimulation to Drug-Resistant Melanoma Cancer Stem Cells. Oncotarget. 2016, 7, 23182–23196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, C.; Gao, S.; Hu, S.; Liu, X.; Li, H.; Dong, J.; Huang, A.; Zhu, L.; Zhou, P.; Li, S.; et al. Self-Assembled Multivalent Aptamer Nanoparticles with Potential CAR-like Characteristics Could Activate T Cells and Inhibit Melanoma Growth. Mol. Ther.-Oncolytics 2020, 17, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Lozano, T.; Soldevilla, M.M.; Casares, N.; Villanueva, H.; Bendandi, M.; Lasarte, J.J.; Pastor, F. Targeting Inhibition of Foxp3 by a CD28 2′-Fluro Oligonucleotide Aptamer Conjugated to P60-Peptide Enhances Active Cancer Immunotherapy. Biomaterials 2016, 91, 73–80. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, S.; Wu, L.; Chen, F.; Qiu, L.; Tan, W. Aptamer-Functionalized Nanodevices for Dynamic Manipulation of Membrane Receptor Signaling in Living Cells. Nano Lett. 2022, 22, 7853–7859. [Google Scholar] [CrossRef]
- Kazemi, Y.; Dehghani, S.; Nosrati, R.; Taghdisi, S.M.; Abnous, K.; Alibolandi, M.; Ramezani, M. Recent Progress in the Early Detection of Cancer Based on CD44 Biomarker; Nano-Biosensing Approaches. Life Sci. 2022, 300, 120593. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Ponta, H. Perspectives of CD44 Targeting Therapies. Arch. Toxicol. 2015, 89, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.W.-S.; Chan, C.K.W.; Yu, J.; He, M.; Choi, C.H.J.; Lau, J.Y.W.; Wong, N. Development of CD44E/s Dual-Targeting DNA Aptamer as Nanoprobe to Deliver Treatment in Hepatocellular Carcinoma. Nanotheranostics 2022, 6, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, J.S.; Kim, W.; Lee, J.H.; Jun, B.-H.; Kim, K.-S.; Kim, D.-E. Aptamer-Conjugated Nano-Liposome for Immunogenic Chemotherapy with Reversal of Immunosuppression. J. Control. Release 2022, 348, 893–910. [Google Scholar] [CrossRef] [PubMed]
- Darabi, F.; Saidijam, M.; Nouri, F.; Mahjub, R.; Soleimani, M. Anti-CD44 and EGFR Dual-Targeted Solid Lipid Nanoparticles for Delivery of Doxorubicin to Triple-Negative Breast Cancer Cell Line: Preparation, Statistical Optimization, and In Vitro Characterization. Biomed Res. Int. 2022, 2022, 6253978. [Google Scholar] [CrossRef] [PubMed]
- Beals, N.; Thiagarajan, P.S.; Soehnlen, E.; Das, A.; Reizes, O.; Lathia, J.D.; Basu, S. Five-Part Pentameric Nanocomplex Shows Improved Efficacy of Doxorubicin in CD44+ Cancer Cells. ACS Omega 2017, 2, 7702–7713. [Google Scholar] [CrossRef]
- Wu, X.; Liu, H.; Han, D.; Peng, B.; Zhang, H.; Zhang, L.; Li, J.; Liu, J.; Cui, C.; Fang, S.; et al. Elucidation and Structural Modeling of CD71 as a Molecular Target for Cell-Specific Aptamer Binding. J. Am. Chem. Soc. 2019, 141, 10760–10769. [Google Scholar] [CrossRef]
- Johnson, M.; El-Khoueiry, A.; Hafez, N.; Lakhani, N.; Mamdani, H.; Rodon, J.; Sanborn, R.E.; Garcia-Corbacho, J.; Boni, V.; Stroh, M.; et al. Phase I, First-in-Human Study of the Probody Therapeutic CX-2029 in Adults with Advanced Solid Tumor Malignancies. Clin. Cancer Res. 2021, 27, 4521–4530. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, C.; Zhang, L.; Peng, B.; Zhang, Y.; Liu, Y.; Li, L.; Ye, M.; Xiong, W.; Tan, W. CD71-Specific Aptamer Conjugated with Monomethyl Auristatin E for the Treatment of Uveal Melanoma. ACS Appl. Mater. Interfaces 2022, 14, 32–40. [Google Scholar] [CrossRef]
- Edling, C.E.; Hallberg, B. C-Kit—A Hematopoietic Cell Essential Receptor Tyrosine Kinase. Int. J. Biochem. Cell Biol. 2007, 39, 1995–1998. [Google Scholar] [CrossRef]
- Hans, C.P.; Finn, W.G.; Singleton, T.P.; Schnitzer, B.; Ross, C.W. Usefulness of Anti-CD117 in the Flow Cytometric Analysis of Acute Leukemia. Am. J. Clin. Pathol. 2002, 117, 301–305. [Google Scholar] [CrossRef]
- Zhao, N.; Pei, S.-N.; Qi, J.; Zeng, Z.; Iyer, S.P.; Lin, P.; Tung, C.-H.; Zu, Y. Oligonucleotide Aptamer-Drug Conjugates for Targeted Therapy of Acute Myeloid Leukemia. Biomaterials 2015, 67, 42–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, S.; Yoon, H.; Yebra, M.; Tang, C.-M.; Gilardi, M.; Narayanan, J.S.S.; White, R.R.; Sicklick, J.K.; Ray, P. Anti-KIT DNA Aptamer for Targeted Labeling of Gastrointestinal Stromal Tumor. Mol. Cancer Ther. 2020, 19, 1173–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shraim, A.S.; Hunaiti, A.; Awidi, A.; Alshaer, W.; Ababneh, N.A.; Abu-Irmaileh, B.; Odeh, F.; Ismail, S. Developing and Characterization of Chemically Modified RNA Aptamers for Targeting Wild Type and Mutated C-KIT Receptor Tyrosine Kinases. J. Med. Chem. 2020, 63, 2209–2228. [Google Scholar] [CrossRef] [PubMed]
- Ulasov, I.V.; Nandi, S.; Dey, M.; Sonabend, A.M.; Lesniak, M.S. Inhibition of Sonic Hedgehog and Notch Pathways Enhances Sensitivity of CD133+ Glioma Stem Cells to Temozolomide Therapy. Mol. Med. 2011, 17, 103–112. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of Human Brain Tumour Initiating Cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Shigdar, S.; Qiao, L.; Zhou, S.-F.; Xiang, D.; Wang, T.; Li, Y.; Lim, L.Y.; Kong, L.; Li, L.; Duan, W. RNA Aptamers Targeting Cancer Stem Cell Marker CD133. Cancer Lett. 2013, 330, 84–95. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Da Won Bae, S.; Nguyen, R.; Huo, X.; Han, S.; Zhang, Z.; Hebbard, L.; Duan, W.; Eslam, M.; Liddle, C.; et al. An Aptamer-Based Drug Delivery Agent (CD133-Apt-Dox) Selectively and Effectively Kills Liver Cancer Stem-like Cells. Cancer Lett. 2021, 501, 124–132. [Google Scholar] [CrossRef]
- Ge, M.H.; Zhu, X.H.; Shao, Y.M.; Wang, C.; Huang, P.; Wang, Y.; Jiang, Y.; Maimaitiyiming, Y.; Chen, E.; Yang, C.; et al. Synthesis and Characterization of CD133 Targeted Aptamer–Drug Conjugates for Precision Therapy of Anaplastic Thyroid Cancer. Biomater. Sci. 2021, 9, 1313–1324. [Google Scholar] [CrossRef]
- Poonaki, E.; Nickel, A.-C.; Ardestani, M.S.; Rademacher, L.; Kaul, M.; Apartsin, E.; Meuth, S.G.; Gorji, A.; Janiak, C.; Kahlert, U.D. CD133-Functionalized Gold Nanoparticles as a Carrier Platform for Telaglenastat (CB-839) against Tumor Stem Cells. Int. J. Mol. Sci. 2022, 23, 5479. [Google Scholar] [CrossRef]
- Pang, L.; Huang, X.; Zhu, L.; Xiao, H.; Li, M.; Guan, H.; Gao, J.; Jin, H. Targeted Killing of CD133+ Lung Cancer Stem Cells Using Paclitaxel-Loaded PLGA-PEG Nanoparticles with CD133 Aptamers. Nan Fang Yi Ke Da Xue Xue Bao 2022, 42, 26–35. [Google Scholar]
- Huang, X.; Huang, J.; Leng, D.; Yang, S.; Yao, Q.; Sun, J.; Hu, J. Gefitinib-Loaded DSPE-PEG2000 Nanomicelles with CD133 Aptamers Target Lung Cancer Stem Cells. World J. Surg. Oncol. 2017, 15, 167. [Google Scholar] [CrossRef] [Green Version]
- Zahiri, M.; Babaei, M.; Abnous, K.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Hybrid Nanoreservoirs Based on Dextran-capped Dendritic Mesoporous Silica Nanoparticles for CD133-targeted Drug Delivery. J. Cell. Physiol. 2020, 235, 1036–1050. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhuang, H.; Zhuang, Z.; Lu, Y.; Xia, R.; Gan, L.; Wu, Y. Development of Docetaxel Liposome Surface Modified with CD133 Aptamers for Lung Cancer Targeting. Artif. Cells Nanomed. Biotechnol. 2017, 46, 1864–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wieduwilt, M.J.; Moasser, M.M. The Epidermal Growth Factor Receptor Family: Biology Driving Targeted Therapeutics. Cell. Mol. Life Sci. 2008, 65, 1566–1584. [Google Scholar] [CrossRef] [Green Version]
- Esposito, C.L.; Passaro, D.; Longobardo, I.; Condorelli, G.; Marotta, P.; Affuso, A.; de Franciscis, V.; Cerchia, L. A Neutralizing RNA Aptamer against EGFR Causes Selective Apoptotic Cell Death. PLoS ONE 2011, 6, e24071. [Google Scholar] [CrossRef]
- Zhou, J.; Bobbin, M.L.; Burnett, J.C.; Rossi, J.J. Current Progress of RNA Aptamer-Based Therapeutics. Front. Genet. 2012, 3, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenkranz, A.A.; Slastnikova, T.A. Epidermal Growth Factor Receptor: Key to Selective Intracellular Delivery. Biochem 2020, 85, 967–993. [Google Scholar] [CrossRef]
- Ulfo, L.; Costantini, P.E.; Di Giosia, M.; Danielli, A.; Calvaresi, M. EGFR-Targeted Photodynamic Therapy. Pharmaceutics 2022, 14, 241. [Google Scholar] [CrossRef]
- Passariello, M.; Camorani, S.; Vetrei, C.; Cerchia, L.; De Lorenzo, C. Novel Human Bispecific Aptamer–Antibody Conjugates for Efficient Cancer Cell Killing. Cancers 2019, 11, 1268. [Google Scholar] [CrossRef] [Green Version]
- Lv, T.; Li, Z.; Xu, L.; Zhang, Y.; Chen, H.; Gao, Y. Chloroquine in Combination with Aptamer-Modified Nanocomplexes for Tumor Vessel Normalization and Efficient Erlotinib/Survivin ShRNA Co-Delivery to Overcome Drug Resistance in EGFR-Mutated Non-Small Cell Lung Cancer. Acta Biomater. 2018, 76, 257–274. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Wang, J.; Wang, L.; Tan, X.; Tu, K.; Tong, X.; Qi, L. Aptamer Functionalized Cisplatin-Albumin Nanoparticles for Targeted Delivery to Epidermal Growth Factor Receptor Positive Cervical Cancer. J. Biomed. Nanotechnol. 2016, 12, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.J.; Jeong, H.Y.; Kim, M.W.; Jeong, I.H.; Choi, M.J.; You, Y.M.; Im, C.S.; Song, I.H.; Lee, T.S.; Park, Y.S. Anti-EGFR Lipid Micellar Nanoparticles Co-Encapsulating Quantum Dots and Paclitaxel for Tumor-Targeted Theranosis. Nanoscale 2018, 10, 19338–19350. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Cao, Y.; Shen, H.; Ma, Q.; Mao, S.; Li, S.; Sun, J. EGFR Aptamer-Conjugated Liposome-Polycation-DNA Complex for Targeted Delivery of SATB1 Small Interfering RNA to Choriocarcinoma Cells. Biomed. Pharmacother. 2018, 107, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shaikh, A.; Yu, Y.; Li, Y.; Ni, S.; Lu, A.; Zhang, G. Potential Diagnostic and Therapeutic Applications of Oligonucleotide Aptamers in Breast Cancer. Int. J. Mol. Sci. 2017, 18, 1851. [Google Scholar] [CrossRef] [Green Version]
- Şahin, S.; Caglayan, M.O.; Üstündağ, Z. Recent Advances in Aptamer-Based Sensors for Breast Cancer Diagnosis: Special Cases for Nanomaterial-Based VEGF, HER2, and MUC1 Aptasensors. Microchim. Acta 2020, 187, 549. [Google Scholar] [CrossRef]
- Dastjerdi, K.; Tabar, G.H.; Dehghani, H.; Haghparast, A. Generation of an Enriched Pool of DNA Aptamers for an HER2-Overexpressing Cell Line Selected by Cell SELEX. Biotechnol. Appl. Biochem. 2011, 58, 226–230. [Google Scholar] [CrossRef]
- Kim, M.Y.; Jeong, S. In Vitro Selection of RNA Aptamer and Specific Targeting of ErbB2 in Breast Cancer Cells. Nucleic Acid Ther. 2011, 21, 173–178. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Nguyen, H.H.; Byrom, M.; Ellington, A.D. Inhibition of Cell Proliferation by an Anti-EGFR Aptamer. PLoS ONE 2011, 6, e20299. [Google Scholar] [CrossRef]
- Chen, C.B.; Chernis, G.A.; Hoang, V.Q.; Landgraf, R. Inhibition of Heregulin Signaling by an Aptamer That Preferentially Binds to the Oligomeric Form of Human Epidermal Growth Factor Receptor-3. Proc. Natl. Acad. Sci. USA 2003, 100, 9226–9231. [Google Scholar] [CrossRef] [Green Version]
- Dassie, J.P.; Liu, X.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O.; Giangrande, P.H. Systemic Administration of Optimized Aptamer-SiRNA Chimeras Promotes Regression of PSMA-Expressing Tumors. Nat. Biotechnol. 2009, 27, 839–846. [Google Scholar] [CrossRef] [Green Version]
- Mahlknecht, G.; Maron, R.; Mancini, M.; Schechter, B.; Sela, M.; Yarden, Y. Aptamer to ErbB-2/HER2 Enhances Degradation of the Target and Inhibits Tumorigenic Growth. Proc. Natl. Acad. Sci. USA 2013, 110, 8170–8175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhritlahre, R.K.; Saneja, A. Recent Advances in HER2-Targeted Delivery for Cancer Therapy. Drug Discov. Today. 2021, 26, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, K.; Chen, Z.; Shi, J.; Zhou, D.; Xie, G. A Fluorescence Biosensor for VEGF Detection Based on DNA Assembly Structure Switching and Isothermal Amplification. Biosens. Bioelectron. 2017, 89, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-L.; Lien, C.-W.; Wang, C.-W.; Harroun, S.G.; Huang, C.-C.; Chang, H.-T. Immobilization of Aptamer-Modified Gold Nanoparticles on BiOCl Nanosheets: Tunable Peroxidase-like Activity by Protein Recognition. Biosens. Bioelectron. 2016, 75, 181–187. [Google Scholar] [CrossRef]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Melone, M.A.B.; Montesarchio, D. Anti-VEGF DNA-based Aptamers in Cancer Therapeutics and Diagnostics. Med. Res. Rev. 2021, 41, 464–506. [Google Scholar] [CrossRef]
- Xie, X.; Zhang, Y.; Ma, W.; Shao, X.; Zhan, Y.; Mao, C.; Zhu, B.; Zhou, Y.; Zhao, H.; Cai, X. Potent Anti-angiogenesis and Anti-tumour Activity of Pegaptanib-loaded Tetrahedral DNA Nanostructure. Cell Prolif. 2019, 52, e12662. [Google Scholar] [CrossRef] [Green Version]
- Fu, B.; Lin, H.-C.; Liu, Y.-C.; Lin, J.-R.; Xiong, W.-M.; Deng, S.-J.; Chen, N.; Liang, R.; Zhao, P. VEGF Aptamer/i-Motif-Grafted Multi-Functional SPION Nanocarrier for Chemotherapeutic/Phototherapeutic Synergistic Research. J. Biomater. Appl. 2022, 36, 1277–1288. [Google Scholar] [CrossRef]
- Macdonald, J.; Henri, J.; Roy, K.; Hays, E.; Bauer, M.; Veedu, R.; Pouliot, N.; Shigdar, S. EpCAM Immunotherapy versus Specific Targeted Delivery of Drugs. Cancers 2018, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Went, P.; Vasei, M.; Bubendorf, L.; Terracciano, L.; Tornillo, L.; Riede, U.; Kononen, J.; Simon, R.; Sauter, G.; Baeuerle, P.A. Frequent High-Level Expression of the Immunotherapeutic Target Ep-CAM in Colon, Stomach, Prostate and Lung Cancers. Br. J. Cancer. 2006, 94, 128–135. [Google Scholar] [CrossRef] [Green Version]
- Munz, M.; Baeuerle, P.A.; Gires, O. The Emerging Role of EpCAM in Cancer and Stem Cell Signaling. Cancer Res. 2009, 69, 5627–5629. [Google Scholar] [CrossRef] [Green Version]
- Shigdar, S.; Lin, J.; Yu, Y.; Pastuovic, M.; Wei, M.; Duan, W. RNA Aptamer against a Cancer Stem Cell Marker Epithelial Cell Adhesion Molecule. Cancer Sci. 2011, 102, 991–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghasemii, K.; Darroudi, M.; Rahimmanesh, I.; Ghomi, M.; Hassanpour, M.; Sharifi, E.; Yousefiasl, S.; Ahmadi, S.; Zarrabi, A.; Borzacchiello, A.; et al. Advances in Aptamer-Based Drug Delivery Vehicles for Cancer Therapy. Biomater. Adv. 2022, 140, 213077. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhao, S.; Yu, X.; Huang, S.; Liu, H.Y. Simultaneous Targeting of CD44 and EpCAM with a Bispecific Aptamer Effectively Inhibits Intraperitoneal Ovarian Cancer Growth. Theranostics 2017, 7, 1373–1388. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Shigdar, S.; Bean, A.G.; Bruce, M.; Yang, W.; Mathesh, M.; Wang, T.; Yin, W.; Tran, P.H.-L.; Al Shamaileh, H.; et al. Transforming Doxorubicin into a Cancer Stem Cell Killer via EpCAM Aptamer-Mediated Delivery. Theranostics 2017, 7, 4071–4086. [Google Scholar] [CrossRef]
- Yoon, S.; Huang, K.-W.; Reebye, V.; Spalding, D.; Przytycka, T.M.; Wang, Y.; Swiderski, P.; Li, L.; Armstrong, B.; Reccia, I.; et al. Aptamer-Drug Conjugates of Active Metabolites of Nucleoside Analogs and Cytotoxic Agents Inhibit Pancreatic Tumor Cell Growth. Mol. Ther.-Nucleic Acids 2017, 6, 80–88. [Google Scholar] [CrossRef]
- Wang, T.; Gantier, M.P.; Xiang, D.; Bean, A.G.; Bruce, M.; Zhou, S.-F.; Khasraw, M.; Ward, A.; Wang, L.; Wei, M.Q.; et al. EpCAM Aptamer-Mediated Survivin Silencing Sensitized Cancer Stem Cells to Doxorubicin in a Breast Cancer Model. Theranostics 2015, 5, 1456–1472. [Google Scholar] [CrossRef] [Green Version]
- Maasch, C.; Vater, A.; Buchner, K.; Purschke, W.G.; Eulberg, D.; Vonhoff, S.; Klussmann, S. Polyetheylenimine-Polyplexes of Spiegelmer NOX-A50 Directed against Intracellular High Mobility Group Protein A1 (HMGA1) Reduce Tumor Growth in Vivo. J. Biol. Chem. 2010, 285, 40012–40018. [Google Scholar] [CrossRef] [Green Version]
- Vater, A.; Klussmann, S. Turning Mirror-Image Oligonucleotides into Drugs: The Evolution of Spiegelmer® Therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef] [Green Version]
- Steurer, M.; Montillo, M.; Scarfò, L.; Mauro, F.R.; Andel, J.; Wildner, S.; Trentin, L.; Janssens, A.; Burgstaller, S.; Frömming, A.; et al. Olaptesed Pegol (NOX-A12) with Bendamustine and Rituximab: A Phase IIa Study in Patients with Relapsed/Refractory Chronic Lymphocytic Leukemia. Haematologica 2019, 104, 2053–2060. [Google Scholar] [CrossRef] [Green Version]
- Charoenphol, P.; Bermudez, H. Aptamer-Targeted DNA Nanostructures for Therapeutic Delivery. Mol. Pharm. 2014, 11, 1721–1725. [Google Scholar] [CrossRef]
- Feng, Y.H.; Chen, B.Z.; Fei, W.M.; Cui, Y.; Zhang, C.Y.; Guo, X.D. Mechanism Studies on the Cellular Internalization of Nanoparticles Using Computer Simulations: A Review. AIChE J. 2022, 68, e17507. [Google Scholar] [CrossRef]
- Wu, T.; Liu, J.; Liu, M.; Liu, S.; Zhao, S.; Tian, R.; Wei, D.; Liu, Y.; Zhao, Y.; Xiao, H.; et al. A Nanobody-Conjugated DNA Nanoplatform for Targeted Platinum-Drug Delivery. Angew. Chem. Int. Ed. 2019, 58, 14224–14228. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Leung, H.M.; Gao, Q.; Wang, F.; Wong, S.W.; Liu, L.S.; Au, Y.J.; Lai, K.W.C.; Lo, P.K. Facile Construction of a DNA Tetrahedron in Unconventional Ladder-like Arrangements at Room Temperature. Nanoscale Adv. 2019, 1, 1240–1248. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Wang, M.; Gao, X.; Yu, Z.; Pan, W.; Wang, H.; Tang, B. A DNA Tetrahedron Nanoprobe with Controlled Distance of Dyes for Multiple Detection in Living Cells and in Vivo. Anal. Chem. 2017, 89, 6670–6677. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, J.P.; Calvert, C.R.; Zhang, D.Y.; Pierce, N.A.; Yin, P. Developmental Self-Assembly of a DNA Tetrahedron. ACS Nano. 2014, 8, 3251–3259. [Google Scholar] [CrossRef]
- Shi, S.; Fu, W.; Lin, S.; Tian, T.; Li, S.; Shao, X.; Zhang, Y.; Zhang, T.; Tang, Z.; Zhou, Y.; et al. Targeted and Effective Glioblastoma Therapy via Aptamer-Modified Tetrahedral Framework Nucleic Acid-Paclitaxel Nanoconjugates That Can Pass the Blood Brain Barrier. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102061. [Google Scholar] [CrossRef]
- Ohtsuki, S.; Matsuzaki, N.; Mohri, K.; Endo, M.; Emura, T.; Hidaka, K.; Sugiyama, H.; Takahashi, Y.; Ishiyama, K.; Kadowaki, N.; et al. Optimal Arrangement of Four Short DNA Strands for Delivery of Immunostimulatory Nucleic Acids to Immune Cells. Nucleic Acid Ther. 2015, 25, 245–253. [Google Scholar] [CrossRef]
- Keum, J.-W.; Ahn, J.-H.; Bermudez, H. Design, Assembly, and Activity of Antisense DNA Nanostructures. Small 2011, 7, 3529–3535. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lytton-Jean, A.K.R.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M.; et al. Molecularly Self-Assembled Nucleic Acid Nanoparticles for Targeted in Vivo SiRNA Delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, X.; Yang, F.; Yuan, R.; Xiang, Y. Targeted Delivery of DNA Framework-Encapsulated Native Therapeutic Protein into Cancer Cells. ACS Appl. Mater. Interfaces 2020, 12, 54489–54496. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Qin, S.; Yang, X.; Wang, Q.; Huang, J.; Wang, K. “Sense-and-Treat” DNA Nanodevice for Synergetic Destruction of Circulating Tumor Cells. ACS Appl. Mater. Interfaces 2016, 8, 26552–26558. [Google Scholar] [CrossRef]
- He, P.; Han, W.; Bi, C.; Song, W.; Niu, S.; Zhou, H.; Zhang, X. Many Birds, One Stone: A Smart Nanodevice for Ratiometric Dual-Spectrum Assay of Intracellular MicroRNA and Multimodal Synergetic Cancer Therapy. ACS Nano 2021, 15, 6961–6976. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Lu, D.-Q.; Liang, H.; Xie, S.; Luo, C.; Hu, M.; Xu, L.; Zhang, X.; Tan, W. Fluorescence Resonance Energy Transfer-Based DNA Tetrahedron Nanotweezer for Highly Reliable Detection of Tumor-Related MRNA in Living Cells. ACS Nano 2017, 11, 4060–4066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Liu, X.; Zhu, J.; Wang, L.; Jiang, W. Quantitative Single-Molecule Detection of Protein Based on DNA Tetrahedron Fluorescent Nanolabels. Talanta 2014, 125, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Özhalıcı-Ünal, H.; Armitage, B.A. Fluorescent DNA Nanotags Based on a Self-Assembled DNA Tetrahedron. ACS Nano 2009, 3, 425–433. [Google Scholar] [CrossRef]
- Zhou, G.; Lin, M.; Song, P.; Chen, X.; Chao, J.; Wang, L.; Huang, Q.; Huang, W.; Fan, C.; Zuo, X. Multivalent Capture and Detection of Cancer Cells with DNA Nanostructured Biosensors and Multibranched Hybridization Chain Reaction Amplification. Anal. Chem. 2014, 86, 7843–7848. [Google Scholar] [CrossRef]
- Jiang, D.; Sun, Y.; Li, J.; Li, Q.; Lv, M.; Zhu, B.; Tian, T.; Cheng, D.; Xia, J.; Zhang, L.; et al. Multiple-Armed Tetrahedral DNA Nanostructures for Tumor-Targeting, Dual-Modality in Vivo Imaging. ACS Appl. Mater. Interfaces 2016, 8, 4378–4384. [Google Scholar] [CrossRef]
- Meng, L.; Ma, W.; Lin, S.; Shi, S.; Li, Y.; Lin, Y. Tetrahedral DNA Nanostructure-Delivered DNAzyme for Gene Silencing to Suppress Cell Growth. ACS Appl. Mater. Interfaces 2019, 11, 6850–6857. [Google Scholar] [CrossRef]
- Engelhardt, F.A.S.; Praetorius, F.; Wachauf, C.H.; Brüggenthies, G.; Kohler, F.; Kick, B.; Kadletz, K.L.; Pham, P.N.; Behler, K.L.; Gerling, T.; et al. Custom-Size, Functional, and Durable DNA Origami with Design-Specific Scaffolds. ACS Nano 2019, 13, 5015–5027. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Ke, Y.; Douglas, S.M.; Liu, M.; Sharma, J.; Cheng, A.; Leung, A.; Liu, Y.; Shih, W.M.; Yan, H. Multilayer DNA Origami Packed on a Square Lattice. J. Am. Chem. Soc. 2009, 131, 15903–15908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bujold, K.E.; Hsu, J.C.C.; Sleiman, H.F. Optimized DNA “Nanosuitcases” for Encapsulation and Conditional Release of SiRNA. J. Am. Chem. Soc. 2016, 138, 14030–14038. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Hidaka, K.; Kato, T.; Namba, K.; Sugiyama, H. DNA Prism Structures Constructed by Folding of Multiple Rectangular Arms. J. Am. Chem. Soc. 2009, 131, 15570–15571. [Google Scholar] [CrossRef]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Hu, C.; Wang, P.; Zhao, B.; Ouyang, X.; Zhou, J.; Liu, R.; He, D.; Fan, C.; Song, S. Growth and Origami Folding of DNA on Nanoparticles for High-Efficiency Molecular Transport in Cellular Imaging and Drug Delivery. Angew. Chem. Int. Ed. 2015, 54, 2431–2435. [Google Scholar] [CrossRef]
- Wei, B.; Dai, M.; Yin, P. Complex Shapes Self-Assembled from Single-Stranded DNA Tiles. Nature 2012, 485, 623–626. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Song, L.; Liu, S.; Jiang, Q.; Liu, Q.; Li, N.; Wang, Z.-G.; Ding, B. A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy. Nano Lett. 2018, 18, 3328–3334. [Google Scholar] [CrossRef]
- Zhao, S.; Duan, F.; Liu, S.; Wu, T.; Shang, Y.; Tian, R.; Liu, J.; Wang, Z.-G.; Jiang, Q.; Ding, B. Efficient Intracellular Delivery of RNase A Using DNA Origami Carriers. ACS Appl. Mater. Interfaces 2019, 11, 11112–11118. [Google Scholar] [CrossRef]
- Sala, L.; Perecko, T.; Mestek, O.; Pinkas, D.; Homola, T.; Kočišek, J. Cisplatin-Cross-Linked DNA Origami Nanostructures for Drug Delivery Applications. ACS Appl. Nano Mater. 2022, 5, 13267–13275. [Google Scholar] [CrossRef]
- Jiang, Q.; Shi, Y.; Zhang, Q.; Li, N.; Zhan, P.; Song, L.; Dai, L.; Tian, J.; Du, Y.; Cheng, Z.; et al. A Self-Assembled DNA Origami-Gold Nanorod Complex for Cancer Theranostics. Small 2015, 11, 5134–5141. [Google Scholar] [CrossRef]
- Du, Y.; Jiang, Q.; Beziere, N.; Song, L.; Zhang, Q.; Peng, D.; Chi, C.; Yang, X.; Guo, H.; Diot, G.; et al. DNA-Nanostructure-Gold-Nanorod Hybrids for Enhanced In Vivo Optoacoustic Imaging and Photothermal Therapy. Adv. Mater. 2016, 28, 10000–10007. [Google Scholar] [CrossRef]
- Li, C. A Targeted Approach to Cancer Imaging and Therapy. Nat. Mater. 2014, 13, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Li, Q.; Wang, L.; Zhang, G.-J.; Fan, C. Framework-Nucleic-Acid-Enabled Biosensor Development. ACS Sens. 2018, 3, 903–919. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Yan, M.; Yang, S. Split Aptamers and Their Applications in Sandwich Aptasensors. TrAC Trends Anal. Chem. 2016, 80, 581–593. [Google Scholar] [CrossRef]
- Funke, J.J.; Ketterer, P.; Lieleg, C.; Schunter, S.; Korber, P.; Dietz, H. Uncovering the Forces between Nucleosomes Using DNA Origami. Sci. Adv. 2016, 2, e1600974. [Google Scholar] [CrossRef] [Green Version]
- Loescher, S.; Groeer, S.; Walther, A. 3D DNA Origami Nanoparticles: From Basic Design Principles to Emerging Applications in Soft Matter and (Bio-)Nanosciences. Angew. Chem. Int. Ed. 2018, 57, 10436–10448. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A Biomimetic DNA-Based Channel for the Ligand-Controlled Transport of Charged Molecular Cargo across a Biological Membrane. Nat. Nanotechnol. 2016, 11, 152–156. [Google Scholar] [CrossRef]
- Dai, Z.; Lo, P.K. Photo-Switchable Patterning of Gold Nanoparticles along 3D DNA Nanotubes. Nanoscale 2018, 10, 5431–5435. [Google Scholar] [CrossRef]
- Endo, M.; Yamamoto, S.; Emura, T.; Hidaka, K.; Morone, N.; Heuser, J.E.; Sugiyama, H. Helical DNA Origami Tubular Structures with Various Sizes and Arrangements. Angew. Chem. Int. Ed. 2014, 53, 7484–7490. [Google Scholar] [CrossRef]
- Burns, J.R.; Göpfrich, K.; Wood, J.W.; Thacker, V.V.; Stulz, E.; Keyser, U.F.; Howorka, S. Lipid-Bilayer-Spanning DNA Nanopores with a Bifunctional Porphyrin Anchor. Angew. Chem. Int. Ed. 2013, 52, 12069–12072. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Ke, Y.; Liu, Y.; Mertig, M.; Gu, J.; Yan, H. Functional DNA Nanotube Arrays: Bottom-Up Meets Top-Down. Angew. Chem. Int. Ed. 2007, 46, 6089–6092. [Google Scholar] [CrossRef]
- Jorgenson, T.D.; Mohammed, A.M.; Agrawal, D.K.; Schulman, R. Self-Assembly of Hierarchical DNA Nanotube Architectures with Well-Defined Geometries. ACS Nano 2017, 11, 1927–1936. [Google Scholar] [CrossRef]
- Ke, Y.; Liu, Y.; Zhang, J.; Yan, H. A Study of DNA Tube Formation Mechanisms Using 4-, 8-, and 12-Helix DNA Nanostructures. J. Am. Chem. Soc. 2006, 128, 4414–4421. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Hariadi, R.F.; Choi, H.M.T.; Winfree, E. Integrating DNA Strand-Displacement Circuitry with DNA Tile Self-Assembly. Nat. Commun. 2013, 4, 1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.-X.; Shaw, A.; Zeng, X.; Benson, E.; Nyström, A.M.; Högberg, B. DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties. ACS Nano 2012, 6, 8684–8691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, X.; Lv, H.; Guo, J.; Ding, C.; Luo, X. A DNA Nanotube–Peptide Biocomplex for MRNA Detection and Its Application in Cancer Diagnosis and Targeted Therapy. Chem.–A Eur. J. 2018, 24, 10171–10177. [Google Scholar] [CrossRef] [PubMed]
- Schüller, V.J.; Heidegger, S.; Sandholzer, N.; Nickels, P.C.; Suhartha, N.A.; Endres, S.; Bourquin, C.; Liedl, T. Cellular Immunostimulation by CpG-Sequence-Coated DNA Origami Structures. ACS Nano 2011, 5, 9696–9702. [Google Scholar] [CrossRef] [Green Version]
- Kuzyk, A.; Schreiber, R.; Fan, Z.; Pardatscher, G.; Roller, E.-M.; Högele, A.; Simmel, F.C.; Govorov, A.O.; Liedl, T. DNA-Based Self-Assembly of Chiral Plasmonic Nanostructures with Tailored Optical Response. Nature 2012, 483, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Lo, P.K.; Karam, P.; Aldaye, F.A.; McLaughlin, C.K.; Hamblin, G.D.; Cosa, G.; Sleiman, H.F. Loading and Selective Release of Cargo in DNA Nanotubes with Longitudinal Variation. Nat. Chem. 2010, 2, 319–328. [Google Scholar] [CrossRef]
- Deng, Y.; Tan, Y.; Zhang, Y.; Zhang, L.; Zhang, C.; Ke, Y.; Su, X. Design of Uracil-Modified DNA Nanotubes for Targeted Drug Release via DNA-Modifying Enzyme Reactions. ACS Appl. Mater. Interfaces 2022, 14, 34470–34479. [Google Scholar] [CrossRef]
- Baig, M.M.F.A.; Ma, J.; Gao, X.; Khan, M.A.; Ali, A.; Farid, A.; Zia, A.W.; Noreen, S.; Wu, H. Exploring the Robustness of DNA Nanotubes Framework for Anticancer Theranostics toward the 2D/3D Clusters of Hypopharyngeal Respiratory Tumor Cells. Int. J. Biol. Macromol. 2023, 236, 123988. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Jungmann, R.; Leifer, A.M.; Li, C.; Levner, D.; Church, G.M.; Shih, W.M.; Yin, P. Submicrometre Geometrically Encoded Fluorescent Barcodes Self-Assembled from DNA. Nat. Chem. 2012, 4, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Han, Z.; Zhao, M.; Zhang, X.; Zhuo, Y.; Chai, Y.; Li, Z.; Yuan, R. Versatile Electrochemical Biosensor Based on the Target-Controlled Capture and Release of DNA Nanotubes for the Ultrasensitive Detection of Multiplexed Biomarkers. Anal. Chem. 2022, 94, 11416–11424. [Google Scholar] [CrossRef]
- Xiang, D.; Zheng, C.; Zhou, S.-F.; Qiao, S.; Tran, P.H.-L.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; et al. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics 2015, 5, 1083–1097. [Google Scholar] [CrossRef] [Green Version]
- do Carmo, F.S.; Pinto, S.R.; Orlando, M.M.C.; de Souza Albernaz, M.; de Souza Junqueira, M.; Bernardes, E.S.; Cerecetto, H.; Moglioni, A.; Kozempel, J.; Szwed, M.; et al. Nano-Aptamer for Breast Cancer Imaging: Initial Considerations. J. Diagn. Imaging Ther. 2015, 2, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Sinitsyna, V.V.; Vetcher, A.A. Nucleic Acid Aptamers in Nanotechnology. Biomedicines 2022, 10, 1079. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Hollenstein, M. Aptamer Chemistry. Adv. Drug Deliv. Rev. 2018, 134, 3–21. [Google Scholar] [CrossRef]
- Diafa, S.; Hollenstein, M. Generation of Aptamers with an Expanded Chemical Repertoire. Molecules 2015, 20, 16643–16671. [Google Scholar] [CrossRef] [Green Version]
- Lapa, S.A.; Chudinov, A.V.; Timofeev, E.N. The Toolbox for Modified Aptamers. Mol. Biotechnol. 2016, 58, 79–92. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three Decades of Nucleic Acid Aptamer Technologies: Lessons Learned, Progress and Opportunities on Aptamer Development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Chakraborty, B.; Das, S.; Gupta, A.; Xiong, Y.; T-V, V.; Kizer, M.E.; Duan, J.; Chandrasekaran, A.R.; Wang, X. Aptamers for Viral Detection and Inhibition. ACS Infect. Dis. 2022, 8, 667–692. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Chen, X. Aptamer-Based Targeted Therapy. Adv. Drug Deliv. Rev. 2018, 134, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Fang, X.; Tan, W. Molecular Aptamer Beacons for Real-Time Protein Recognition. Biochem. Biophys. Res. Commun. 2002, 292, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Pu, Y.; Zhu, Z.; Liu, H.; Zhang, J.; Liu, J.; Tan, W. Using Aptamers to Visualize and Capture Cancer Cells. Anal. Bioanal. Chem. 2010, 397, 3225–3233. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.S.M.; Matthews, C.S.; Missailidis, S. DNA Aptamers That Bind to MUC1 Tumour Marker: Design and Characterization of MUC1-Binding Single-Stranded DNA Aptamers. Tumor Biol. 2006, 27, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Cheng, Y.; Wang, J.; Lu, J.; Zhang, B.; Zhao, Y.; Gu, Z. Aptamer-Functionalized Barcode Particles for the Capture and Detection of Multiple Types of Circulating Tumor Cells. Adv. Mater. 2014, 26, 7333–7338. [Google Scholar] [CrossRef]
- Lin, Z.; Ma, Q.; Fei, X.; Zhang, H.; Su, X. A Novel Aptamer Functionalized CuInS2 Quantum Dots Probe for Daunorubicin Sensing and near Infrared Imaging of Prostate Cancer Cells. Anal. Chim. Acta. 2014, 818, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Tang, Z.; Kim, Y.; Nie, H.; Huang, Y.F.; He, X.; Deng, K.; Wang, K.; Tan, W. In Vivo Fluorescence Imaging of Tumors Using Molecular Aptamers Generated by Cell-SELEX. Chem.-Asian J. 2010, 5, 2209–2213. [Google Scholar] [CrossRef]
- Shi, H.; Cui, W.; He, X.; Guo, Q.; Wang, K.; Ye, X.; Tang, J. Whole Cell-SELEX Aptamers for Highly Specific Fluorescence Molecular Imaging of Carcinomas In Vivo. PLoS ONE 2013, 8, e70476. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; He, X.; Wang, K.; Wu, X.; Ye, X.; Guo, Q.; Tan, W.; Qing, Z.; Yang, X.; Zhou, B. Activatable Aptamer Probe for Contrast-Enhanced in Vivo Cancer Imaging Based on Cell Membrane Protein-Triggered Conformation Alteration. Proc. Natl. Acad. Sci. USA 2011, 108, 3900–3905. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Ye, X.; He, X.; Wang, K.; Cui, W.; He, D.; Li, D.; Jia, X. Au@Ag/Au Nanoparticles Assembled with Activatable Aptamer Probes as Smart “Nano-Doctors” for Image-Guided Cancer Thermotherapy. Nanoscale 2014, 6, 8754. [Google Scholar] [CrossRef]
- Kim, J.K.; Choi, K.-J.; Lee, M.; Jo, M.; Kim, S. Molecular Imaging of a Cancer-Targeting Theragnostics Probe Using a Nucleolin Aptamer- and MicroRNA-221 Molecular Beacon-Conjugated Nanoparticle. Biomaterials 2012, 33, 207–217. [Google Scholar] [CrossRef]
- Gong, P.; Shi, B.; Zheng, M.; Wang, B.; Zhang, P.; Hu, D.; Gao, D.; Sheng, Z.; Zheng, C.; Ma, Y.; et al. PEI Protected Aptamer Molecular Probes for Contrast-Enhanced in Vivo Cancer Imaging. Biomaterials 2012, 33, 7810–7817. [Google Scholar] [CrossRef] [PubMed]
- Yigit, M.V.; Mazumdar, D.; Kim, H.-K.; Lee, J.H.; Odintsov, B.; Lu, Y. Smart “Turn-on” Magnetic Resonance Contrast Agents Based on Aptamer-Functionalized Superparamagnetic Iron Oxide Nanoparticles. ChemBioChem 2007, 8, 1675–1678. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, A.; Sun, J.; Li, X.; Gao, F.; Wu, L.; Fang, Y.; Yang, H.; An, L.; Wu, H.; et al. Aptamer-Conjugated Mn3O4@SiO2 Core–Shell Nanoprobes for Targeted Magnetic Resonance Imaging. Nanoscale 2013, 5, 10447. [Google Scholar] [CrossRef] [PubMed]
- Rockey, W.M.; Huang, L.; Kloepping, K.C.; Baumhover, N.J.; Giangrande, P.H.; Schultz, M.K. Synthesis and Radiolabeling of Chelator–RNA Aptamer Bioconjugates with Copper-64 for Targeted Molecular Imaging. Bioorg. Med. Chem. 2011, 19, 4080–4090. [Google Scholar] [CrossRef] [Green Version]
- Kichkailo, A.S.; Narodov, A.A.; Komarova, M.A.; Zamay, T.N.; Zamay, G.S.; Kolovskaya, O.S.; Erakhtin, E.E.; Glazyrin, Y.E.; Veprintsev, D.V.; Moryachkov, R.V.; et al. Development of DNA Aptamers for Visualization of Glial Brain Tumors and Detection of Circulating Tumor Cells. Mol. Ther. Nucleic Acids 2023, in press. [Google Scholar] [CrossRef]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression Profiling Reveals Off-Target Gene Regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. SiRNAs Can Function as MiRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in Mice Due to Oversaturation of Cellular MicroRNA/Short Hairpin RNA Pathways. Nature 2006, 441, 537–541. [Google Scholar] [CrossRef]
- Kovacevic, K.D.; Gilbert, J.C.; Jilma, B. Pharmacokinetics, Pharmacodynamics and Safety of Aptamers. Adv. Drug Deliv. Rev. 2018, 134, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, Biodistribution and Cell Uptake of Antisense Oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shemesh, C.S.; Yu, R.Z.; Gaus, H.J.; Seth, P.P.; Swayze, E.E.; Bennett, F.C.; Geary, R.S.; Henry, S.P.; Wang, Y. Pharmacokinetic and Pharmacodynamic Investigations of ION-353382, a Model Antisense Oligonucleotide: Using Alpha-2-Macroglobulin and Murinoglobulin Double-Knockout Mice. Nucleic Acid Ther. 2016, 26, 223–235. [Google Scholar] [CrossRef]
- Sahay, G.; Querbes, W.; Alabi, C.; Eltoukhy, A.; Sarkar, S.; Zurenko, C.; Karagiannis, E.; Love, K.; Chen, D.; Zoncu, R.; et al. Efficiency of SiRNA Delivery by Lipid Nanoparticles Is Limited by Endocytic Recycling. Nat. Biotechnol. 2013, 31, 653–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashrafuzzaman, M. Aptamers as Both Drugs and Drug-Carriers. Biomed Res. Int. 2014, 2014, 697923. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.-R.; Kim, H.Y.; Lee, Y.-D.; Ha, J.S.; Kang, J.H.; Jeong, H.; Bang, D.; Ko, Y.T.; Kim, S.; Lee, H.; et al. Self-Assembled Mirror DNA Nanostructures for Tumor-Specific Delivery of Anticancer Drugs. J. Control. Release 2016, 243, 121–131. [Google Scholar] [CrossRef]
- Jiang, S.; Ge, Z.; Mou, S.; Yan, H.; Fan, C. Designer DNA Nanostructures for Therapeutics. Chem 2021, 7, 1156–1179. [Google Scholar] [CrossRef]
- Coleridge, E.L.; Dunn, K.E. Assessing the Cost-Effectiveness of DNA Origami Nanostructures for Targeted Delivery of Anti-Cancer Drugs to Tumours. Biomed. Phys. Eng. Express 2020, 6, 065030. [Google Scholar] [CrossRef]
- Li, S.; Tian, T.; Zhang, T.; Cai, X.; Lin, Y. Advances in Biological Applications of Self-Assembled DNA Tetrahedral Nanostructures. Mater. Today 2019, 24, 57–68. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA Origami as an In Vivo Drug Delivery Vehicle for Cancer Therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Benson, E.; Fördős, F.; Lolaico, M.; Baars, I.; Fang, T.; Teixeira, A.I.; Högberg, B. DNA Origami Penetration in Cell Spheroid Tissue Models Is Enhanced by Wireframe Design. Adv. Mater. 2021, 33, 2008457. [Google Scholar] [CrossRef]
- Bouchard, P.R.; Hutabarat, R.M.; Thompson, K.M. Discovery and Development of Therapeutic Aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, J. Complement Activation-Related Pseudoallergy: A Stress Reaction in Blood Triggered by Nanomedicines and Biologicals. Mol. Immunol. 2014, 61, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Y.; Yu, T.; Clifford, C.; Liu, Y.; Yan, H.; Chang, Y. A DNA Nanostructure Platform for Directed Assembly of Synthetic Vaccines. Nano Lett. 2012, 12, 4254–4259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Oh, N. Endocytosis and Exocytosis of Nanoparticles in Mammalian Cells. Int. J. Nanomedicine 2014, 9 (Suppl. S1), 51–63. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.-Y.; Yuan, W.-F.; Ai, W.-B.; Ai, Y.-W.; Wang, J.-J.; Chu, L.-Y.; Zhang, Y.-Q.; Wu, J.-F. An Exploration of Aptamer Internalization Mechanisms and Their Applications in Drug Delivery. Expert Opin. Drug Deliv. 2019, 16, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L. The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef]
- Alamudi, S.H.; Kimoto, M.; Hirao, I. Uptake Mechanisms of Cell-Internalizing Nucleic Acid Aptamers for Applications as Pharmacological Agents. RSC Med. Chem. 2021, 12, 1640–1649. [Google Scholar] [CrossRef]
- El-Sayed, A.; Harashima, H. Endocytosis of Gene Delivery Vectors: From Clathrin-Dependent to Lipid Raft-Mediated Endocytosis. Mol. Ther. 2013, 21, 1118–1130. [Google Scholar] [CrossRef] [Green Version]
- Shapira, A.; Livney, Y.D.; Broxterman, H.J.; Assaraf, Y.G. Nanomedicine for Targeted Cancer Therapy: Towards the Overcoming of Drug Resistance. Drug Resist. Updat. 2011, 14, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Li, Q.; Wang, L.; Guo, Q.; Liu, S.; Zhu, S.; Sun, Y.; Fan, Y.; Sun, Y.; Li, H.; et al. Advances in Regenerative Medicine Applications of Tetrahedral Framework Nucleic Acid-Based Nanomaterials: An Expert Consensus Recommendation. Int. J. Oral Sci. 2022, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, D.; Wilkens, G.D.; Heddle, J.G. Delivering DNA Origami to Cells. Nanomedicine 2019, 14, 911–925. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Sharma, D.; Garg, M.; Kumar, A.; Baliyan, A.; Rani, R.; Kumar, V. Current Understanding of Biological Interactions and Processing of DNA Origami Nanostructures: Role of Machine Learning and Implications in Drug Delivery. Biotechnol. Adv. 2022, 61, 108052. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Rahman, M.A.; Zhao, Z.; Weiss, K.; Zhang, C.; Chen, Z.; Hurwitz, S.J.; Chen, Z.G.; Shin, D.M.; Ke, Y. Visualization of the Cellular Uptake and Trafficking of DNA Origami Nanostructures in Cancer Cells. J. Am. Chem. Soc. 2018, 140, 2478–2484. [Google Scholar] [CrossRef]
- Udomprasert, A.; Kangsamaksin, T. DNA Origami Applications in Cancer Therapy. Cancer Sci. 2017, 108, 1535–1543. [Google Scholar] [CrossRef] [Green Version]
- Hahn, J.; Wickham, S.F.J.; Shih, W.M.; Perrault, S.D. Addressing the Instability of DNA Nanostructures in Tissue Culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef] [Green Version]
- Mei, Q.; Wei, X.; Su, F.; Liu, Y.; Youngbull, C.; Johnson, R.; Lindsay, S.; Yan, H.; Meldrum, D. Stability of DNA Origami Nanoarrays in Cell Lysate. Nano Lett. 2011, 11, 1477–1482. [Google Scholar] [CrossRef] [Green Version]
- Harris, M.A.; Kuang, H.; Schneiderman, Z.; Shiao, M.L.; Crane, A.T.; Chrostek, M.R.; Tăbăran, A.-F.; Pengo, T.; Liaw, K.; Xu, B.; et al. SsDNA Nanotubes for Selective Targeting of Glioblastoma and Delivery of Doxorubicin for Enhanced Survival. Sci. Adv. 2021, 7, eabl5872. [Google Scholar] [CrossRef]
- Shen, X.; Jiang, Q.; Wang, J.; Dai, L.; Zou, G.; Wang, Z.-G.; Chen, W.-Q.; Jiang, W.; Ding, B. Visualization of the Intracellular Location and Stability of DNA Origami with a Label-Free Fluorescent Probe. Chem. Commun. 2012, 48, 11301. [Google Scholar] [CrossRef]
- Chopra, A.; Krishnan, S.; Simmel, F.C. Electrotransfection of Polyamine Folded DNA Origami Structures. Nano Lett. 2016, 16, 6683–6690. [Google Scholar] [CrossRef]
- Chen, G.; Liu, D.; He, C.; Gannett, T.R.; Lin, W.; Weizmann, Y. Enzymatic Synthesis of Periodic DNA Nanoribbons for Intracellular PH Sensing and Gene Silencing. J. Am. Chem. Soc. 2015, 137, 3844–3851. [Google Scholar] [CrossRef]
- Aye, S.; Sato, Y. Therapeutic Applications of Programmable DNA Nanostructures. Micromachines 2022, 13, 315. [Google Scholar] [CrossRef]
- Hicke, B.J.; Stephens, A.W. Escort Aptamers: A Delivery Service for Diagnosis and Therapy. J. Clin. Investig. 2000, 106, 923–928. [Google Scholar] [CrossRef] [Green Version]
- Aljohani, M.M.; Cialla-May, D.; Popp, J.; Chinnappan, R.; Al-Kattan, K.; Zourob, M. Aptamers: Potential Diagnostic and Therapeutic Agents for Blood Diseases. Molecules 2022, 27, 383. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.P.; Gleeson, P.A. Macropinocytosis: An Endocytic Pathway for Internalising Large Gulps. Immunol. Cell Biol. 2011, 89, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Rossi, J.J. Aptamers: Uptake Mechanisms and Intracellular Applications. Adv. Drug Deliv. Rev. 2018, 134, 22–35. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A New Paradigm for Aptamer Therapeutic AS1411 Action: Uptake by Macropinocytosis and Its Stimulation by a Nucleolin-Dependent Mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotula, J.W.; Pratico, E.D.; Ming, X.; Nakagawa, O.; Juliano, R.L.; Sullenger, B.A. Aptamer-Mediated Delivery of Splice-Switching Oligonucleotides to the Nuclei of Cancer Cells. Nucleic Acid Ther. 2012, 22, 187–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, M.L.; Wagstaff, K.M. Internalized Functional DNA Aptamers as Alternative Cancer Therapies. Front. Pharmacol. 2020, 11, 1115. [Google Scholar] [CrossRef]
- Talbot, L.J.; Mi, Z.; Bhattacharya, S.D.; Kim, V.; Guo, H.; Kuo, P.C. Pharmacokinetic Characterization of an RNA Aptamer against Osteopontin and Demonstration of in Vivo Efficacy in Reversing Growth of Human Breast Cancer Cells. Surgery 2011, 150, 224–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.; Nixon, R.L.; Liu, W.; Wang, R. The Applications of Functionalized DNA Nanostructures in Bioimaging and Cancer Therapy. Biomaterials 2021, 268, 120560. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhan, Y.; Zhang, Y.; Shao, X.; Xie, X.; Mao, C.; Cui, W.; Li, Q.; Shi, J.; Li, J.; et al. An Intelligent DNA Nanorobot with in Vitro Enhanced Protein Lysosomal Degradation of HER2. Nano Lett. 2019, 19, 4505–4517. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Ouyang, Y.; Sohn, Y.S.; Nechushtai, R.; Pikarsky, E.; Fan, C.; Willner, I. PH- and MiRNA-Responsive DNA-Tetrahedra/Metal–Organic Framework Conjugates: Functional Sense-and-Treat Carriers. ACS Nan. 2021, 15, 6645–6657. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Ma, W.; Zhang, Y.; Mao, C.; Shao, X.; Xie, X.; Wang, F.; Liu, X.; Li, Q.; Lin, Y. DNA-Based Nanomedicine with Targeting and Enhancement of Therapeutic Efficacy of Breast Cancer Cells. ACS Appl. Mater. Interfaces 2019, 11, 15354–15365. [Google Scholar] [CrossRef]
- Xu, X.; Zhao, Y.; Lu, H.; Fu, C.; Li, X.; Jiang, L.; Li, S. G4-Tetra DNA Duplex Induce Lung Cancer Cell Apoptosis in A549 Cells. Nanoscale Res. Lett. 2016, 11, 437. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wu, L.; Wang, L.; Jiang, W. A Dual-Targeting DNA Tetrahedron Nanocarrier for Breast Cancer Cell Imaging and Drug Delivery. Talanta 2018, 179, 356–363. [Google Scholar] [CrossRef]
- Dong, J.; Dong, H.; Dai, W.; Meng, X.; Zhang, K.; Cao, Y.; Yang, F.; Zhang, X. Functional DNA Hexahedron for Real-Time Detection of Multiple MicroRNAs in Living Cells. Anal. Chim. Acta 2019, 1078, 176–181. [Google Scholar] [CrossRef]
- Wang, Q.; He, Z.; Zhu, H.; Gao, W.; Zhang, N.; Li, J.; Yan, J.; He, B.; Ye, X. Targeting Drug Delivery and Efficient Lysosomal Escape for Chemo-Photodynamic Cancer Therapy by a Peptide/DNA Nanocomplex. J. Mater. Chem. B 2022, 10, 438–449. [Google Scholar] [CrossRef]
- Lopes-Nunes, J.; Lifante, J.; Shen, Y.; Ximendes, E.C.; Jaque, D.; Iglesias-de la Cruz, M.C.; Cruz, C. Biological Studies of an ICG-Tagged Aptamer as Drug Delivery System for Malignant Melanoma. Eur. J. Pharm. Biopharm. 2020, 154, 228–235. [Google Scholar] [CrossRef]
- Li, X.; Yu, Y.; Ji, Q.; Qiu, L. Targeted Delivery of Anticancer Drugs by Aptamer AS1411 Mediated Pluronic F127/Cyclodextrin-Linked Polymer Composite Micelles. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Song, C.; Jiang, C.; Shen, X.; Qiao, Q.; Hu, Y. Nucleolin Targeting AS1411 Modified Protein Nanoparticle for Antitumor Drugs Delivery. Mol. Pharm. 2013, 10, 3555–3563. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yang, W.; Che, C.; Liu, J.; Si, M.; Gong, Z.; Gao, R.; Yang, G. A Targeted Nano Drug Delivery System of AS1411 Functionalized Graphene Oxide Based Composites. ChemistryOpen 2021, 10, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wang, Y.; Shi, H.; Dong, M.; Han, H.; Li, Q. Nucleolin-Targeting AS1411 Aptamer-Modified Micelle for the Co-Delivery of Doxorubicin and MiR-519c to Improve the Therapeutic Efficacy in Hepatocellular Carcinoma Treatment. Int. J. Nanomed. 2021, 16, 2569–2584. [Google Scholar] [CrossRef]
- Khatami, F.; Matin, M.M.; Danesh, N.M.; Bahrami, A.R.; Abnous, K.; Taghdisi, S.M. Targeted Delivery System Using Silica Nanoparticles Coated with Chitosan and AS1411 for Combination Therapy of Doxorubicin and AntimiR-21. Carbohydr. Polym. 2021, 266, 118111. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Zhang, Q.; Lu, D.; Li, S.; Li, J.; Yang, G.; Shan, Y. Dynamics of Delivering Aptamer Targeted Nano-Drugs into Cells. J. Mater. Chem. B 2021, 9, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Yan, Z.; Jin, K.; Pang, Q.; Jiang, T.; Lu, H.; Liu, X.; Pang, Z.; Yu, L.; Jiang, X. Precise Glioblastoma Targeting by AS1411 Aptamer-Functionalized Poly (l-γ-Glutamylglutamine)–Paclitaxel Nanoconjugates. J. Colloid Interface Sci. 2017, 490, 783–796. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Z.; Zhang, G.; Lin, F.; Liu, Y.; Zhang, Y.; Feng, J.; Chen, W.; Meng, Q.; Chen, L. AS1411 Aptamer/Hyaluronic Acid-Bifunctionalized Microemulsion Co-Loading Shikonin and Docetaxel for Enhanced Antiglioma Therapy. J. Pharm. Sci. 2019, 108, 3684–3694. [Google Scholar] [CrossRef]
- Alibolandi, M.; Taghdisi, S.M.; Ramezani, P.; Hosseini Shamili, F.; Farzad, S.A.; Abnous, K.; Ramezani, M. Smart AS1411-Aptamer Conjugated Pegylated PAMAM Dendrimer for the Superior Delivery of Camptothecin to Colon Adenocarcinoma in Vitro and in Vivo. Int. J. Pharm. 2017, 519, 352–364. [Google Scholar] [CrossRef]
- Li, L.; Hou, J.; Liu, X.; Guo, Y.; Wu, Y.; Zhang, L.; Yang, Z. Nucleolin-Targeting Liposomes Guided by Aptamer AS1411 for the Delivery of SiRNA for the Treatment of Malignant Melanomas. Biomaterials 2014, 35, 3840–3850. [Google Scholar] [CrossRef]
- Lopes-Nunes, J.; Agonia, A.S.; Rosado, T.; Gallardo, E.; Palmeira-de-Oliveira, R.; Palmeira-de-Oliveira, A.; Martinez-de-Oliveira, J.; Fonseca-Moutinho, J.; Campello, M.P.C.; Paiva, A.; et al. Aptamer-Functionalized Gold Nanoparticles for Drug Delivery to Gynecological Carcinoma Cells. Cancers 2021, 13, 4038. [Google Scholar] [CrossRef] [PubMed]
- Barzegar Behrooz, A.; Nabavizadeh, F.; Adiban, J.; Shafiee Ardestani, M.; Vahabpour, R.; Aghasadeghi, M.R.; Sohanaki, H. Smart Bomb AS1411 Aptamer-Functionalized/PAMAM Dendrimer Nanocarriers for Targeted Drug Delivery in the Treatment of Gastric Cancer. Clin. Exp. Pharmacol. Physiol. 2017, 44, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hu, Y.; Zheng, X.; Cao, X.; Li, Q.; Wei, Z.; Zhu, Z.; Zhang, S. Self-Assembled Peptide Nanoparticles with Endosome Escaping Permits for Co-Drug Delivery. Talanta 2021, 221, 121572. [Google Scholar] [CrossRef]
- Charbgoo, F.; Soltani, F.; Alibolandi, M.; Taghdisi, S.M.; Abnous, K.; Ramezani, P.; Ramezani, M. Ladder-like Targeted and Gated Doxorubicin Delivery Using Bivalent Aptamer in Vitro and in Vivo. Mater. Sci. Eng. C 2021, 119, 111618. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, A.; Yaghoobi, E.; Azizollahi, H.; Shojaee, S.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. Design and Synthesis of a Star-like Polymeric Micelle Modified with AS1411 Aptamer for Targeted Delivery of Camptothecin for Cancer Therapy. Int. J. Pharm. 2022, 611, 121346. [Google Scholar] [CrossRef]
- Zhao, D.; Yang, G.; Liu, Q.; Liu, W.; Weng, Y.; Zhao, Y.; Qu, F.; Li, L.; Huang, Y. A Photo-Triggerable Aptamer Nanoswitch for Spatiotemporal Controllable SiRNA Delivery. Nanoscale 2020, 12, 10939–10943. [Google Scholar] [CrossRef]
- Aravind, A.; Jeyamohan, P.; Nair, R.; Veeranarayanan, S.; Nagaoka, Y.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. AS1411 Aptamer Tagged PLGA-Lecithin-PEG Nanoparticles for Tumor Cell Targeting and Drug Delivery. Biotechnol. Bioeng. 2012, 109, 2920–2931. [Google Scholar] [CrossRef]
- Xu, L.; He, X.-Y.; Liu, B.-Y.; Xu, C.; Ai, S.-L.; Zhuo, R.-X.; Cheng, S.-X. Aptamer-Functionalized Albumin-Based Nanoparticles for Targeted Drug Delivery. Colloids Surf. B Biointerfaces 2018, 171, 24–30. [Google Scholar] [CrossRef]
- Zhuang, Y.; Deng, H.; Su, Y.; He, L.; Wang, R.; Tong, G.; He, D.; Zhu, X. Aptamer-Functionalized and Backbone Redox-Responsive Hyperbranched Polymer for Targeted Drug Delivery in Cancer Therapy. Biomacromolecules 2016, 17, 2050–2062. [Google Scholar] [CrossRef]
- Zhuang, J.; Chen, S.; Hu, Y.; Yang, F.; Huo, Q.; Xie, N. Tumour-Targeted and Redox-Responsive Mesoporous Silica Nanoparticles for Controlled Release of Doxorubicin and an SiRNA Against Metastatic Breast Cancer. Int. J. Nanomed. 2021, 16, 1961–1976. [Google Scholar] [CrossRef]
- Yu, S.; Bi, X.; Yang, L.; Wu, S.; Yu, Y.; Jiang, B.; Zhang, A.; Lan, K.; Duan, S. Co-Delivery of Paclitaxel and PLK1-Targeted SiRNA Using Aptamer-Functionalized Cationic Liposome for Synergistic Anti-Breast Cancer Effects In Vivo. J. Biomed. Nanotechnol. 2019, 15, 1135–1148. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Tian, B.; Liu, J.; Yang, L.; Zeng, L.; Chen, T.; Hong, A.; Wang, X. Nucleolin-Targeted Extracellular Vesicles as a Versatile Platform for Biologics Delivery to Breast Cancer. Theranostics 2017, 7, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhuang, Q.; Ji, T.; Zhang, Y.; Li, C.; Wang, Y.; Li, H.; Jia, H.; Liu, Y.; Du, L. Multi-Functionalized Chitosan Nanoparticles for Enhanced Chemotherapy in Lung Cancer. Carbohydr. Polym. 2018, 195, 311–320. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Hu, X.; Peng, R.; Li, J.; Zhang, X.; Tan, W. Multicolor Two-Photon Nanosystem for Multiplexed Intracellular Imaging and Targeted Cancer Therapy. Angew. Chem. Int. Ed. 2021, 60, 12569–12576. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Peng, Y.; Li, Y.; Xie, X.; Wei, X.; Yang, G.; Zhang, H.; Li, N.; Li, T.; Qin, X.; et al. Aptamer-Dendrimer Functionalized Magnetic Nano-Octahedrons: Theranostic Drug/Gene Delivery Platform for Near-Infrared/Magnetic Resonance Imaging-Guided Magnetochemotherapy. ACS Nano 2021, 15, 16683–16696. [Google Scholar] [CrossRef] [PubMed]
- Thorat, N.D.; Bauer, J.; Tofail, S.A.M.; Gascón Pérez, V.; Bohara, R.A.; Yadav, H.M. Silica Nano Supra-Assembly for the Targeted Delivery of Therapeutic Cargo to Overcome Chemoresistance in Cancer. Colloids Surf. B Biointerfaces 2020, 185, 110571. [Google Scholar] [CrossRef]
- Zhang, H.-J.; Zhao, X.; Chen, L.-J.; Yang, C.-X.; Yan, X.-P. Dendrimer Grafted Persistent Luminescent Nanoplatform for Aptamer Guided Tumor Imaging and Acid-Responsive Drug Delivery. Talanta 2020, 219, 121209. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, Y.; Sun, Q.; Cheng, J.; Yue, C.; Liu, Y.; Song, J.; Jin, W.; Ding, X.; de la Fuente, J.M.; et al. Au-SiRNA@ Aptamer Nanocages as a High-Efficiency Drug and Gene Delivery System for Targeted Lung Cancer Therapy. J. Nanobiotechnol. 2021, 19, 54. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, J.; Zhang, Y.; Zhang, J.; Tang, A.; Kong, D. A ZnO-Gated Porphyrinic Metal–Organic Framework-Based Drug Delivery System for Targeted Bimodal Cancer Therapy. J. Mater. Chem. B 2018, 6, 7898–7907. [Google Scholar] [CrossRef]
- Wang, L.; Niu, M.; Zheng, C.; Zhao, H.; Niu, X.; Li, L.; Hu, Y.; Zhang, Y.; Shi, J.; Zhang, Z. A Core-Shell Nanoplatform for Synergistic Enhanced Sonodynamic Therapy of Hypoxic Tumor via Cascaded Strategy. Adv. Healthc. Mater. 2018, 7, 1800819. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Yazdian-Robati, R.; Abnous, K. A Novel AS1411 Aptamer-Based Three-Way Junction Pocket DNA Nanostructure Loaded with Doxorubicin for Targeting Cancer Cells in Vitro and in Vivo. Mol. Pharm. 2018, 15, 1972–1978. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-C.; Li, W.-T.; Madanayake, T.W.; Tao, C.; Niu, Q.; Yan, S.-Q.; Gao, B.-A.; Ping, Z. Aptamer-Guided Upconversion Nanoplatform for Targeted Drug Delivery and near-Infrared Light-Triggered Photodynamic Therapy. J. Biomater. Appl. 2020, 34, 875–888. [Google Scholar] [CrossRef]
- Qi, J.; Zhang, Y.; Gou, Y.; Lee, P.; Wang, J.; Chen, S.; Zhou, Z.; Wu, X.; Yang, F.; Liang, H. Multidrug Delivery Systems Based on Human Serum Albumin for Combination Therapy with Three Anticancer Agents. Mol. Pharm. 2016, 13, 3098–3105. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, K.; Sathiyaseelan, A.; Mariadoss, A.V.A.; Jeevithan, E.; Hu, X.; Shin, S.; Wang, M.-H. Dual Stimuli-Responsive Release of Aptamer AS1411 Decorated Erlotinib Loaded Chitosan Nanoparticles for Non-Small-Cell Lung Carcinoma Therapy. Carbohydr. Polym. 2020, 245, 116407. [Google Scholar] [CrossRef] [PubMed]
- Kong, N.; Deng, M.; Sun, X.-N.; Chen, Y.-D.; Sui, X.-B. Polydopamine-Functionalized CA-(PCL-Ran-PLA) Nanoparticles for Target Delivery of Docetaxel and Chemo-Photothermal Therapy of Breast Cancer. Front. Pharmacol. 2018, 9, 125. [Google Scholar] [CrossRef] [Green Version]
- Zahiri, M.; Taghdisi, S.M.; Abnous, K.; Ramezani, M.; Alibolandi, M. Fabrication of Versatile Targeted Lipopolymersomes for Improved Camptothecin Efficacy against Colon Adenocarcinoma in Vitro and in Vivo. Expert Opin. Drug Deliv. 2021, 18, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, X.; Yang, H.; Li, L.; Ye, Z.; Rao, Y. A Nuclear Targeted Dox-Aptamer Loaded Liposome Delivery Platform for the Circumvention of Drug Resistance in Breast Cancer. Biomed. Pharmacother. 2019, 117, 109072. [Google Scholar] [CrossRef]
- Hao, Y.; Zheng, C.; Song, Q.; Chen, H.; Nan, W.; Wang, L.; Zhang, Z.; Zhang, Y. Pressure-Driven Accumulation of Mn-Doped Mesoporous Silica Nanoparticles Containing 5-Aza-2-Deoxycytidine and Docetaxel at Tumours with a Dry Cupping Device. J. Drug Target. 2021, 29, 900–909. [Google Scholar] [CrossRef]
- Yu, Z.; Li, X.; Duan, J.; Yang, X.-D. Targeted Treatment of Colon Cancer with Aptamer-Guided Albumin Nanoparticles Loaded with Docetaxel. Int. J. Nanomed. 2020, 15, 6737–6748. [Google Scholar] [CrossRef]
- Shiao, Y.-S.; Chiu, H.-H.; Wu, P.-H.; Huang, Y.-F. Aptamer-Functionalized Gold Nanoparticles As Photoresponsive Nanoplatform for Co-Drug Delivery. ACS Appl. Mater. Interfaces 2014, 6, 21832–21841. [Google Scholar] [CrossRef]
- Taghavi, S.; Abnous, K.; Babaei, M.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Synthesis of Chimeric Polymersomes Based on PLA-b-PHPMA and PCL-b-PHPMA for Nucleoline Guided Delivery of SN38. Nanomed. Nanotechnol. Biol. Med. 2020, 28, 102227. [Google Scholar] [CrossRef]
- Sun, G.-Y.; Du, Y.-C.; Cui, Y.-X.; Wang, J.; Li, X.-Y.; Tang, A.-N.; Kong, D.-M. Terminal Deoxynucleotidyl Transferase-Catalyzed Preparation of PH-Responsive DNA Nanocarriers for Tumor-Targeted Drug Delivery and Therapy. ACS Appl. Mater. Interfaces 2019, 11, 14684–14692. [Google Scholar] [CrossRef]
- Latorre, A.; Posch, C.; Garcimartín, Y.; Celli, A.; Sanlorenzo, M.; Vujic, I.; Ma, J.; Zekhtser, M.; Rappersberger, K.; Ortiz-Urda, S.; et al. DNA and Aptamer Stabilized Gold Nanoparticles for Targeted Delivery of Anticancer Therapeutics. Nanoscale 2014, 6, 7436–7442. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Wang, Z.; Zhang, Y.; Ren, J.; Qu, X. Metal–Organic Framework-Based Nanoplatform for Intracellular Environment-Responsive Endo/Lysosomal Escape and Enhanced Cancer Therapy. ACS Appl. Mater. Interfaces 2018, 10, 31998–32005. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Li, Z.; Guo, C.; Huang, X.; Song, Y.; Zhou, N.; Wang, M.; Zhang, Z.; He, L.; Du, M. A γ-Cyclodextrin-Based Metal–Organic Framework Embedded with Graphene Quantum Dots and Modified with PEGMA via SI-ATRP for Anticancer Drug Delivery and Therapy. Nanoscale 2019, 11, 20956–20967. [Google Scholar] [CrossRef] [PubMed]
- Ayatollahi, S.; Salmasi, Z.; Hashemi, M.; Askarian, S.; Oskuee, R.K.; Abnous, K.; Ramezani, M. Aptamer-Targeted Delivery of Bcl-XL ShRNA Using Alkyl Modified PAMAM Dendrimers into Lung Cancer Cells. Int. J. Biochem. Cell Biol. 2017, 92, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Yan, J.; Xiong, H.; Wu, Q.; Xing, H.; Liu, Y.; Liu, S.; Liu, Z. Aptamer-Functionalized Molybdenum Disulfide Nanosheets for Tumor Cell Targeting and Lysosomal Acidic Environment/NIR Laser Responsive Drug Delivery to Realize Synergetic Chemo-Photothermal Therapeutic Effects. Int. J. Pharm. 2020, 590, 119948. [Google Scholar] [CrossRef]
- Pei, W.; Liu, M.; Wu, Y.; Zhao, Y.; Liu, T.; Sun, B.; Liu, Y.; Wang, Q.; Han, J. High Payload and Targeted Release of Anthracyclines by Aptamer-Tethered DNA Nanotrains—Thermodynamic and Release Kinetic Study. Eur. J. Pharm. Sci. 2020, 148, 105319. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zhu, Q.; Ruan, J.; Sun, T.; Han, L. Polyplexes by Polymerized Dequalinium and Bifunctional Aptamer for Mitochondrial Targeting Drug Release to Overcome Drug Resistance. ACS Appl. Bio Mater. 2020, 3, 5182–5192. [Google Scholar] [CrossRef]
- Peng, L.-H.; Zhang, Y.-H.; Han, L.-J.; Zhang, C.-Z.; Wu, J.-H.; Wang, X.-R.; Gao, J.-Q.; Mao, Z.-W. Cell Membrane Capsules for Encapsulation of Chemotherapeutic and Cancer Cell Targeting in Vivo. ACS Appl. Mater. Interfaces 2015, 7, 18628–18637. [Google Scholar] [CrossRef]
- Liu, G.; Gao, N.; Zhou, Y.; Nie, J.; Cheng, W.; Luo, M.; Mei, L.; Zeng, X.; Deng, W. Polydopamine-Based “Four-in-One” Versatile Nanoplatforms for Targeted Dual Chemo and Photothermal Synergistic Cancer Therapy. Pharmaceutics 2019, 11, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Qian, J.; Cao, S.; Yang, Z.; Pang, Z.; Pan, S.; Fan, L.; Xi, Z.; Jiang, X.; Zhang, Q. Precise Glioma Targeting of and Penetration by Aptamer and Peptide Dual-Functioned Nanoparticles. Biomaterials 2012, 33, 5115–5123. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Hu, H.; Zhang, M.G.; Song, J.; Nie, L.; Wang, S.; Niu, G.; Huang, P.; Lu, G.; Chen, X. An Aptamer-Targeting Photoresponsive Drug Delivery System Using “off–on” Graphene Oxide Wrapped Mesoporous Silica Nanoparticles. Nanoscale 2015, 7, 6304–6310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Liu, B.; Xu, X.; Zhuang, B.; Li, H.; Yin, J.; Cong, M.; Xu, W.; Lu, A. Toward Targeted Therapy in Chemotherapy-Resistant Pancreatic Cancer with a Smart Triptolide Nanomedicine. Oncotarget 2016, 7, 8360–8372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Chen, Q.; Zhang, Y.; Zhou, W.; Li, X.; Li, C.; Zhang, Y.; Chen, H.; Liu, P.; Chu, Y.; et al. Click-Nucleic-Acid-Containing Codelivery System Inducing Collapse of Cellular Homeostasis for Tumor Therapy through Bidirectional Regulation of Autophagy and Glycolysis. ACS Appl. Mater. Interfaces 2020, 12, 57757–57767. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Hu, X.; Shanmugam, S.; Chelliah, R.; Sekar, P.; Oh, D.-H.; Vijayakumar, S.; Kathiresan, K.; Wang, M.-H. Enhanced Cancer Therapy with PH-Dependent and Aptamer Functionalized Doxorubicin Loaded Polymeric (Poly D, L-Lactic-Co-Glycolic Acid) Nanoparticles. Arch. Biochem. Biophys. 2019, 671, 143–151. [Google Scholar] [CrossRef]
- Vandghanooni, S.; Eskandani, M.; Barar, J.; Omidi, Y. Antisense LNA-Loaded Nanoparticles of Star-Shaped Glucose-Core PCL-PEG Copolymer for Enhanced Inhibition of OncomiR-214 and Nucleolin-Mediated Therapy of Cisplatin-Resistant Ovarian Cancer Cells. Int. J. Pharm. 2020, 573, 118729. [Google Scholar] [CrossRef]
- Ma, Y.; Zhu, Y.; Wang, C.; Pan, D.; Liu, S.; Yang, M.; Xiao, Z.; Yang, X.; Zhao, W.; Zhou, X.; et al. Annealing Novel Nucleobase-Lipids with Oligonucleotides or Plasmid DNA Based on H-Bonding or π-π Interaction: Assemblies and Transfections. Biomaterials 2018, 178, 147–157. [Google Scholar] [CrossRef]
- Zhao, F.; Zhou, J.; Su, X.; Wang, Y.; Yan, X.; Jia, S.; Du, B. A Smart Responsive Dual Aptamers-Targeted Bubble-Generating Nanosystem for Cancer Triplex Therapy and Ultrasound Imaging. Small 2017, 13, 1603990. [Google Scholar] [CrossRef]
- Sanati, S.; Taghavi, S.; Abnous, K.; Taghdisi, S.M.; Babaei, M.; Ramezani, M.; Alibolandi, M. Fabrication of Anionic Dextran-Coated Micelles for Aptamer Targeted Delivery of Camptothecin and Survivin-ShRNA to Colon Adenocarcinoma. Gene Ther. 2022, 29, 55–68. [Google Scholar] [CrossRef]
- Tao, W.; Xu, G.; Yu, X.; Zhang, J.; Sheng, Y.; Liu, G.; Mei, L. Robust Aptamer–Polydopamine-Functionalized M-PLGA–TPGS Nanoparticles for Targeted Delivery of Docetaxel and Enhanced Cervical Cancer Therapy. Int. J. Nanomed. 2016, 11, 2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Tian, Q.; Sun, Y.; Xu, J.-J.; Ye, D.; Chen, H.-Y. ATP-Activatable Photosensitizer Enables Dual Fluorescence Imaging and Targeted Photodynamic Therapy of Tumor. Anal. Chem. 2017, 89, 13610–13617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Luo, Z.; Liu, J.; Ding, X.; Li, J.; Cai, K. Cytochrome c End-Capped Mesoporous Silica Nanoparticles as Redox-Responsive Drug Delivery Vehicles for Liver Tumor-Targeted Triplex Therapy in Vitro and in Vivo. J. Control. Release 2014, 192, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Mosafer, J.; Teymouri, M.; Abnous, K.; Tafaghodi, M.; Ramezani, M. Study and Evaluation of Nucleolin-Targeted Delivery of Magnetic PLGA-PEG Nanospheres Loaded with Doxorubicin to C6 Glioma Cells Compared with Low Nucleolin-Expressing L929 Cells. Mater. Sci. Eng. C 2017, 72, 123–133. [Google Scholar] [CrossRef]
- Chen, D.; Li, B.; Cai, S.; Wang, P.; Peng, S.; Sheng, Y.; He, Y.; Gu, Y.; Chen, H. Dual Targeting Luminescent Gold Nanoclusters for Tumor Imaging and Deep Tissue Therapy. Biomaterials 2016, 100, 1–16. [Google Scholar] [CrossRef] [PubMed]
- He, Z.-M.; Zhang, P.-H.; Li, X.; Zhang, J.-R.; Zhu, J.-J. A Targeted DNAzyme-Nanocomposite Probe Equipped with Built-in Zn2+ Arsenal for Combined Treatment of Gene Regulation and Drug Delivery. Sci. Rep. 2016, 6, 22737. [Google Scholar] [CrossRef] [Green Version]
- Joshi, M.; Choi, J.-S.; Park, J.-W.; Doh, K.-O. Combination of Doxorubicin with Gemcitabine-Incorporated GQuadruplex Aptamer Showed Synergistic and Selective Anticancer Effect in Breast Cancer Cells. J. Microbiol. Biotechnol. 2019, 29, 1799–1805. [Google Scholar] [CrossRef]
- Dai, F.; Zhang, Y.; Zhu, X.; Shan, N.; Chen, Y. Anticancer Role of MUC1 Aptamer–MiR-29b Chimera in Epithelial Ovarian Carcinoma Cells through Regulation of PTEN Methylation. Target. Oncol. 2012, 7, 217–225. [Google Scholar] [CrossRef]
- Ranjbar-Navazi, Z.; Fathi, M.; Abdolahinia, E.D.; Omidi, Y.; Davaran, S. MUC-1 Aptamer Conjugated InP/ZnS Quantum Dots/Nanohydrogel Fluorescent Composite for Mitochondria-Mediated Apoptosis in MCF-7 Cells. Mater. Sci. Eng. C 2021, 118, 111469. [Google Scholar] [CrossRef]
- Si, P.; Shi, J.; Zhang, P.; Wang, C.; Chen, H.; Mi, X.; Chu, W.; Zhai, B.; Li, W. MUC-1 Recognition-Based Activated Drug Nanoplatform Improves Doxorubicin Chemotherapy in Breast Cancer. Cancer Lett. 2020, 472, 165–174. [Google Scholar] [CrossRef]
- Tan, Y.; Li, Y.; Qu, Y.-X.; Su, Y.; Peng, Y.; Zhao, Z.; Fu, T.; Wang, X.-Q.; Tan, W. Aptamer-Peptide Conjugates as Targeted Chemosensitizers for Breast Cancer Treatment. ACS Appl. Mater. Interfaces 2021, 13, 9436–9444. [Google Scholar] [CrossRef] [PubMed]
- Alves, L.N.; Missailidis, S.; Lage, C.A.S.; De Almeida, C.E.B. Anti-MUC1 Aptamer as Carrier Tool of the Potential Radiosensitizer 1,10 Phenanthroline in MCF-7 Breast Cancer Cells. Anticancer. Res. 2019, 39, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Lee, S.H.; Hwang, Y.; Yoo, H.; Jung, H.; Kim, S.H.; Mok, H. Multivalent Aptamer-RNA Conjugates for Simple and Efficient Delivery of Doxorubicin/SiRNA into Multidrug-Resistant Cells. Macromol. Biosci. 2017, 17, 1600343. [Google Scholar] [CrossRef] [PubMed]
- Yazdian-Robati, R.; Arab, A.; Ramezani, M.; Rafatpanah, H.; Bahreyni, A.; Nabavinia, M.S.; Abnous, K.; Taghdisi, S.M. Smart Aptamer-Modified Calcium Carbonate Nanoparticles for Controlled Release and Targeted Delivery of Epirubicin and Melittin into Cancer Cells in Vitro and in Vivo. Drug Dev. Ind. Pharm. 2019, 45, 603–610. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Dehestani, S.; Hashemi, M.; Taghdisi, S.M.; Abnous, K. Smart Delivery of Epirubicin to Cancer Cells Using Aptamer-Modified Ferritin Nanoparticles. J. Drug Target. 2022, 30, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Charbgoo, F.; Alibolandi, M.; Taghdisi, S.M.; Abnous, K.; Soltani, F.; Ramezani, M. MUC1 Aptamer-Targeted DNA Micelles for Dual Tumor Therapy Using Doxorubicin and KLA Peptide. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 685–697. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Lavaee, P.; Jalalian, S.H.; Robati, R.Y.; Abnous, K. Double Targeting and Aptamer-Assisted Controlled Release Delivery of Epirubicin to Cancer Cells by Aptamers-Based Dendrimer in Vitro and in Vivo. Eur. J. Pharm. Biopharm. 2016, 102, 152–158. [Google Scholar] [CrossRef]
- Liu, B.-Y.; He, X.-Y.; Zhuo, R.-X.; Cheng, S.-X. Reversal of Tumor Malignization and Modulation of Cell Behaviors through Genome Editing Mediated by a Multi-Functional Nanovector. Nanoscale 2018, 10, 21209–21218. [Google Scholar] [CrossRef]
- Shahrad, S.; Rajabi, M.; Javadi, H.; Karimi Zarchi, A.A.; Darvishi, M.H. Targeting Lung Cancer Cells with MUC1 Aptamer-Functionalized PLA-PEG Nanocarriers. Sci. Rep. 2022, 12, 4718. [Google Scholar] [CrossRef]
- Aghanejad, A.; Babamiri, H.; Adibkia, K.; Barar, J.; Omidi, Y. Mucin-1 Aptamer-Armed Superparamagnetic Iron Oxide Nanoparticles for Targeted Delivery of Doxorubicin to Breast Cancer Cells. BioImpacts 2018, 8, 117–127. [Google Scholar] [CrossRef]
- Nassireslami, E.; Ajdarzade, M. Gold Coated Superparamagnetic Iron Oxide Nanoparticles as Effective Nanoparticles to Eradicate Breast Cancer Cells via Photothermal Therapy. Adv. Pharm. Bull. 2018, 8, 201–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacko, K.; Thangavel, K.; Shoyele, S.A. Codelivery of Genistein and MiRNA-29b to A549 Cells Using Aptamer-Hybrid Nanoparticle Bioconjugates. Nanomaterials 2019, 9, 1052. [Google Scholar] [CrossRef] [Green Version]
- Jafari, R.; Majidi Zolbanin, N.; Majidi, J.; Atyabi, F.; Yousefi, M.; Jadidi-Niaragh, F.; Aghebati-Maleki, L.; Shanehbandi, D.; Soltani Zangbar, M.S.; Rafatpanah, H. Anti-Mucin1 Aptamer-Conjugated Chitosan Nanoparticles for Targeted Co-Delivery of Docetaxel and IGF-1R SiRNA to SKBR3 Metastatic Breast Cancer Cells. Iran Biomed J. 2019, 23, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Jha, P.; Singh, V.; Sinha, K.; Hussain, S.; Singh, M.K.; Das, P. A Quantum Dot–MUC1 Aptamer Conjugate for Targeted Delivery of Protoporphyrin IX and Specific Photokilling of Cancer Cells through ROS Generation. Integr. Biol. 2016, 8, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Pascual, L.; Cerqueira-Coutinho, C.; García-Fernández, A.; de Luis, B.; Bernardes, E.S.; Albernaz, M.S.; Missailidis, S.; Martínez-Máñez, R.; Santos-Oliveira, R.; Orzaez, M.; et al. MUC1 Aptamer-Capped Mesoporous Silica Nanoparticles for Controlled Drug Delivery and Radio-Imaging Applications. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2495–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayari, E.; Dinarvand, M.; Amini, M.; Azhdarzadeh, M.; Mollarazi, E.; Ghasemi, Z.; Atyabi, F. MUC1 Aptamer Conjugated to Chitosan Nanoparticles, an Efficient Targeted Carrier Designed for Anticancer SN38 Delivery. Int. J. Pharm. 2014, 473, 304–315. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, J.; Ma, Y.; Zhu, X.; Zhang, C. Aptamers Entirely Built from Therapeutic Nucleoside Analogues for Targeted Cancer Therapy. J. Am. Chem. Soc. 2022, 144, 1493–1497. [Google Scholar] [CrossRef]
- Mohapatra, S.; Saha, A.; Mondal, P.; Jana, B.; Ghosh, S.; Biswas, A.; Ghosh, S. Synergistic Anticancer Effect of Peptide-Docetaxel Nanoassembly Targeted to Tubulin: Toward Development of Dual Warhead Containing Nanomedicine. Adv. Healthc. Mater. 2017, 6, 1600718. [Google Scholar] [CrossRef]
- Wei, H.; Zhao, Z.; Wang, Y.; Zou, J.; Lin, Q.; Duan, Y. One-Step Self-Assembly of Multifunctional DNA Nanohydrogels: An Enhanced and Harmless Strategy for Guiding Combined Antitumor Therapy. ACS Appl. Mater. Interfaces 2019, 11, 46479–46489. [Google Scholar] [CrossRef]
- Chuang, E.-Y.; Lin, C.-C.; Chen, K.-J.; Wan, D.-H.; Lin, K.-J.; Ho, Y.-C.; Lin, P.-Y.; Sung, H.-W. A FRET-Guided, NIR-Responsive Bubble-Generating Liposomal System for in Vivo Targeted Therapy with Spatially and Temporally Precise Controlled Release. Biomaterials 2016, 93, 48–59. [Google Scholar] [CrossRef]
- Majidi Zolbanin, N.; Jafari, R.; Majidi, J.; Atyabi, F.; Yousefi, M.; Jadidi-Niaragh, F.; Aghebati-Maleki, L.; Shanehbandi, D.; Soltani Zangbar, M.-S.; Nayebi, A.M. Targeted Co-Delivery of Docetaxel and CMET SiRNA for Treatment of Mucin1 Overexpressing Breast Cancer Cells. Adv. Pharm. Bull. 2018, 8, 383–393. [Google Scholar] [CrossRef]
- Moosavian, S.A.; Abnous, K.; Akhtari, J.; Arabi, L.; Gholamzade Dewin, A.; Jafari, M. 5TR1 Aptamer-PEGylated Liposomal Doxorubicin Enhances Cellular Uptake and Suppresses Tumour Growth by Targeting MUC1 on the Surface of Cancer Cells. Artif. Cells Nanomed. Biotechnol. 2017, 46, 2054–2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.; Wang, S.; Luo, N.; Chen, F.; Hu, C.; Zhang, K. The Application of Aptamer 5TR1 in Triple Negative Breast Cancer Target Therapy. J. Cell. Biochem. 2018, 119, 896–908. [Google Scholar] [CrossRef]
- Taghavi, S.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. Chitosan-Modified PLGA Nanoparticles Tagged with 5TR1 Aptamer for in Vivo Tumor-Targeted Drug Delivery. Cancer Lett. 2017, 400, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jalalian, S.H.; Taghdisi, S.M.; Shahidi Hamedani, N.; Kalat, S.A.M.; Lavaee, P.; ZandKarimi, M.; Ghows, N.; Jaafari, M.R.; Naghibi, S.; Danesh, N.M.; et al. Epirubicin Loaded Super Paramagnetic Iron Oxide Nanoparticle-Aptamer Bioconjugate for Combined Colon Cancer Therapy and Imaging in Vivo. Eur. J. Pharm. Sci. 2013, 50, 191–197. [Google Scholar] [CrossRef]
- Zhen, S.; Takahashi, Y.; Narita, S.; Yang, Y.-C.; Li, X. Targeted Delivery of CRISPR/Cas9 to Prostate Cancer by Modified GRNA Using a Flexible Aptamer-Cationic Liposome. Oncotarget 2017, 8, 9375–9387. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Zhao, H.; Guo, L.; Wang, Y.; Song, J.; Zhao, X.; Li, C.; Hao, L.; Wang, D.; Tang, J. Ultrasound-Mediated Nanobubble Destruction (UMND) Facilitates the Delivery of A10-3.2 Aptamer Targeted and SiRNA-Loaded Cationic Nanobubbles for Therapy of Prostate Cancer. Drug Deliv. 2018, 25, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Tong, R.; Coyle, V.J.; Yin, Q.; Pondenis, H.; Borst, L.B.; Cheng, J.; Fan, T.M. Targeting Tumor Vasculature with Aptamer-Functionalized Doxorubicin–Polylactide Nanoconjugates for Enhanced Cancer Therapy. ACS Nano 2015, 9, 5072–5081. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Shi, D.; Shang, M.; Sun, X.; Meng, D.; Liu, X.; Zhou, X.; Li, J. Utilizing RNA Nanotechnology to Construct Negatively Charged and Ultrasound-Responsive Nanodroplets for Targeted Delivery of SiRNA. Drug Deliv. 2022, 29, 316–327. [Google Scholar] [CrossRef]
- Wen, J.; Tao, W.; Hao, S.; Iyer, S.P.; Zu, Y. A Unique Aptamer-Drug Conjugate for Targeted Therapy of Multiple Myeloma. Leukemia 2016, 30, 987–991. [Google Scholar] [CrossRef]
- Alshaer, W.; Hillaireau, H.; Vergnaud, J.; Ismail, S.; Fattal, E. Functionalizing Liposomes with Anti-CD44 Aptamer for Selective Targeting of Cancer Cells. Bioconjugate Chem. 2015, 26, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Natesh, J.; Chandola, C.; Meeran, S.M.; Neerathilingam, M. Targeted Delivery of Doxorubicin through CD44 Aptamer to Cancer Cells. Therapeutic Delivery. 2021, 12, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Hillaireau, H.; Vergnaud, J.; Mura, S.; Deloménie, C.; Sauvage, F.; Ismail, S.; Fattal, E. Aptamer-Guided SiRNA-Loaded Nanomedicines for Systemic Gene Silencing in CD-44 Expressing Murine Triple-Negative Breast Cancer Model. J. Control. Release 2018, 271, 98–106. [Google Scholar] [CrossRef]
- Fan, W.; Wang, X.; Ding, B.; Cai, H.; Wang, X.; Fan, Y.; Li, Y.; Liu, S.; Nie, S.; Lu, Q. Thioaptamer-Conjugated CD44-Targeted Delivery System for the Treatment of Breast Cancer in Vitro and in Vivo. J. Drug Target. 2016, 24, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Behrooz, A.B.; Vazifehmand, R.; Tajudin, A.A.; Masarudin, M.J.; Sekawi, Z.; Masomian, M.; Syahir, A. Tailoring Drug Co-Delivery Nanosystem for Mitigating U-87 Stem Cells Drug Resistance. Drug Deliv. Transl. Res. 2022, 12, 1253–1269. [Google Scholar] [CrossRef]
- Alibolandi, M.; Abnous, K.; Anvari, S.; Mohammadi, M.; Ramezani, M.; Taghdisi, S.M. CD133-Targeted Delivery of Self-Assembled PEGylated Carboxymethylcellulose-SN38 Nanoparticles to Colorectal Cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Qian, W.; Tao, W.; Zhou, Y.; Xue, B. Delivery Of Curcumin Nanoliposomes Using Surface Modified With CD133 Aptamers For Prostate Cancer. Drug Des. Dev. Ther. 2019, 13, 4021–4033. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhu, X.; Gao, J.; Liu, D.; Dong, C.; Jin, X. PLGA Nanoparticles with CD133 Aptamers for Targeted Delivery and Sustained Release of Propranolol to Hemangioma. Nanomedicine 2017, 12, 2611–2624. [Google Scholar] [CrossRef]
- Yu, Z.; Ni, M.; Xiong, M.; Zhang, X.; Cai, G.; Chen, H.; Zeng, Q. Poly(Lactic-Co-Glycolic Acid) Nanoparticles Conjugated with CD133 Aptamers for Targeted Salinomycin Delivery to CD133+ Osteosarcoma Cancer Stem Cells. Int. J. Nanomed. 2015, 10, 2537. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Chen, H.; Yu, C.; Zhang, Y.; Chen, M.; Tian, S.; Sun, C. The Promotion of Salinomycin Delivery to Hepatocellular Carcinoma Cells through EGFR and CD133 Aptamers Conjugation by PLGA Nanoparticles. Nanomedicine 2015, 10, 1863–1879. [Google Scholar] [CrossRef]
- Smiley, S.B.; Yun, Y.; Ayyagari, P.; Shannon, H.E.; Pollok, K.E.; Vannier, M.W.; Das, S.K.; Veronesi, M.C. Development of CD133 Targeting Multi-Drug Polymer Micellar Nanoparticles for Glioblastoma—In Vitro Evaluation in Glioblastoma Stem Cells. Pharm. Res. 2021, 38, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Gui, K.; Zhang, X.; Chen, F.; Ge, Z.; Zhang, S.; Qi, X.; Sun, J.; Yu, Z. Lipid-Polymer Nanoparticles with CD133 Aptamers for Targeted Delivery of All-Trans Retinoic Acid to Osteosarcoma Initiating Cells. Biomed. Pharmacother. 2019, 111, 751–764. [Google Scholar] [CrossRef]
- Li, W.; Wang, Z.; Gao, T.; Sun, S.; Xu, M.; Pei, R. Selection of CD133-Targeted DNA Aptamers for the Efficient and Specific Therapy of Colorectal Cancer. J. Mater. Chem. B 2022, 10, 2057–2066. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Xu, H.; Ye, B.-C. Membrane-Decorated Exosomes for Combination Drug Delivery and Improved Glioma Therapy. Langmuir 2022, 38, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Dong, C.; Liu, Q.; Zhu, X.; Zuo, S.; Zhang, H. The Sustained and Targeted Treatment of Hemangiomas by Propranolol-Loaded CD133 Aptamers Conjugated Liposomes-in-Microspheres. Biomed. Pharmacother. 2019, 114, 108823. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.Y.; Kim, H.; Lee, M.; Hong, J.; Lee, J.H.; Kim, J.; Choi, M.J.; Park, Y.S.; Kim, S.-C. Development of HER2-Specific Aptamer-Drug Conjugate for Breast Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 9764. [Google Scholar] [CrossRef]
- Xue, L.; Maihle, N.J.; Yu, X.; Tang, S.-C.; Liu, H.Y. Synergistic Targeting HER2 and EGFR with Bivalent Aptamer-SiRNA Chimera Efficiently Inhibits HER2-Positive Tumor Growth. Mol. Pharm. 2018, 15, 4801–4813. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Li, M.; Liu, T.; Liu, J.; Xie, Y.; Zhang, J.; Xu, S.; Liu, H. Dual-Functional HER2 Aptamer-Conjugated, PH-Activated Mesoporous Silica Nanocarrier-Based Drug Delivery System Provides in Vitro Synergistic Cytotoxicity in HER2-Positive Breast Cancer Cells. Int. J. Nanomed. 2019, 14, 4029–4044. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Sung, H.J.; Lee, Y.M.; Choi, S.I.; Kim, Y.-H.; Heo, K.; Kim, I.-H. Therapeutic Application of Drug-Conjugated HER2 Oligobody (HER2-DOligobody). Int. J. Mol. Sci. 2020, 21, 3286. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, J.; Hao, W.; Wang, T.; Liu, J.; Xie, Y.; Xu, S.; Liu, H. Copolymer Micelles Function as PH-Responsive Nanocarriers to Enhance the Cytotoxicity of a HER2 Aptamer in HER2-Positive Breast Cancer Cells. Int. J. Nanomed. 2018, 13, 537–553. [Google Scholar] [CrossRef] [Green Version]
- Saleh, T.; Soudi, T.; Shojaosadati, S.A. Aptamer Functionalized Curcumin-Loaded Human Serum Albumin (HSA) Nanoparticles for Targeted Delivery to HER-2 Positive Breast Cancer Cells. Int. J. Biol. Macromol. 2019, 130, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Han, L.; Guo, Y.; Zheng, G.; Fan, L.; Shen, Z.; Zhao, R.; Shao, J. A Carrier-Free Dual-Drug Nanodelivery System Functionalized with Aptamer Specific Targeting HER2-Overexpressing Cancer Cells. J. Mater. Chem. B 2017, 5, 9121–9129. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.; Chaudhry, S.; Hall, N.; Olverson, G.; Zhang, Q.-J.; Mandal, T.; Dash, S.; Kundu, A. Targeted Delivery of Doxorubicin Liposomes for Her-2+ Breast Cancer Treatment. AAPS PharmSciTech 2020, 21, 202. [Google Scholar] [CrossRef]
- Powell, D.; Chandra, S.; Dodson, K.; Shaheen, F.; Wiltz, K.; Ireland, S.; Syed, M.; Dash, S.; Wiese, T.; Mandal, T.; et al. Aptamer-Functionalized Hybrid Nanoparticle for the Treatment of Breast Cancer. Eur. J. Pharm. Biopharm. 2017, 114, 108–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, F.; Zeng, Q.; Qian, H.; Zhang, D.; Wei, Y.; Wang, Y.; Chai, C.; Cheng, W.; Ding, S.; Chen, T. Dual-Targeted Self-Assembled DNA Hydrogels Decorated With Multivalent Aptamers Loaded With DOX for Anticancer Therapy. Front. Pharmacol. 2022, 13, 8. [Google Scholar] [CrossRef]
- Lee, H.; Dam, D.H.M.; Ha, J.W.; Yue, J.; Odom, T.W. Enhanced Human Epidermal Growth Factor Receptor 2 Degradation in Breast Cancer Cells by Lysosome-Targeting Gold Nanoconstructs. ACS Nano 2015, 9, 9859–9867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohiya, G.; Katti, D.S. Carboxylated Chitosan-Mediated Improved Efficacy of Mesoporous Silica Nanoparticle-Based Targeted Drug Delivery System for Breast Cancer Therapy. Carbohydr. Polym. 2022, 277, 118822. [Google Scholar] [CrossRef]
- Mashreghi, M.; Zamani, P.; Karimi, M.; Mehrabian, A.; Arabsalmani, M.; Zarqi, J.; Moosavian, S.A.; Jaafari, M.R. Anti-Epithelial Cell Adhesion Molecule RNA Aptamer-conjugated Liposomal Doxorubicin as an Efficient Targeted Therapy in Mice Bearing Colon Carcinoma Tumor Model. Biotechnol. Prog. 2021, 37, e3116. [Google Scholar] [CrossRef]
- Shahriari, M.; Taghdisi, S.M.; Abnous, K.; Ramezani, M.; Alibolandi, M. Self-Targeted Polymersomal Co-Formulation of Doxorubicin, Camptothecin and FOXM1 Aptamer for Efficient Treatment of Non-Small Cell Lung Cancer. J. Control. Release 2021, 335, 369–388. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular Assembly of an Aptamer-Drug Conjugate for Targeted Drug Delivery to Tumor Cells. ChemBioChem 2009, 10, 862–868. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Tiemann, K.; Chomchan, P.; Alluin, J.; Swiderski, P.; Burnett, J.; Zhang, X.; Forman, S.; Chen, R.; Rossi, J. Dual Functional BAFF Receptor Aptamers Inhibit Ligand-Induced Proliferation and Deliver SiRNAs to NHL Cells. Nucleic Acids Res. 2013, 41, 4266–4283. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, C.; Pei, H.; Shi, J.; Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. [Google Scholar] [CrossRef]
- Wang, D.-X.; Wang, J.; Wang, Y.-X.; Du, Y.-C.; Huang, Y.; Tang, A.-N.; Cui, Y.-X.; Kong, D.-M. DNA Nanostructure-Based Nucleic Acid Probes: Construction and Biological Applications. Chem. Sci. 2021, 12, 7602–7622. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Linko, V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed. 2020, 59, 15818–15833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Q.; Nie, C.; Hu, Y.; Yi, J.; Liu, C.; Zhang, J.; He, M.; He, M.; Chen, T.; Chu, X. Aptamer-Functionalized DNA Origami for Targeted Codelivery of Antisense Oligonucleotides and Doxorubicin to Enhance Therapy in Drug-Resistant Cancer Cells. ACS Appl. Mater. Interfaces 2020, 12, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Angell, C.; Xie, S.; Zhang, L.; Chen, Y. DNA Nanotechnology for Precise Control over Drug Delivery and Gene Therapy. Small 2016, 12, 1117–1132. [Google Scholar] [CrossRef]
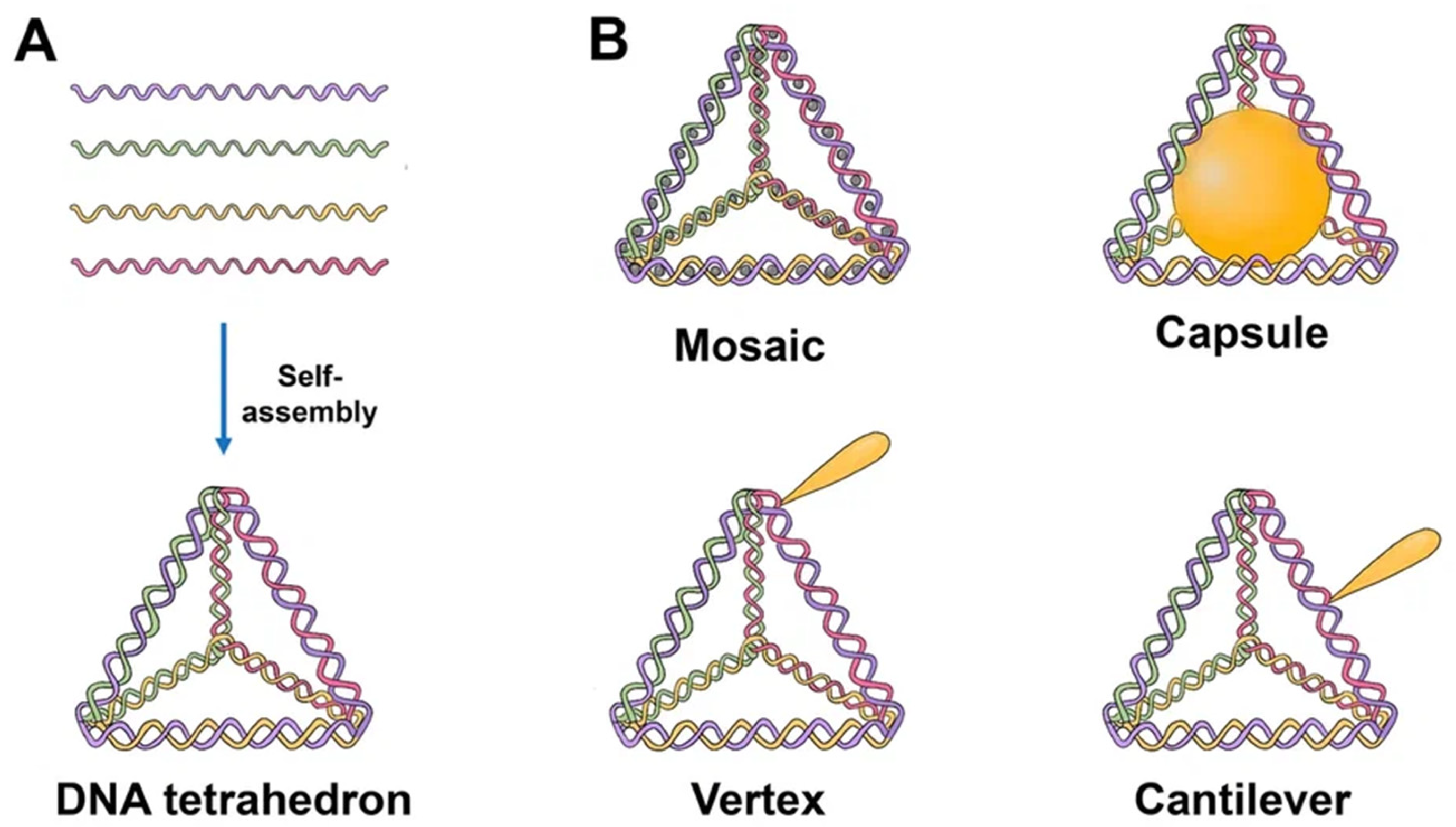

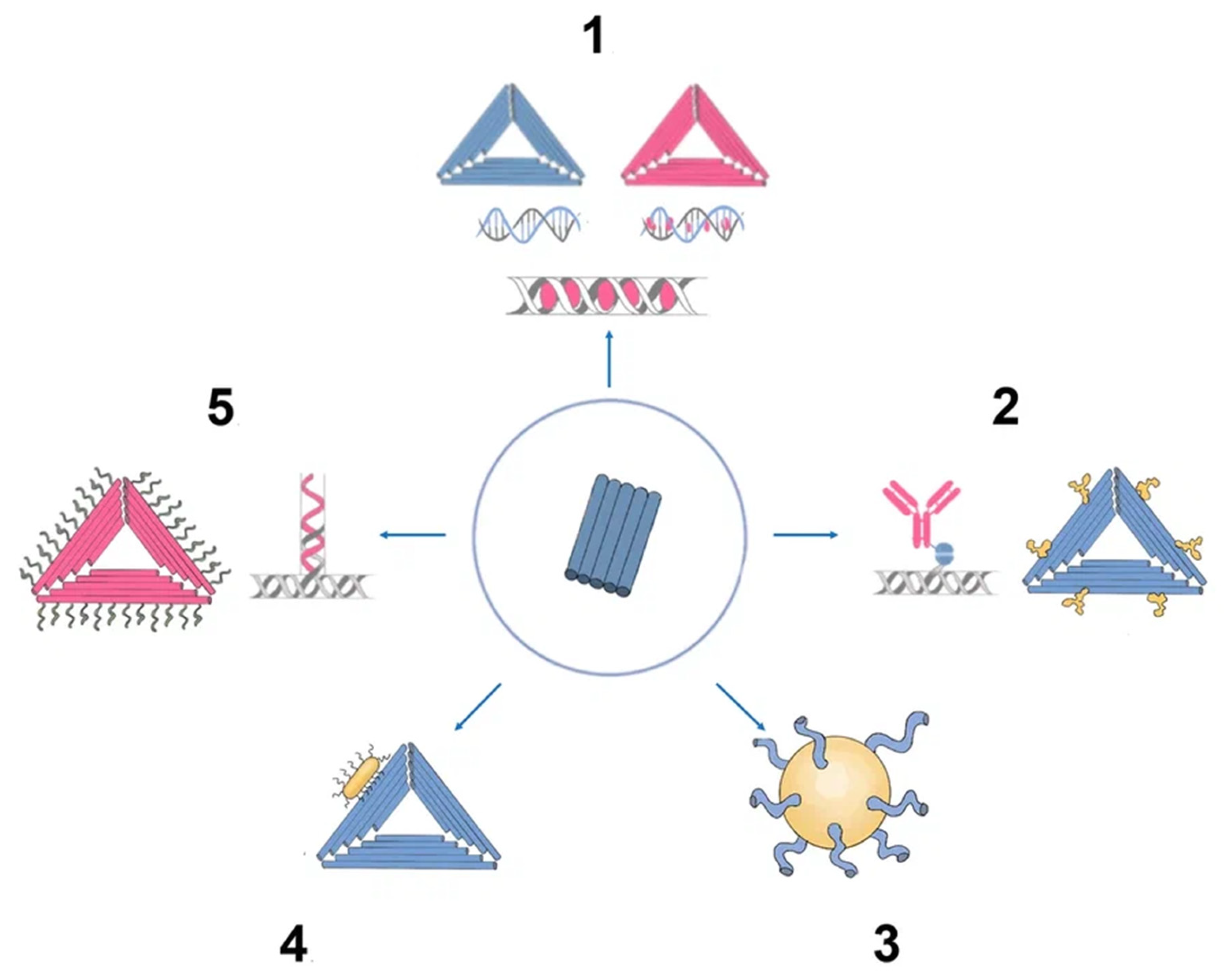
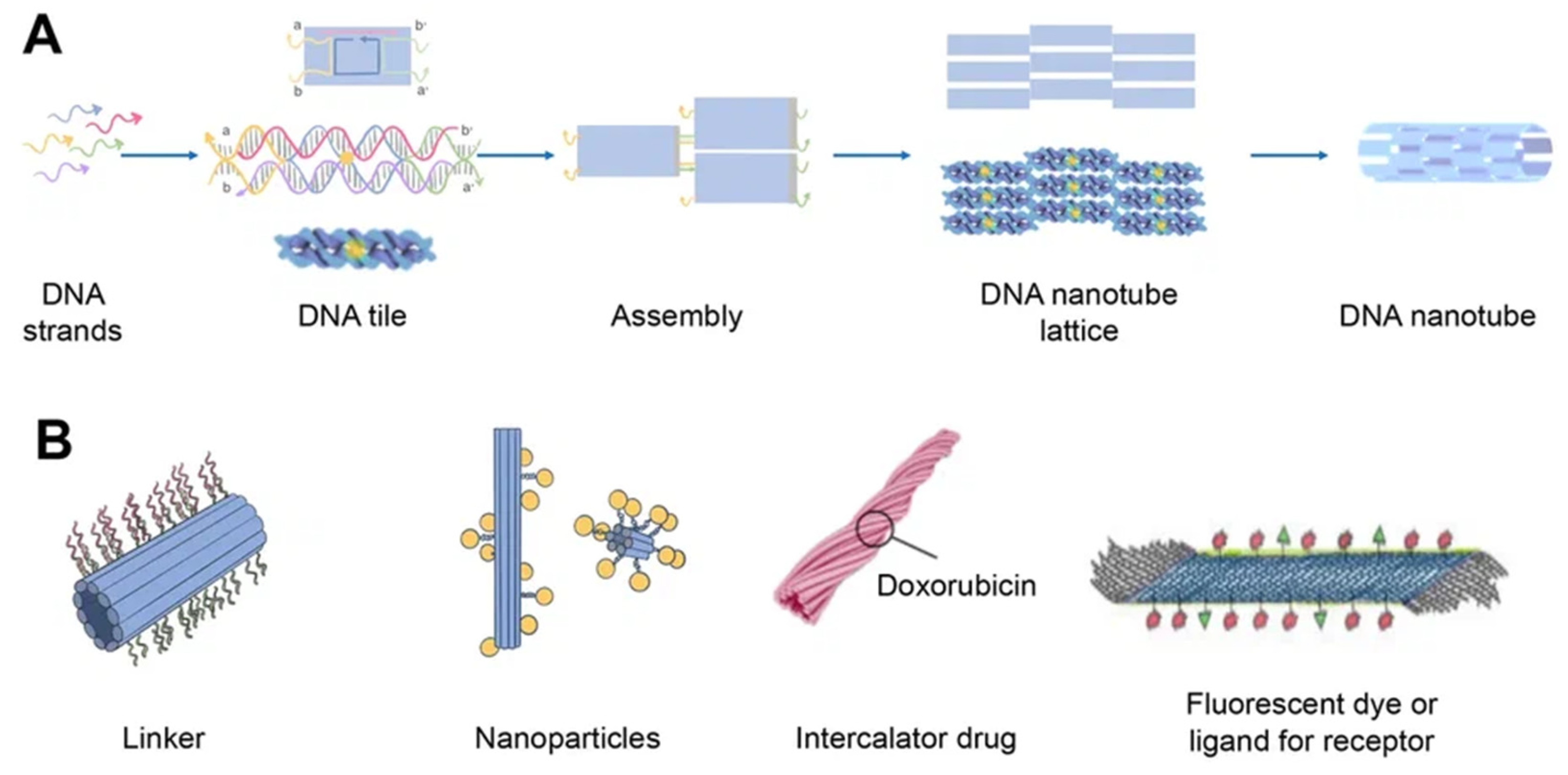
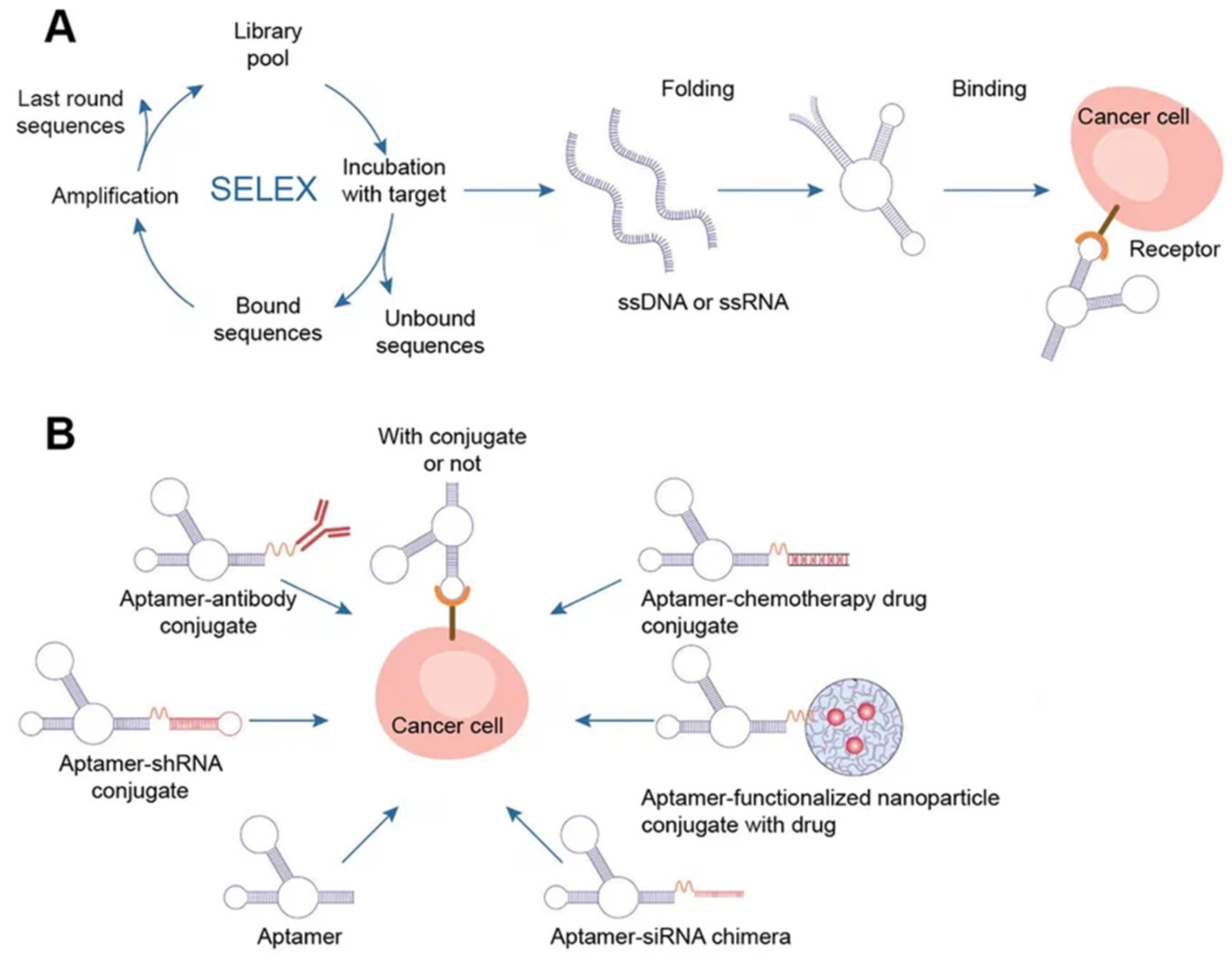

| DNA Nanostructure | Size | Structure | Synthesis Method and Assembly | Applications for Cancer Therapy |
|---|---|---|---|---|
| DNA tetrahedron | 7 nm [150]– 20 nm [151]. | Tetrahedral shape, double-boundle TDN [152]. | Single-step synthesis method: TDN synthesized by mixing four single-stranded DNAs in one pot after a quick thermal annealing process; self-assembly [153]. Two-step synthesis method: self-assembly of TDN with fluorophore and amido bond formation with drug [154]. Isothermal synthesis of a DNA tetrahedron: 3D wireframe TDN synthesized by ring forming reactions in the presence of an initiator [155]. | Drug delivery carriers: small molecules—doxorubicin [19], paclitaxel [156]; nucleic acid drugs: CpG [157], ASOs [158], siRNA [159], aptamers [150], RNase A [160]. Photodynamic therapy: carrier for iridium catalyst [25], therapy circulating tumor cells [161]. Cancer theranostics: [162]. Cancer cell detection and in vivo imaging: [150,163,164,165,166,167,168]. |
| DNA origami | 2D DNA origami ~100 nm [28], 3D origami ~ size up to 1000 nm [169]. | 2D DNA origami (rectangles, triangles, five-pointed star, etc.) [28], 3D DNA origami: tube [35], honeycomb [170], square [171] lattices, origami cage structure [172], 3D prism structures [173], cube [174], DNA nanoribbons [175]. | Scaffolded self-assembly of DNA strands: single-stranded scaffold and over 200 short oligonucleotide “staple” strands are mixed and self-assembled in a single step in desired shapes [28]. Single-stranded tile assembly: single strands of DNA containing four domains associate into staggered duplexes, resulting in DNA lattices [176]. Simulated annealing algorithm for automatic generation of DNA origami: computes unique designs by utilizing shape annealing, by integration of shape grammars and the simulated annealing algorithm [32]. | Immune engineering: encapsulates antibodies to antigen protein expressed on the surface of leukemia cells [174]. Drug delivery carriers: doxorubicin [35], p53 gene and doxorubicin [177], daunorubicin [37], MUC-1 aptamer and RNase A [178], cisplatin [179]. Photodynamic therapy: Au nanorods [180,181], Au NPs [175], BMEPC [38]. Cancer imaging and therapy: [182]. Biosensors: [183,184,185,186]. |
| DNA nanotube | 9 nm [187]–29.5 µm [188] | Tubular [189,190,191], triangular and square-shaped DNA nanotubes [43], L-, T-, and Y-shaped [192]. | Single-stranded DNA tiles: ssDNA containing several domains hybridized with each other to form tiles [42,191]. Multi-crossover DNA tiles: Hybridization of DNA tiles and wrapping by intrinsic or external factors [193,194]. «Scaffolded» DNA Origami: The formation of nanotubes from DNA origami occurs via either direct self-assembly [171]. Multi-rungs: nanotubes are formed from single-stranded and cyclic DNA templates, when rigid vertices helped the cyclic DNA to create “rungs” and then assembled using perpendicular linkers [43]. | Drug delivery carriers: doxorubicin [195], doxorubicin or paclitaxel with MUC-1 aptamer [196], CpG [197], cytochrome c [48], aptamer and ricin A [49], AuNPs [198,199], enzyme-responsive DNA nanotubes with targeting DOX release [200]. Cancer Diagnosis and Targeted Therapy: DNA nanotube–peptide biocomplex [196], anti-EGFR targeting with a pH-responsive controlled release of TW-37 [201]. Fluorescent Dyes: Cy3 [47], Blue-Red-Green [202]. Biosensors: detection of multiplexed biomarkers—microRNA-21 (miRNA-21) and glutathione (GSH) [203]. |
| Aptamers | EpCAM aptamer ~2.09 nm [204]–PLA-MUC1aptamer nanoparticle 200 nm [205]. | single-stranded DNA or RNA molecule; unique due to their secondary and tertiary structure [206] | Automated solid-phase synthesis [207], enzymatic synthesis (for evolutionary selection of modified aptamers) [208,209]. SELEX is process for generation aptamers, including initial library design, target preparation, PCR optimization, and single strand DNA separation [210]. Types of SELEX: Cell SELEX, Complex–Target SELEX, Genomic SELEX, Microfluidic SELEX (M-SELEX), Magnetic Bead-Based Microfluidic SELEX, Capillary Electrophoretic (CE) SELEX, Sol–Gel Method, AFM-SELEX, Toggle-SELEX [211]. | Aptamer and aptamer–drug conjugates for targeted therapy: AS1411 (clinical study) [63]; chemotherapy, gene therapy, immunotherapy, radiotherapy, phototherapy [212]. Targeted real-time imaging and diagnostics: aptamer-based protein recognition [213], detection of cancer cells [214], detection of MUC-1 positive cells [215], capture and detection of circulating tumor cells [216,217], in vivo imaging tumors [218,219], prostate cancer detection [217], probe for contrast-enhanced in vivo cancer imaging [220], image-guided cancer thermotherapy [221], cancer-targeting therangostics probe [222], monitor DNA degradation [223], positron emission tomography [224], magnetic resonance imaging [225], PET imaging [226], CT imaging and therapy of cancer [82], PET/CT tumor visualization [227]. |
| DNA-Based Nanomaterial | Method and/or Strategy for Delivering Drug | Target | Cell Line/Cancer | Payload | Reference |
|---|---|---|---|---|---|
| DNA tetrahedron | aptamer HER2 was anchored on tetrahedral framework nucleic acid via covalent bond | HER2 | SKBR-3, MCF7, MCF-10A | HApt- tetrahedral DNA | [273] |
| DNA tetrahedron | TDNs conjugated with pegaptanib | VEGF | HUVECs, Cal27 | Pegatinib | [136] |
| DNA tetrahedron | miR-21 or miR-155 or AS1411 modified DNA tetrahedron/metal–organic framework conjugates | nucleolin | MDA-MB-23, HepG2 | Doxorubicin or camptothecin or rhodamine 6G or fluorescein | [274] |
| DNA tetrahedron | Methylene blue loaded into TDN via intercalation | cell apoptosis | B6F10, SCC7, MDA-MB231 | Methylene Blue, PDT | [23] |
| DNA tetrahedron | PTX loaded into TDN via intercalation | MDR cell apoptosis | A549, PTX-resistant A549/T | Paclitaxel | [18] |
| DNA tetrahedron | ASOs loaded into DNA tetrahedrons conjugated with NLS peptide | c-raf | A549 | ASO | [26] |
| DNA tetrahedron | One of the strands mirror TDN was biotinylated and conjugated to streptavidin. Various biotinylated enzymes can be loaded onto the streptavidin subunit of the hybrid. | apoptosis, gene modification, and carbohydrate hydrolysis | HeLa, Nano fibroblast cells | Caspase 3; Cre recombinase; β-galactosidase | [24] |
| DNA tetrahedron | Cy5, 5-FU, and AS1411 were conjugated with TDN | nucleolin | MCF10A, MCF7 | 5-Fluorouracil | [275] |
| DNA tetrahedron | AS1411 was linked with TDN | nucleolin | A549 | AS1411 | [276] |
| double-bundle DNA tetrahedron | 56MESS was loaded into TDN through intercalation | EGFR | A431, A549, A549/DDP, MCF-7, and A2780 | platinum-based DNA intercalator, 56MESS | [152] |
| DNA tetrahedron | TDN modified with MUC1 and AS1411aptamer; DOX-loaded via intercalation | MUC1, nucleolin | MCF7, HL-7702 | Doxorubicin | [277] |
| FUdR-DNA-affibody chimera | 10 FUdR molecules were attached to 5′ end of each DNA strand | HER2 | BT474, MCF-7 | 5-Fluorodeoxyuridine | [27] |
| DNA mini-hexahedron (DMH) | hibridization of AS1411 sequence with one strand of DMH | nucleolin | A549 | miRNA-1246 and miRNA-21 | [278] |
| DNA tetrahedron | DOX and TMPyP4 were loaded via intercalation into AS1411-modified TDN | nucleolin | HeLa | DOX and TMPyP4 | [279] |
| DNA origami | intercalation | cell apoptosis | MDA-MB-231, MDA-MB-468, MCF-7 | Doxorubicin | [195] |
| DNA origami | RNase A-DNA origami conjugate | MUC1 | MCF-7 | MUC1-modified RNase A | [178] |
| DNA origami | intercalation | cell apoptosis | MCF-7 | BMEPC (3,6-bis [2-(1-methylpyridinium) ethynyl]-9-pentyl-carbazole diiodide), PDT | [38] |
| DNA origami | intercalation | MDR cell apoptosis | HL-60/ADR | Daunorubicin | [37] |
| DNA nanotubes | cholesterol-modified DNA nanotubes conjugated with cytochrome c | Cell apoptosis | HeLa | Cytochrome C | [48] |
| self-assembled DNA nanotubes | Cy3-labeled and folate-conjugated nanotubes | FR (folate receptor) | Nasopharyngeal epidermal carcinoma KB cells | Cy3 | [47] |
| DNA nanotube | DOX and PTX were loaded into the modified with MUC-1 capsulated DNA nanotube–peptide | MUC-1, integrin receptors (αv β3) | MCF-7 | Doxorubicin or paclitaxel | [196] |
| AS1411 aptamer | conjugation of aptamer AS1411 with indocyanine green (ICG) or with acridine orange ligand C8 | nucleolin | B16 | C8—acridine orange derivative or ICG | [280] |
| AS1411 | AS1411-conjugated pluronic F127 mixed micelles encapsulating doxorubicin | nucleolin | MCF-7 | Doxorubicin | [281] |
| AS1411 | AS1411 was conjugated with self-assembled HSA-paclitaxel nanoparticle (HSA—Human serum albumin) | nucleolin | MCF-7, MCF-10A and 3T3 | Paclitaxel | [282] |
| AS1411 | Aminated AS1411 covalently modify graphene oxide-chitosan oligosaccharide-γ-polyglutamic acid nanocarrier loaded with doxorubicin | nucleolin | Beas-2B, HeLa | Doxorubicin | [283] |
| AS1411 | AS1411 aptamer-functionalized PEG-PLA micelle were constructed through an emulsion/solvent evaporation strategy for the codelivery of doxorubicin and miR-519c | nucleolin | HepG2 | Doxorubicin and miR-519c | [284] |
| AS1411 | AS1411 attached onto the surface chitosan-coated MSNs | nucleolin | C26, MCF-7, 4T1, CHO | Doxorubicin and antimiR-21 | [285] |
| AS1411 | AS1411 is covalently conjugated to the PAMAM dendrimer loaded with camptothecin | nucleolin | A549 | Camptothecin | [286] |
| AS1411 | AS1411-PGG-PTX conjugate (PGG: L-γ-glutamyl-glutamine) | nucleolin | U87 MG, and HUVEC | Paclitaxel | [287] |
| AS1411 | Aptamer/hyaluronic acid-bifunctionalized microemulsion | Nucleolin, CD44 | U87, HUVEC | Docetaxel + Shikonin | [288] |
| AS1411 | AS1411 conjugated with PEGylated PAMAM loaded with camptothecin | nucleolin | HT29, C26, CHO | Camptothecin | [289] |
| AS1411 | AS1411-conjugated DC-Chol/DOPE liposomes (ASLP). The ASLP/siRNA complex was formed through electrostatic interaction between ASLP and siRNA | nucleolin | A375, MDA-MB-231, A549, C6, HeLa, A375 tumor xenograft mice | anti-BRAF siRNA | [290] |
| AS1411 | Aptamer was covalently bound to Au nanoparticles | nucleolin | HeLa, NHDF and HEC-1-A | C8 or Imiquimod | [291] |
| AS1411 | AS1411 was conjugated with PAMAM-PEG encapsulated wth 5-fluorouracil | nucleolin | MKN45 | 5-Fluorouracil | [292] |
| AS1411 | AS1411, influenza hemagglutinin peptide and clofarabine were conjugated into self-assembled peptide nanoparticles with siRNA and doxorubicin | nucleolin | MCF-7, L0-2 | Doxorubicin, siRNA(TK1), clofarabine | [293] |
| ATP-AS1411 bivalent aptamer | ATP-AS1411 was conjugated to DOX-loaded Si nanoparticles | ATP and nucleolin | CHO | Doxorubicin | [294] |
| AS1411 | Camptothecin was encapsulated in β-CD-(PCL-PAEMA)21 micelle modified with AS1411 aptamer | nucleolin | MCF-7, 4T1, L929 | Camptothecin | [295] |
| AS1411 | siRNA with conjugate AS1411-photo-sensitive oligonucleotide (OliP) | nucleolin | 4T1 | siRNA, irridation with 365 nm light | [296] |
| AS1411 | AS1411-conjugated PLGA-lecithin-PEG nanoparticles loaded with paclitaxel and Nile red | nucleolin | MCF-7, GI-1, L929, HMEC | Paclitaxel + Nile red | [297] |
| AS1411 | Doxorubicin-loaded AS1411-conjugated nanoparticles | nucleolin | MCF-7 | Doxorubicin | [298] |
| AS1411 | AS1411-conjugated HPAEG polimer could form self-assembed nanoparticles loaded with doxorubicin | nucleolin | MCF-7 | Doxorubicin | [299] |
| AS1411 | AS1411 and siRNA attached to the surfaces of MSNs nanoparticles loaded doxorubicin | nucleolin | MDA-MB-231 | Doxorubicin + siRNA(TIE2) | [300] |
| AS1411 | AS1411 was conjugate with DOPE-sphingomyelin-cholesterol-DSPE-PEG2000 liposome loaded with paclitaxel and siRNA | nucleolin | MCF-7 | Paclitaxel + siRNA | [301] |
| AS1411 | AS1411 was conjugated with extracellular vesicles loaded with microRNA precursor let-7 or VEGF siRNA | nucleolin | MDA-MB-231 | miRNA let-7 or siRNA (VEGF) | [302] |
| AS1411 | Electrostatic interaction between fluorescent gold nanocluster-conjugated chitosan and AS1411 | nucleolin | A549 | Methotrexate | [303] |
| AS1411 | AS1411-capped FRET-based two-photon MSNs loaded with doxorubicin | nucleolin | MCF-7, HEK293 | Doxorubicin | [304] |
| AS1411 | AS1411-PAMAM-PEG-fluorescent tags modified magnetic zinc-doped iron oxide nanoparicle with the loading doxorubicin and siRNAs | nucleolin | MDA-MB-231, 4T1, MCF-10A | Doxorubicin + siRNA HSP70 or HSP90. NIR/MR | [305] |
| AS1411 | Silica nano supra-assembly nanoparicle with conjugating DOX, cytochrome c and AS1411 | nucleolin | HCT116 | Doxorubicin | [306] |
| AS1411 | AS1411-conjugated PAMAM grafted persistent luminescence nanoparticles loaded with doxorubicin | nucleolin | HeLa, 3T3 | Doxorubicin | [307] |
| AS1411 | AS1411- absorptio, dsDNA and MMP-2 cleavable peptide-fabricated gold nanocage vehicle which could load doxorubicin and siRNAs | nucleolin | NCI-H889 | Doxorubicin and siRNAs | [308] |
| AS1411 | AS1411 decorated via the absorption of a single-stranded thymidine (T)-rich tail of oligonucleotide T20 onto the surface ZnO-gated porphyrinic metal–organic framework loaded with doxorubicin | nucleolin | HeLa, NIH3T3 | Doxorubicin, PDT | [309] |
| AS1411 | AS1411 was anchored onto the surface of the MnO2-coated and loaded with acriflavine and HMME liposomes | nucleolin | SKOV3, HL-7702 | Acriflavine, HMME. Sonodynamic therapy (SDT) | [310] |
| AS1411 | AS1411 aptamer-based three-way junction pocket DNA nanostructure loaded with doxorubicin | nucleolin | PC-3, 4T1, CHO | Doxorubicin | [311] |
| AS1411 | The protoporphyrin IX loading AS1411r-conjugated upconversion nanoparticle | nucleolin | MCF-7, HeLa | Protoporphyrin IX, PDT, NIR | [312] |
| AS1411 | As1411-conjugated HSA nanoparticle loaded with 5- Fluorouracil and BpT (2-benzoylpyridine thiosemicarbazide copper II) | nucleolin | Bel-7402 | 5-Fluorouracil, BpT | [313] |
| AS1411 | As1411 was conjugated with chitosan nanoparticles loaded with erlotinib | nucleolin | A549 | Erlotinib | [314] |
| AS1411 | As1411 was conjugated with polydopamine-functionalized CA-(PCL-ran-PLA) nanoparticles loaded with docetaxel | nucleolin | MCF-7 | Docetaxel | [315] |
| AS1411 | As1411 was conjugated with PEG-PLA-DPPC lipopolymersome loaded with camptothecin | nucleolin | HT29, C26 | Camptothecin | [316] |
| AS1411 | Doxorubicin inserted in the aptamer AS1411 was encapsulated in the aqueous interior of liposome | nucleolin | MCF-7/Adr | Doxorubicin | [317] |
| AS1411 | As1411 was conjugated with Mn-doped mesoporous silica nanoparticles containing 5-aza-2-deoxycytidine and docetaxel | nucleolin | MCF-7 | 5-aza-2-deoxycytidine + docetaxel | [318] |
| AS1411 | AS1411-conjugated albumin nanoparticles loaded with docetaxel | nucleolin | CT26 | Docetaxel | [319] |
| AS1411 | TMPyP4 and doxorubicin were then physically attached to the AS1411-conjugated Au NPsAu nanoparticles | nucleolin | HeLa, Dox-resistant MCF-7R | TMPyP4 + Doxorubicin, PDT | [320] |
| AS1411 | AS1411-conjugated chimeric self-assembled polymersomes loaded with SN38 | nucleolin | HT29, CHO | SN38 (7-ethyl-10-hydroxycamptothecin) | [321] |
| AS1411 | AS1411 immobilized on the surface DNA-modified gold nanoparticles loaded with doxorubicin | nucleolin | HeLa, NIH-3T3 | Doxorubicin | [322] |
| AS1411 | AS1411-conjugated gold nanoparticles loaded with doxorubicin or AZD8055 | nucleolin | MCF-7, OMM1.3, Mel202 | Doxorubicin or AZD8055 | [323] |
| AS1411 | AS1411 was introduced into zeolitic imidazolate framework-8 nanoparticles loaded with doxorubicin | nucleolin | HeLa, HEK 293T | Doxorubicin | [324] |
| AS1411 | AS1411 was immobilizing over composite [γ-cyclodextrin-based metal–organic framework embedded with graphene quantum dots and modified with PEGMA]. Doxorubicin was encapsulated within this composite | nucleolin | MCF-7, L929 | Doxorubicin | [325] |
| AS1411 | AS1411-conjugated PAMAM-10C-10C-PEG nanoparticles loaded with bcl-xl shRNA (10C—10-bromodecanoic acid) | nucleolin | A549, L929 | bcl-xl shRNA | [326] |
| AS1411 | AS1411 and PEG were conjugated with molybdenum disulfide nanosheets loaded with doxorubicin and coated with polydopamine (PDA) layer | nucleolin | MCF-7 | Doxorubicin, NIR | [327] |
| AS1411 | AS1411 aptamer-tethered DNA nanotrains loaded with anthracycline drugs | nucleolin | HeLa, L02 | Doxorubicin or, epirubicin, or daunorubicin | [328] |
| AS1411-ATP fusion aptamer | Polyplexes by doxorubicin -loaded AS1411-ATP fusion aptamer and PEG-pDQA | nucleolin | MCF-7/DOX | Doxorubicin | [329] |
| AS1411 | AS1411 was conjugated onto cell membrane capsules loaded with doxorubicin | nucleolin | QGY-7703, Hepli | Doxorubicin | [330] |
| AS1411 | AS1411-PEG was conjugated with PDA-coated CA-PLGA nanoparticles loaded with docetaxel | nucleolin | MCF-7 | Docetaxel | [331] |
| AS1411 | AS1411 and TGN peptide modified PEG-PCL nanoparticles loaded with docetaxel | nucleolin | C6, bEnd.3 | Docetaxel | [332] |
| Cy5.5-AS1411 | Covalent assembly of Cy5.5-AS1411 aptamer conjugate on the surface of graphene oxide wrapped doxorubicin-loaded mesoporous silica nanoparticles | nucleolin | MCF-7 | Doxorubicin | [333] |
| AS1411 | AS1411 aptamer was conjugated with PEG-PDLLA micelle loaded with triptolide | nucleolin | MIA PaCa-2 | Triptolide | [334] |
| AS1411 | Click-nucleic-acid containing platform for codelivery of rapamycin, anti-PFKFB4 siRNA and aptamer AS1411 | PFKFB4, nucleolin | HEK 293, T1-luci | Rapamycin and anti-PFKFB4 siRNA | [335] |
| AS1411 | AS1411-conjugated PLGA-PVP nanoparticles loaded with doxorubicin | nucleolin | A549 | Doxorubicin | [336] |
| AS1411 | AS1411-coniugated star-shaped glucose-core PCL-PEG nanoparticles containing LNA-anti-miR-214 | Nucleolin, miR-214 | A2780 | anti-miR-214 | [337] |
| AS1411 | DNCA lipid-AS1411 nanoparticles. AS1411 interact via Watson–Crick and p-stacking with DNCA | nucleolin | A549, MCF-7, K562 | AS1411 | [338] |
| AS1411 and S2.2 aptamers | AS1411 and S2.2 aptamers were conjugated to the surface of liposomes coated with gold nanoshells and loaded with docetaxel | Nucleolin, MUC1 | MCF-7 | Docetaxel | [339] |
| AS1411 | AS1411 was covalently attached to the surface of the dextran-coated PLA-PEI nanoparticles which co-loaded with camptothecin and survivin-shRNA | nucleolin | C26, CHO | Camptothecin, survivin-shRNA | [340] |
| AS1411-polydopamine | AS1411 was conjugated wth polydopamine-modified M-PLGA–TPGS nanoparticles loaded with docetaxel | nucleolin | HeLa | Docetaxel | [341] |
| AS1411 | AS1411 and a BHQ2-labeled ATP aptamers incorporated into a hybrid micellar nanoparticle | nucleolin, ATP | HeLa | BHQ2, PDT | [342] |
| AS1411 | AS1411 and cytochrome c were conjugated with mesoporous silica nanoparticles loaded with doxorubicin | nucleolin | HepG2 | Doxorubicin | [343] |
| AS1411 | AS1411 was conjugated with magnetic PLGA-PEG nanospheres loaded with doxorubicin | nucleolin | C6, L929 | Doxorubicin | [344] |
| AS1411 | AS1411 and cRGD were conjugated with gold nanocluster, functionalized with NIR dye; doxorubicin was linked with gold nanoconjugate | nucleolin | U87MG, MCF-7, L02, A549 | Doxorubicin, NIR | [345] |
| AS1411 | Dual-functional probe composed of gold nanoparticles, catalytic Zn2+-dependent DNAzyme, doxorubicin, targeted AS1411 aptamer and acid-decomposable ZnO quantum dots. The Zn2+-dependent ligation DNAzyme and AS1411 aptamer were assembled onto the gold nanoparticles (GNPs) via Au-S bonding. | nucleolin | HeLa | Doxorubicin and miRNA-21 | [346] |
| Apta 12 | DOX-APTA12 conjugate | nucleolin | MCF-10A MDA-MB-231 | Doxorubicin + gemcitabine | [347] |
| MUC1 aptamer | MUC1 aptamer–siR-29b chimera was sinthesized | MUC-1, PTEN | OVCAR-3 | miRNA-29b | [348] |
| MUC1 aptamer | MUC-1 aptamer was conjugated InP/ZnS quantum dots/nanohydrogel fluorescent composite loaded with paclitaxel and sodium oxamate | MUC-1 | MCF-7 | Paclitaxel + sodium oxamate | [349] |
| MUC1 aptamer | MUC-1-responsive DNA motif with hairpin structure (smart sensor) was conjugated with doxorubicin-loaded MSNs | MUC-1 | MCF-7, Hs578bst | Doxorubicin | [350] |
| MUC1 aptamer | MUC1 aptamer-peptide anti-HSP70 peptides (P8 and P17) conjugates | MUC-1 | MCF-7/ADR, A549, HepG2 | Doxorubicin | [351] |
| MUC1 aptamer (aptA) | 1,10-phenanthroline can be intercalated within aptA when complexed with Fe(II) ions | MUC-1 | MCF-7 | 1,10-phenanthroline | [352] |
| MUC1 aptamer | Doxorubicin-incorporated multivalent aptamer–siRNA conjugate | MUC-1 | MCF-7 | BCL2-specific siRNA + doxorubicin | [353] |
| MUC1 aptamer | MUC1-dimer aptamer-calcium carbonate complexes loaded with epirubicin and melittin | MUC-1 | MCF7, C26, HepG2 | Epirubicin and melittin | [354] |
| MUC1 aptamer | MUC1aptamer was conjugared with ferritin nanoparticles loaded with epirubicin | MUC-1 | colon cancer cells | Epirubicin | [355] |
| MUC1 aptamer | Conjugate of KLA–MUC1 aptamer loaded with doxorubicin to the surface DNA nanoparticles | MUC-1 | MCF-7 | Doxorubicin and KLA peptide | [356] |
| MUC1, AS1411 and ATP aptamers | Aptamers-dendrimer conjugate | MUC-1, nucleolin, ATP | MCF-7, C26, CHO | Epirubicin | [357] |
| MUC1 aptamer and AS1411 | MUC1 aptamer incorporated heparin and AS1411 incorporated heparin are decorated on the surface of protamine sulfate/CaCO3 nanoparticle loaded with CRISPR–Cas9 plasmid | MUC-1, nucleolin | HeLa, MCF-7, HEK 293T | CRISPR–Cas9 plasmid (for FAK knockout) | [358] |
| MUC1 aptamer | MUC1 aptamer was conjugated to surface PLA-PEG nanoparticles loaded with doxorubicin | MUC-1 | A-549 | Doxorubicin | [359] |
| MUC1 aptamer | Doxorubicin-conjugated MUC-1 aptamer-armed PEGylated SPIONs | MUC-1 | MDA-MB-231 | Doxorubicin | [360] |
| MUC1 aptamer | Gold coated Fe2O3 nanoparticles conjugated with MUC-1 aptamer | MUC-1 | MCF-7, CHO | Photothermal therapy | [361] |
| MUC1 aptamer | MUC1 aptamer was conjugated with genistein-miRNA-29b-loaded hybrid nanoparticles | MUC-1 | A549 | Genistein and miRNA-29b | [362] |
| MUC1 aptamer | MUC1 aptamer-conjugated chitosan nanoparticles loaded with siRNA and docetaxel | MUC-1 | SKBR3 | IGF-1R siRNA and docetaxel | [363] |
| MUC1 aptamer | ZnSe/ZnS quantum dot-Protoporphyrin IX-MUC1 aptamer conjugate | MUC-1 | HeLa, RAW | Protoporphyrin IX, PDT | [364] |
| MUC1 aptamer | MSNs loaded with safranin O or with doxorubicin and conjuugated with MUC1 aptamer | MUC-1 | MDA-MB-231, MCF-10-A | Safranin O or doxorubicin | [365] |
| MUC1 aptamer | MUC1 aptamer was conjugated to chitosan-SN38 nanoparticles | MUC-1 | HT-29, CHO | SN38 | [366] |
| MUC1 aptamer | MUC1 was conjugated with chitosan-coated albumin nanoparticles loaded with paclitaxel | MUC-1 | MCF7, T47D | Paclitaxel | [68] |
| MUC1 aptamer | Clofarabine, ara-guanosine, gemcitabine, and floxuridine to replace all natural nucleosides in aptamer sequences | MUC-1 | Therapeutic Nucleoside Analogues | [367] | |
| MUC1 aptamer | Aptamer-conjugated liposome containing docetaxel | Tubulin, MUC-1 | MCF 7, MCF 10A, MDA-MB-231, HeLa, Hep G2 | Docetaxel | [368] |
| MUC1 aptamer | Doxorubicin was intercalated into self-assembled CpG-MUC1-hydrogel | MUC-1 | MCF-7, A549, HepG-2, RAW264.7 | Doxorubicin | [369] |
| MUC1 aptamer | MUC1 aptamer was conjugated with liposomes (sDPPC, cholesterol, PEG 2000-DSPE and Mal-PEG 2000-DSPE) encapsulated gold nanocages and doxorubicin | MUC-1 | MCF-7 | Doxorubicin, gold nanocages + NIR light irradiation | [370] |
| MUC1 aptamer | MUC1 aptamer-conjugated chitosan nanoparticles loaded with docetaxel and siRNA | MUC-1 | SKBR3, CHO | Docetaxel and cMET siRNA | [371] |
| 5TR1-aptamer | 5TR1 aptamer conjugated to surface PEGylated liposome loading doxorubicin | MUC-1 | C26 | Doxorubicin | [372] |
| 5TR1-aptamer | Doxorubicin was loaded on the modified 5TR1-GC (5TR1 to add a GC loop) | MUC-1 | MDA-MB-231, MCF-10A | Doxorubicin | [373] |
| 5TR1-aptamer | 5TR1 aptamer was conjugated with chitosan-modified PLGA nanoparticles | MUC-1 | MCF7, C26 | Epirubicin | [374] |
| 5TR1 aptamer | SPION-5TR1 aptamer conjugate loading epirubicin | MUC-1 | C26, CHO-K1 | Epirubicin | [375] |
| PSMA aptamer | PSMA aptamer-anchored PLGA-b-PEG-COOH nanoparticles loaded with docetaxel | PSMA | LNCaP | Docetaxel | [78] |
| PSMA RNA aptamer | PSMA aptamer-conjugated liposome loaded with doxorubicin | PSMA | LNCaP | Doxorubicin | [79] |
| PSMA aptamer (A10) | A10 was conjugated with DSPE-PEG2000 liposome-CRISPR/Cas9 chimeras | PSMA, PLK-1 | LNCaP, PC-3 | CRISPR/Cas9 gRNA target PLK1 | [376] |
| A10 aptamer | A10-3.2 aptamer was conjugated with FoxM1 siRNA-loaded cationic nanobubbles | PSMA | LNCaP | FoxM1 siRNA | [377] |
| PSMA aptamer | PSMA aptamer was conjugated with doxorubicin-loaded H40-PLA-PEG micelles | PSMA | CWR22Rν1 | Doxorubicin | [80] |
| A10 aptamer | A10 aptamer-functionalized doxorubicin-polylactide conjugate | PSMA | LNCaP, PC-3 | Doxorubicin | [378] |
| A10-3.2 aptamer | A10-3.2 aptamer-modified ultrasound-responsive nanodroplets loading siRNA | PSMA, CAT-1 | 22RV1, PC-3, 16HBE | siRNA against siCAT-1 transporter | [379] |
| MRP1-CD28 aptamer | MRP1-CD28 aptamer conjugate | MRP1, CD28 | B16F10, chemotherapy-resistant tumors | Translational whole-cell vaccine (Aptvax) has been developed based on this MRP1-CD28 aptamer conjugate | [86] |
| CD28 aptamer | CAR-like multivalent aptamer nanoparticles (X-polymers): murine CD28Apt7 + the tetramer of CTLA-4 + RNA aptamer (Del60) + folic acid labeled ssDNA | CD28, CTLA-4 | mouse melanoma B16 cell | CAR-T-cell immunotherapy | [87] |
| CD38 aptamer | CD38 aptamer-doxorubicin conjugate | CD38 | MM cells, CD38-negative cells, HDLM2, Jeko1, L428, K299 | Doxorubicin | [380] |
| CD44 aptamer | CD44 aptamer was conjugated to the surface of PEGylated liposomes | CD44 | A549, MDA-MB-231, the CD44(-) cell line, NIH/3T3 | - | [381] |
| CD44 aptamer | CD44 aptamer-doxorubicin conjugate | CD44 | Breast cancer cells | Doxorubicin | [382] |
| CD44 aptamer | CD44 aptamer was conjugated with PEGylated liposomes loaded with siRNA | CD44 | MDA-MB-231-Luc2-GFP | siRNA | [383] |
| CD44 thioaptamer | CD44 thioaptamer was conjugated with PAMAM-PEG/miRNA-145 nanoparticles | CD44 | MDA-MB-231, MCF-7 | miRNA-145 | [384] |
| CD44-anti-PD-L1 aptamer | Liposomes have been loaded with loaded DOX and IDO1 siRNA and conjugated to CD44 and anti-PD-L1 DNA aptamers | CD44, PD-1/PD-L1 interaction | MDA-MB-231 cells; mouse: 4T1 cells | IDO1 siRNA, Doxorubicin | [93] |
| Anti-EGFR-CD44 aptamer | An anti-EGFR-CD44 aptamer was conjugated with solid lipid dexamethasone-modified nanoparticles modified loaded with doxorubicin. | EGFR, CD44 | MDA-MB-468 cell | Doxorubicin | [94] |
| CD44 DNA aptamer | Five-Part Pentameric nanocomplex incorporating CD44 DNA aptamer for cellular targeting and thiolated hyaluronic acid to stabilize Au NPs | CD44 | HeLa, SKOV-3, C13, NIH3T3, SH-SY5Y | Doxorubicin | [95] |
| CD71- aptamer | CD71 aptamer was conjuugated with XQ-2d-MMAE | CD71 | OCM-1 | Monomethyl auristatin E (MMAE) | [98] |
| CD117 aptamer | CD117 aptamer-methotrexate conjugates | CD117 | CD117-expressing HEL cells | Methotrexate | [101] |
| B19 aptamer | B19 aptamer-conjugated PAMAM G4C12 dendrimer nanoparticles loaded with paclitaxel and temozolomide | CD133 | U-87, C6 | Paclitaxel + temozolomide | [385] |
| CD133 aptamer | CD133 aptamer-PLGA-PEG micelle loaded with paclitaxel | CD133 | A549 | Paclitaxel | [110] |
| CD133 aptamer | PEGylated CD133aptamer-doxorubicin conjugate | CD133 | Hep3B, Huh7, HEK293T | Doxorubicin | [107] |
| CD133 aptamer | CD133 was conjugated covalently to the self-assembled PEGylated carboxymethylcellulose-SN38-conjugate nanoparticles | CD133 | HT29, CHO | SN38 | [386] |
| Thiolated CD133 aptamer A15 | A15 aptamers were conjugated with curcumin-loaded liposomes (EPC, CHOL and DSPE-PEG2000-MAL) | CD133 | DU145 | Curcumin | [387] |
| CD133 aptamer | CD133 aptamers were conjugated propranolol-loaded poly(lactic-co-glycolic acid) nanoparticle | CD133 | HemSC | Propranolol | [388] |
| CD133 aptamer | CD133 aptamer was conjugated with salinomycin-loaded PEGylated nanoparticles | CD133 | Saos-2, U-2 OS, MG-63 | Salinomycin | [389] |
| CD133 aptamer | Gefitinib-loaded nanomicelles conjugated with CD133 aptamers | CD133 | A549, A431 | Gefitinib | [111] |
| CD133 aptamer (A15) + EGFR (CL4) | Salinomycin-loaded poly(lactic-co-glycolic acid) nanoparticles A15 or CL4-conjugted or nontargeted salinomycin-loaded nanoparticles. | CD133, EGFR | Huh7, Hep3B | Salinomycin | [390] |
| CD133 aptamer | CD133 aptamer was conjugated with polymer-micellar NPs composed of poly(styrene-b-ethylene oxide) (PS-b-PEO) and PLGA and labeling with 89Zr | CD133 | Human GBM cancer stem cells | Temozolomide, idasanutlin | [391] |
| CD133 aptamer | Conjugating CD133 aptamers to DTX liposomes (DSPE-PEG2000-CHOL-SPC) | CD133 | A549 | Docetaxel | [113] |
| CD133 aptamer | All-trans retinoic acid (ATRA)-loaded lipid-polymer nanoparticles conjugated with CD133 aptamers | CD133 | Saos-2, U-2 OS | ATRA | [392] |
| CD133 aptamer | CD133 aptamer shortened form (AP-1-M) was conjugated with doxorubicin | CD133 | FRO cells | Doxorubicin | [108] |
| Cs5 aptamer | Doxorubicin was loaded into the Cs5 aptamer to form a chimera. | CD133 | HCT116 | Doxorubicin | [393] |
| CD133 aptamer + angiopep-2 (An2) | Angiopep-2 and CD133 RNA aptamers were conjugated on exosomes as vehicles loaded with temozolomide and O6-benzylguanine | CD133, An2 | U87MG, GSC | Temozolomide + O6-benzylguanine | [394] |
| CD133 aptamer | Propranolol-loaded CD133 aptamers conjugated liposomes-in-microspheres | CD133 | HemSCs | Propranolol | [395] |
| HER2 aptamer | HER2 aptamer was conjugated with mertansine | HER2 | BT-474, MDA-MB-231 | Mertansine | [396] |
| HER2-EGFR aptamer | Bivalent HER2 aptamer-EGFR siRNA chimera | HER2, EGFR | BT474, SKBR3, MDA-MB-231, MCF7, Hs578 T | siRNA | [397] |
| HER2 aptamer | HER2 aptamer-functionalized pH-sensitive β-cyclodextrin -capped doxorubicin-loaded MSNs | HER2 | SKBR3, MCF7, HEK-293T | Doxorubicin | [398] |
| HER2 aptamer | MMAE-conjugated HER2 oligobody (the cotinine (cot)-body, cot-linker, aptamer and MMAE) | HER2 | SKBR3, NCI-N87 | MMAE | [399] |
| HER2 aptamer | pH-responsive micelle-like nanoparticles carrying a Texas red-fluorescently labeled HER2 aptamer | HER2 | SKBR3, MCF7, HeLa | HER2-aptamer | [400] |
| HER2 aptamer | HER2 was conjugated to the surface curcumin-loaded HSA nanoparticles | HER2 | SKBR3 | Curcumin | [401] |
| HER2 aptamer | HER2 aptamer/Ursolic acid + doxorubicin nanoparticles constructed as carrier-free nanodrugs | HER2 | BT474 | Ursolic acid + doxorubicin | [402] |
| Aptamer A6 | Aptamer-labeled liposomal nanoparticle using different saturated (HSPC and DPPC) and unsaturated (POPC and DOPC) lipids | HER2 | SKBR3, MCF7, MDA-MB-231 | Doxorubicin | [403] |
| Aptamer A6 | Aptamer-labeled P-gp siRNA encapsulated PLGA or PLGA-PEG nanoparticles | HER2, P-gp | SKBR-3, 4T1-R, MDA MB-231, MCF-7 | Aptamer-labeled P-gp siRNA | [404] |
| HER2 aptamer and AS1411 aptamer | Dual aptamer-decorated DNA hydrogel | HER2 + nucleolin | SKBR-3, MCF7, MDA-MB-23 | Doxorubicin | [405] |
| HER2 aptamer | HER2 densely grafted on gold nanostars | HER2 | SKBR-3, MCF-10A | HApt-AuNS | [406] |
| HER2 aptamer | Carboxylated chitosan-coated (pH-responsive), doxorubicin-loaded aptamer- MSN bioconjugates | HER2, EGFR | SKBR-3, MCF7 | Doxorubicin | [407] |
| VEGF aptamer | DNM and TMPyP were physically assembled with aptamer of VEGF + cytosine (C)-rich DNA fragment (gc-34) | VEGF | MCF-7 | Daunomycin + TMPyP, PDT | [61] |
| EpCAM RNA aptamer | RNA aptamer-conjugated PEGylated liposome loaded with doxorubicin | EpCAM | CHO-K1, C26 | Doxorubicin | [408] |
| EpCAM | EpCAM aptamer-survivin siRNA fusion, combined with doxorubicin treatment doxorubicin-resistant subline of the MCF-7 cells | EpCAM | MCF-7 | Doxorubicin and survivin siRNA | [146] |
| FOXM1 aptamer | FOXM1 aptamer conjugated with DOX-CPT-loaded HA-b-PCL nanopolymersomes | FOXM1 | A549, SK-MES-1 | Doxorubicin, camptothecin | [409] |
| Sgc8c aptamer | Aptamer sgc8c–doxorubicin conjugate | PTK7 | NB-4, Ramos | Doxorubicin | [410] |
| Aptamer BAFF-R | BAFF-R aptamer–siRNA conjugate | BAFF-R | Jeko-1, Z138 | siRNA | [411] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shishparenok, A.N.; Furman, V.V.; Zhdanov, D.D. DNA-Based Nanomaterials as Drug Delivery Platforms for Increasing the Effect of Drugs in Tumors. Cancers 2023, 15, 2151. https://doi.org/10.3390/cancers15072151
Shishparenok AN, Furman VV, Zhdanov DD. DNA-Based Nanomaterials as Drug Delivery Platforms for Increasing the Effect of Drugs in Tumors. Cancers. 2023; 15(7):2151. https://doi.org/10.3390/cancers15072151
Chicago/Turabian StyleShishparenok, Anastasiya N., Vitalina V. Furman, and Dmitry D. Zhdanov. 2023. "DNA-Based Nanomaterials as Drug Delivery Platforms for Increasing the Effect of Drugs in Tumors" Cancers 15, no. 7: 2151. https://doi.org/10.3390/cancers15072151
APA StyleShishparenok, A. N., Furman, V. V., & Zhdanov, D. D. (2023). DNA-Based Nanomaterials as Drug Delivery Platforms for Increasing the Effect of Drugs in Tumors. Cancers, 15(7), 2151. https://doi.org/10.3390/cancers15072151