Exosomal DNA: Role in Reflecting Tumor Genetic Heterogeneity, Diagnosis, and Disease Monitoring
Abstract
:Simple Summary
Abstract
1. Introduction
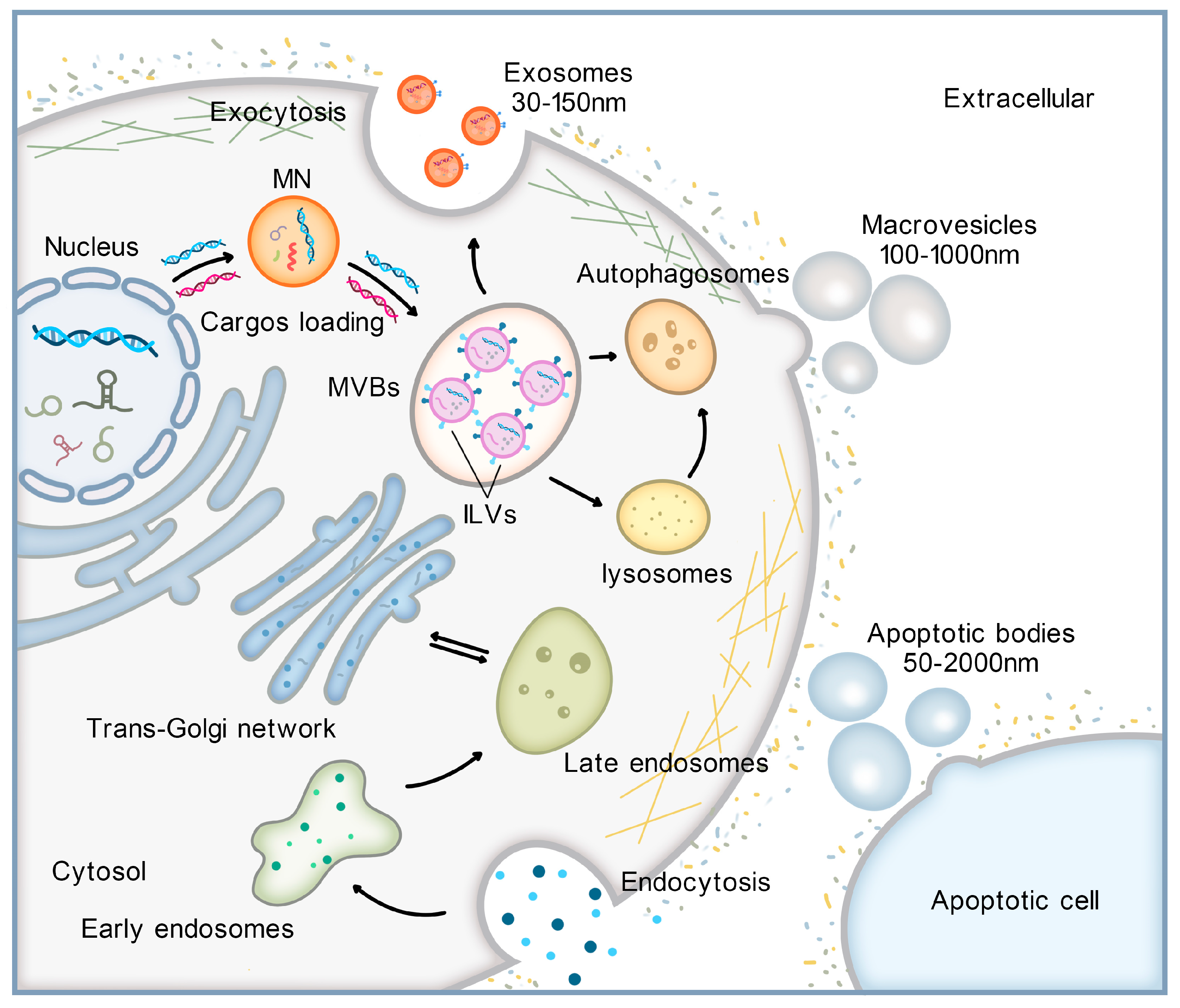
2. Formation of EVs and Mechanisms of exoDNA Loading
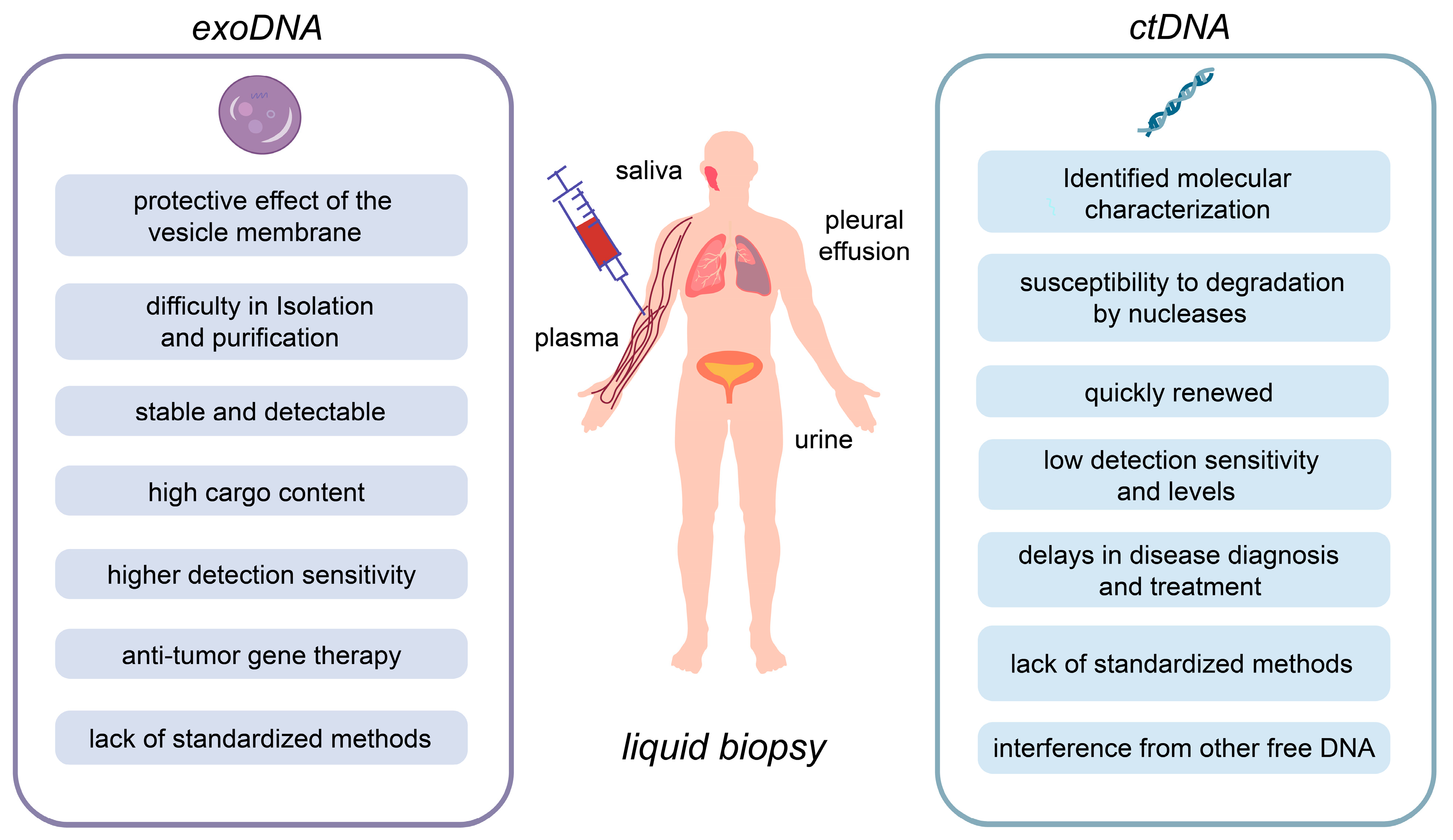
3. Research Progress on the Application of exoDNA in LB
3.1. MtDNA as a Potential Biomarker for Tumor Diagnosis and Treatment
3.1.1. Prostate Cancer (PCa)
3.1.2. Breast Cancer
3.1.3. Glioblastoma
3.1.4. Ovarian Cancers
3.2. Known Relationships between Nuclear DNA Mutations and Clinical Tumors
3.2.1. Pancreatic Ductal Adenocarcinoma (PDAC)
3.2.2. Gliomas
3.2.3. HCC
3.2.4. Neuroblastoma (NB)
3.2.5. Bladder Cancer (BC)
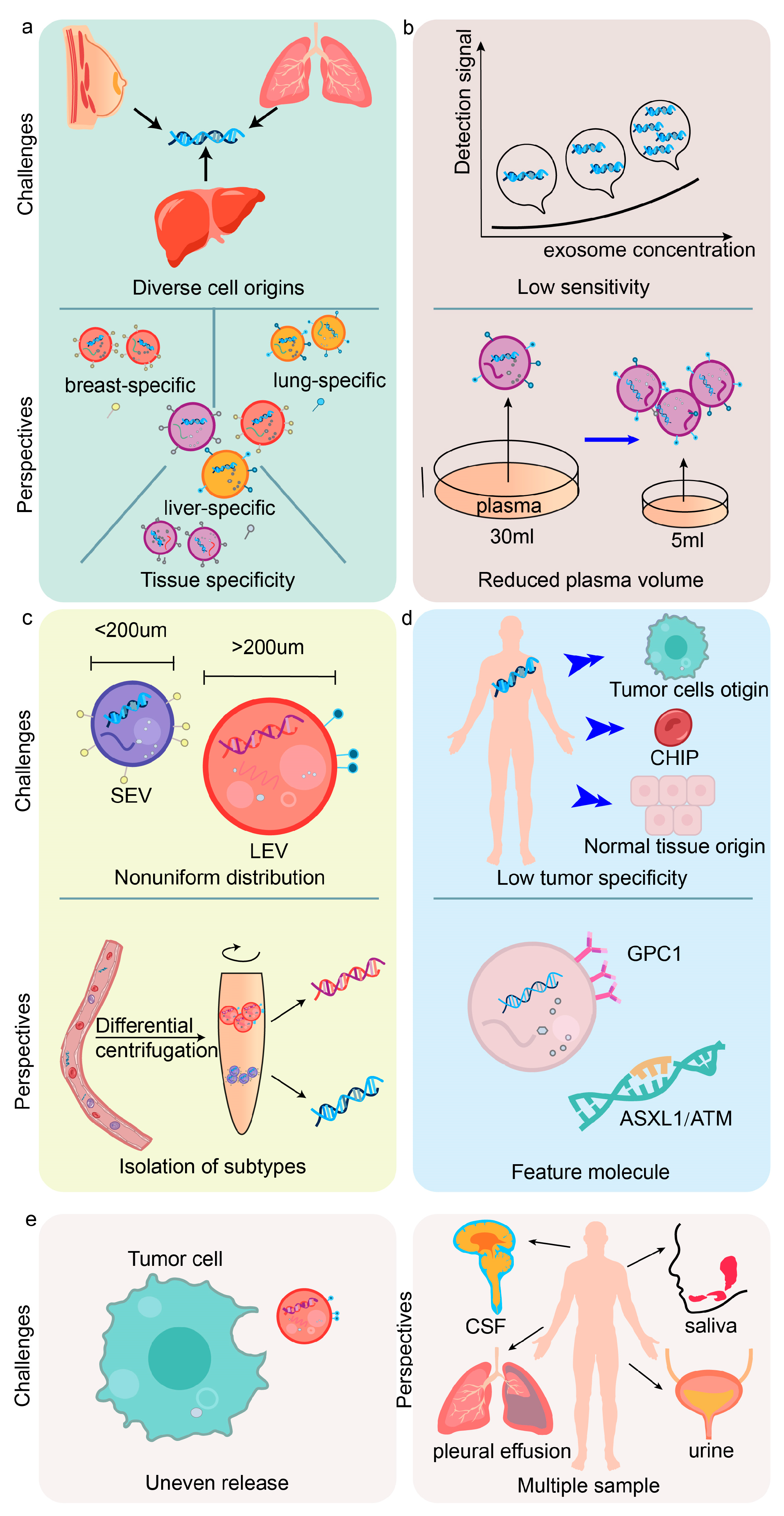
4. Current Challenges and Future Perspectives of exoDNA
4.1. Challenges and Limitations
4.2. Therapeutic Applications
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am. J. Physiol.-Cell Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Li, L.; Wang, Z.; Irfan, M.; Qu, F. Recent advances of aptasensors for exosomes detection. Biosens. Bioelectron. 2020, 160, 112213. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Szatmári, T.; Balázs, K.; Csordás, I.B.; Sáfrány, G.; Lumniczky, K. Effect of radiotherapy on the DNA cargo and cellular uptake mechanisms of extracellular vesicles. Strahlenther. Und Onkol. 2023, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Elzanowska, J.; Semira, C.; Costa-Silva, B. DNA in extracellular vesicles: Biological and clinical aspects. Mol. Oncol. 2020, 15, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- San Lucas, F.A.; Allenson, K.; Bernard, V.; Castillo, J.; Kim, D.U.; Ellis, K.; Ehli, E.A.; Davies, G.E.; Petersen, J.L.; Li, D.; et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann. Oncol. 2016, 27, 635–641. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Kan, S.; Zhu, Y.; Feng, S.; Feng, W.; Gao, S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomed. 2018, 13, 585–599. [Google Scholar] [CrossRef]
- Guo, X.; Sui, R.; Piao, H. Tumor-derived small extracellular vesicles: Potential roles and mechanism in glioma. J. Nanobiotechnology 2022, 20, 383. [Google Scholar] [CrossRef]
- Sharma, A.; Johnson, A. Exosome DNA: Critical regulator of tumor immunity and a diagnostic biomarker. J. Cell. Physiol. 2019, 235, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Lichá, K.; Pastorek, M.; Repiská, G.; Celec, P.; Konečná, B. Investigation of the Presence of DNA in Human Blood Plasma Small Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 5915. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Li, B.; Cao, Y.; Sun, M.; Feng, H. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. Faseb J. 2021, 35, e21916. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, H.; Fu, J.; Wu, X.; Liang, X.Y.; Liu, X.Y.; Wu, X.; Cao, L.L.; Xu, Z.Y.; Dong, M. Microfluidic-based exosome isolation and highly sensitive aptamer exosome membrane protein detection for lung cancer diagnosis. Biosens. Bioelectron. 2022, 214, 114487. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.L.; Liu, J.Y.; Chen, G. Small extracellular vesicle PD-L1 in cancer: The knowns and unknowns. NPJ Precis. Oncol. 2022, 6, 42. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, Z.L.; Wu, M.; Ren, J.G.; Xia, H.F.; Sa, G.L.; Zhu, J.Y.; Pang, D.W.; Zhao, Y.F.; Chen, G. Magnetic and Folate Functionalization Enables Rapid Isolation and Enhanced Tumor-Targeting of Cell-Derived Microvesicles. ACS Nano 2017, 11, 277–290. [Google Scholar] [CrossRef]
- Xia, H.F.; Yu, Z.L.; Zhang, L.J.; Liu, S.L.; Zhao, Y.; Huang, J.; Fu, D.D.; Xie, Q.H.; Liu, H.M.; Zhang, Z.L.; et al. Real-Time Dissection of the Transportation and miRNA-Release Dynamics of Small Extracellular Vesicles. Adv. Sci. 2023, 10, e2205566. [Google Scholar] [CrossRef] [PubMed]
- Movahedpour, A.; Khatami, S.H.; Karami, N.; Vakili, O.; Naeli, P.; Jamali, Z.; Shabaninejad, Z.; Tazik, K.; Behrouj, H.; Ghasemi, H. Exosomal noncoding RNAs in prostate cancer. Clin. Chim. Acta 2022, 537, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, M.; Sugaya, K. DNA Associated with Circulating Exosomes as a Biomarker for Glioma. Genes 2020, 11, 1276. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kumar, S.; Jayachandran, M.; Herrera Hernandez, L.P.; Wang, S.; Wilson, E.M.; Lieske, J.C. Excretion of urine extracellular vesicles bearing markers of activated immune cells and calcium/phosphorus physiology differ between calcium kidney stone formers and non-stone formers. BMC Nephrol. 2021, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hatch, E.M.; Fischer, A.H.; Deerinck, T.J.; Hetzer, M.W. Catastrophic Nuclear Envelope Collapse in Cancer Cell Micronuclei. Cell 2013, 154, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef] [PubMed]
- Karn, V.; Ahmed, S.; Tsai, L.-W.; Dubey, R.; Ojha, S.; Singh, H.; Kumar, M.; Gupta, P.; Sadhu, S.; Jha, N.; et al. Extracellular Vesicle-Based Therapy for COVID-19: Promises, Challenges and Future Prospects. Biomedicines 2021, 9, 1373. [Google Scholar] [CrossRef]
- Lin, Z.; Jayachandran, M.; Haskic, Z.; Kumar, S.; Lieske, J.C. Differences of Uric Acid Transporters Carrying Extracellular Vesicles in the Urine from Uric Acid and Calcium Stone Formers and Non-Stone Formers. Int. J. Mol. Sci. 2022, 23, 10010. [Google Scholar] [CrossRef]
- Jayachandran, M.; Yuzhakov, S.V.; Kumar, S.; Larson, N.B.; Enders, F.T.; Milliner, D.S.; Rule, A.D.; Lieske, J.C. Specific populations of urinary extracellular vesicles and proteins differentiate type 1 primary hyperoxaluria patients without and with nephrocalcinosis or kidney stones. Orphanet J. Rare Dis. 2020, 15, 1–14. [Google Scholar] [CrossRef]
- Yu, Z.L.; Zhao, Y.; Miao, F.; Wu, M.; Xia, H.F.; Chen, Z.K.; Liu, H.M.; Zhao, Y.F.; Chen, G. In Situ Membrane Biotinylation Enables the Direct Labeling and Accurate Kinetic Analysis of Small Extracellular Vesicles in Circulation. Anal. Chem. 2021, 93, 10862–10870. [Google Scholar] [CrossRef]
- Xu, R.; Yu, Z.L.; Liu, X.C.; Xie, Q.H.; Wu, M.; Chen, G. Aptamer-Assisted Traceless Isolation of PD-L1-Positive Small Extracellular Vesicles for Dissecting Their Subpopulation Signature and Function. Anal. Chem. 2023, 95, 1016–1026. [Google Scholar] [CrossRef]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Szilágyi, M.; Pös, O.; Márton, É.; Buglyó, G.; Soltész, B.; Keserű, J.; Penyige, A.; Szemes, T.; Nagy, B. Circulating Cell-Free Nucleic Acids: Main Characteristics and Clinical Application. Int. J. Mol. Sci. 2020, 21, 6827. [Google Scholar] [CrossRef] [PubMed]
- Balachander, K.; Roy, A.; Priyadharsini, J.V.; Murugan, S.; Paramasivam, A. Mitochondrial DNA in circulating exosomes: A novel biomarker and potential therapeutic target for oral cancer. Oral. Oncol. 2022, 128, 105857. [Google Scholar] [CrossRef] [PubMed]
- Degli Esposti, C.; Iadarola, B.; Maestri, S.; Beltrami, C.; Lavezzari, D.; Morini, M.; De Marco, P.; Erminio, G.; Garaventa, A.; Zara, F.; et al. Exosomes from Plasma of Neuroblastoma Patients Contain Doublestranded DNA Reflecting the Mutational Status of Parental Tumor Cells. Int. J. Mol. Sci. 2021, 22, 3667. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Ibáñez, E.; Lässer, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; García-Rodríguez, A.; Lötvall, J. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell. Vesicles 2019, 8, 1656993. [Google Scholar] [CrossRef] [PubMed]
- Soltész, B.; Urbancsek, R.; Pös, O.; Hajas, O.; Forgács, I.N.; Szilágyi, E.; Nagy-Baló, E.; Szemes, T.; Csanádi, Z.; Nagy, B. Quantification of peripheral whole blood, cell-free plasma and exosome encapsulated mitochondrial DNA copy numbers in patients with atrial fibrillation. J. Biotechnol. 2019, 299, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Philley, J.V.; Kannan, A.; Qin, W.; Sauter, E.R.; Ikebe, M.; Hertweck, K.L.; Troyer, D.A.; Semmes, O.J.; Dasgupta, S. Complex-I Alteration and Enhanced Mitochondrial Fusion Are Associated With Prostate Cancer Progression. J. Cell. Physiol. 2016, 231, 1364–1374. [Google Scholar] [CrossRef] [PubMed]
- Akoto, T.; Saini, S. Role of Exosomes in Prostate Cancer Metastasis. Int. J. Mol. Sci. 2021, 22, 3528. [Google Scholar] [CrossRef]
- Albertsen, P.C. PSA testing, cancer treatment, and prostate cancer mortality reduction: What is the mechanism? Urol. Oncol. 2023, 41, 78–81. [Google Scholar] [CrossRef]
- Liu, Z.; Jin, Q.; Yan, T.; Wo, Y.; Liu, H.; Wang, Y. Exosome-mediated transduction of mechanical force regulates prostate cancer migration via microRNA. Biochem. Biophys. Rep. 2022, 31, 101299. [Google Scholar] [CrossRef]
- Sekhoacha, M.; Riet, K.; Motloung, P.; Gumenku, L.; Adegoke, A.; Mashele, S. Prostate Cancer Review: Genetics, Diagnosis, Treatment Options, and Alternative Approaches. Molecules 2022, 27, 5730. [Google Scholar] [CrossRef]
- Nguyen-Nielsen, M.; Borre, M. Diagnostic and Therapeutic Strategies for Prostate Cancer. Semin. Nucl. Med. 2016, 46, 484–490. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, J.; Donovan, M.J.; Margolis, E.; Partin, A.; Carter, B.; Brown, G.; Torkler, P.; Noerholm, M.; Skog, J.; Shore, N.; et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2–10 ng/mL at Initial Biopsy. Eur. Urol. 2018, 74, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Wilt, T.J.; Dahm, P. PSA Screening for Prostate Cancer: Why Saying No is a High-Value Health Care Choice. J. Natl. Compr. Canc. Netw. 2015, 13, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Reznik, E.; Miller, M.L.; Şenbabaoğlu, Y.; Riaz, N.; Sarungbam, J.; Tickoo, S.K.; Al-Ahmadie, H.A.; Lee, W.; Seshan, V.E.; Hakimi, A.A.; et al. Mitochondrial DNA copy number variation across human cancers. Elife 2016, 5, e10769. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, Z.; Li, L.; Zhang, Z.; Jin, X.; Wu, P.; Sun, S.; Pan, J.; Su, K.; Jia, F.; et al. Aged neutrophils form mitochondria-dependent vital NETs to promote breast cancer lung metastasis. J. Immunother Cancer 2021, 9, e002875. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed]
- Herzog, S.K.; Fuqua, S.A.W. ESR1 mutations and therapeutic resistance in metastatic breast cancer: Progress and remaining challenges. Br. J. Cancer 2022, 126, 174–186. [Google Scholar] [CrossRef]
- Najim, O.; Seghers, S.; Sergoynne, L.; Van Gaver, H.; Papadimitriou, K.; Wouters, K.; Trinh, X.B.; Huizing, M.T.; Tjalma, W. The association between type of endocrine therapy and development of estrogen receptor-1 mutation(s) in patients with hormone-sensitive advanced breast cancer: A systematic review and meta-analysis of randomized and non-randomized trials. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 188315. [Google Scholar] [CrossRef]
- Dahlgren, M.; George, A.M.; Brueffer, C.; Gladchuk, S.; Chen, Y.; Vallon-Christersson, J.; Hegardt, C.; Häkkinen, J.; Rydén, L.; Malmberg, M.; et al. Preexisting Somatic Mutations of Estrogen Receptor Alpha (ESR1) in Early-Stage Primary Breast Cancer. JNCI Cancer Spectr. 2021, 5, pkab028. [Google Scholar] [CrossRef]
- Le Rhun, E.; Preusser, M.; Roth, P.; Reardon, D.A.; van den Bent, M.; Wen, P.; Reifenberger, G.; Weller, M. Molecular targeted therapy of glioblastoma. Cancer Treat Rev. 2019, 80, 101896. [Google Scholar] [CrossRef]
- Rong, L.; Li, N.; Zhang, Z. Emerging therapies for glioblastoma: Current state and future directions. J. Exp. Clin. Cancer Res. 2022, 41, 142. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Xia, Q.; Muhammad, T.; Liu, L.; Meng, X.; Bars-Cortina, D.; Khan, A.A.; Huang, Y.; Dong, L. Glioblastoma Therapy: Rationale for a Mesenchymal Stem Cell-based Vehicle to Carry Recombinant Viruses. Stem. Cell Rev. Rep. 2022, 18, 523–543. [Google Scholar] [CrossRef] [PubMed]
- Habashy, K.J.; Mansour, R.; Moussalem, C.; Sawaya, R.; Massaad, M.J. Challenges in glioblastoma immunotherapy: Mechanisms of resistance and therapeutic approaches to overcome them. Br. J. Cancer 2022, 127, 976–987. [Google Scholar] [CrossRef]
- Ronvaux, L.; Riva, M.; Coosemans, A.; Herzog, M.; Rommelaere, G.; Donis, N.; D’Hondt, L.; Douxfils, J. Liquid Biopsy in Glioblastoma. Cancers 2022, 14, 3394. [Google Scholar] [CrossRef] [PubMed]
- Eibl, R.H.; Schneemann, M. Liquid biopsy and glioblastoma. Explor. Target Antitumor. Ther. 2023, 4, 28–41. [Google Scholar] [CrossRef]
- Duraj, T.; García-Romero, N.; Carrión-Navarro, J.; Madurga, R.; Mendivil, A.O.; Prat-Acin, R.; Garcia-Cañamaque, L.; Ayuso-Sacido, A. Beyond the Warburg Effect: Oxidative and Glycolytic Phenotypes Coexist within the Metabolic Heterogeneity of Glioblastoma. Cells 2021, 10, 202. [Google Scholar] [CrossRef]
- Tang, P.L.Y.; Méndez Romero, A.; Jaspers, J.P.M.; Warnert, E.A.H. The potential of advanced MR techniques for precision radiotherapy of glioblastoma. Magma 2022, 35, 127–143. [Google Scholar] [CrossRef]
- Vidone, M.; Clima, R.; Santorsola, M.; Calabrese, C.; Girolimetti, G.; Kurelac, I.; Amato, L.B.; Iommarini, L.; Trevisan, E.; Leone, M.; et al. A comprehensive characterization of mitochondrial DNA mutations in glioblastoma multiforme. Int. J. Biochem. Cell Biol. 2015, 63, 46–54. [Google Scholar] [CrossRef]
- Gatto, L.; Franceschi, E.; Di Nunno, V.; Tosoni, A.; Lodi, R.; Brandes, A.A. Liquid Biopsy in Glioblastoma Management: From Current Research to Future Perspectives. Oncologist 2021, 26, 865–878. [Google Scholar] [CrossRef]
- Soltész, B.; Pös, O.; Wlachovska, Z.; Budis, J.; Hekel, R.; Strieskova, L.; Liptak, J.B.; Krampl, W.; Styk, J.; Németh, N.; et al. Mitochondrial DNA copy number changes, heteroplasmy, and mutations in plasma-derived exosomes and brain tissue of glioblastoma patients. Mol. Cell. Probes 2022, 66, 101875. [Google Scholar] [CrossRef]
- Keserű, J.S.; Soltész, B.; Lukács, J.; Márton, É.; Szilágyi-Bónizs, M.; Penyige, A.; Póka, R.; Nagy, B. Detection of cell-free, exosomal and whole blood mitochondrial DNA copy number in plasma or whole blood of patients with serous epithelial ovarian cancer. J. Biotechnol. 2019, 298, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Vadakedath, S.; Kandi, V.; Ca, J.; Vijayan, S.; Achyut, K.C.; Uppuluri, S.; Reddy, P.K.K.; Ramesh, M.; Kumar, P.P. Mitochondrial Deoxyribonucleic Acid (mtDNA), Maternal Inheritance, and Their Role in the Development of Cancers: A Scoping Review. Cureus 2023, 15, e39812. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol. Cancer 2019, 18, 124. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Xia, M.; Zhang, Y.; Luo, H.; Dong, D.; Sun, L. Mitochondrial integration and ovarian cancer chemotherapy resistance. Exp. Cell Res. 2021, 401, 112549. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, V.D.B. Ovarian Cancer Biomarkers: Moving Forward in Early Detection. Adv. Exp. Med. Biol. 2020, 1219, 355–363. [Google Scholar] [CrossRef]
- Shi, S.; Yu, Z.-L.; Jia, J. The Roles of Exosomes in the Diagnose, Development and Therapeutic Resistance of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 1968. [Google Scholar] [CrossRef]
- Liu, J.Y.; Yu, Z.L.; Fu, Q.Y.; Zhang, L.Z.; Li, J.B.; Wu, M.; Liu, B.; Chen, G. Immunosuppressive effect of small extracellular vesicle PD-L1 is restricted by co-expression of CD80. Br. J. Cancer 2023, 129, 925–934. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of Double-stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and p53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; San Lucas, F.A.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118.e4. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Allenson, K.; Castillo, J.; San Lucas, F.A.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutantKRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Azizian, A.; Rühlmann, F.; Krause, T.; Bernhardt, M.; Jo, P.; König, A.; Kleiß, M.; Leha, A.; Ghadimi, M.; Gaedcke, J. CA19-9 for detecting recurrence of pancreatic cancer. Sci. Rep. 2020, 10, 1332. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.S.; Stoita, A. Biomarkers in the diagnosis of pancreatic cancer: Are we closer to finding the golden ticket? World J. Gastroenterol. 2021, 27, 4045–4087. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Villanueva, M.; Ruiz-Rebollo, L.; Montes, M.; Rodriguez-Lopez, M.; Francisco, M.; Cubiella, J.; Iyo, E.; Garabitos, E.; Martínez Moneo, E.; Martos, M.; et al. CA19-9 capability as predictor of pancreatic cancer resectability in a Spanish cohort. Mol. Biol. Rep. 2020, 47, 1583–1588. [Google Scholar] [CrossRef]
- Shi, W.; Lv, C.; Qi, J.; Zhao, W.; Wu, X.; Jing, R.; Wu, X.; Ju, S.; Chen, J. Prognostic Value of Free DNA Quantification in Serum and Cerebrospinal Fluid in Glioma Patients. J. Mol. Neurosci. 2011, 46, 470–475. [Google Scholar] [CrossRef]
- Saenz, A.; Auzmendi, I.; Carrasco, G.; Moreno, C.; Ruiz, I.; Villanua, J.; Egaña, L.; Otaegui, D.; Samprón, N.; Matheu, A. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers 2019, 11, 950. [Google Scholar] [CrossRef]
- Piazza, A.; Rosa, P.; Ricciardi, L.; Mangraviti, A.; Pacini, L.; Calogero, A.; Raco, A.; Miscusi, M. Circulating Exosomal-DNA in Glioma Patients: A Quantitative Study and Histopathological Correlations—A Preliminary Study. Brain Sci. 2022, 12, 500. [Google Scholar] [CrossRef]
- Barthel, L.; Hadamitzky, M.; Dammann, P.; Schedlowski, M.; Sure, U.; Thakur, B.K.; Hetze, S. Glioma: Molecular signature and crossroads with tumor microenvironment. Cancer Metastasis Rev. 2022, 41, 53–75. [Google Scholar] [CrossRef]
- Noemí García-Romero, J.C.-N.; Susana, E.-R.; Elisa, L.-I.; María, P.-C. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 2017, 8, 1416–1428. [Google Scholar] [CrossRef]
- de la Fuente, M.I. Targeting IDH1/IDH2 mutations in gliomas. Curr. Opin. Neurol. 2022, 35, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. Epidemiology and Overview of Gliomas. Semin. Oncol. Nurs. 2018, 34, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Śledzińska, P.; Bebyn, M.G.; Furtak, J.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Biomarkers in Gliomas. Int. J. Mol. Sci. 2021, 22, 10373. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Pfister, S.M.; Jones, D.T.W. Pediatric Gliomas: Current Concepts on Diagnosis, Biology, and Clinical Management. J. Clin. Oncol. 2017, 35, 2370–2377. [Google Scholar] [CrossRef] [PubMed]
- El Touny, L.H.; Hose, C.; Connelly, J.; Harris, E.; Monks, A.; Dull, A.B.; Wilsker, D.F.; Hollingshead, M.G.; Gottholm-Ahalt, M.; Alcoser, S.Y.; et al. ATR inhibition reverses the resistance of homologous recombination deficient MGMT(low)/MMR(proficient) cancer cells to temozolomide. Oncotarget 2021, 12, 2114–2130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Stevens, M.F.; Bradshaw, T.D. Temozolomide: Mechanisms of action, repair and resistance. Curr. Mol. Pharmacol. 2012, 5, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Herrlinger, U.; Tzaridis, T.; Mack, F.; Steinbach, J.P.; Schlegel, U.; Sabel, M.; Hau, P.; Kortmann, R.D.; Krex, D.; Grauer, O.; et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): A randomised, open-label, phase 3 trial. Lancet 2019, 393, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Schiff, D. Treatment considerations for MGMT-unmethylated glioblastoma. Curr. Neurol. Neurosci. Rep. 2015, 15, 507. [Google Scholar] [CrossRef]
- Oldrini, B.; Vaquero-Siguero, N.; Mu, Q.; Kroon, P.; Zhang, Y.; Galán-Ganga, M.; Bao, Z.; Wang, Z.; Liu, H.; Sa, J.K.; et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat. Commun. 2020, 11, 3883. [Google Scholar] [CrossRef]
- Salkeni, M.A.; Zarzour, A.; Ansay, T.Y.; McPherson, C.M.; Warnick, R.E.; Rixe, O.; Bahassi, E.M. Detection of EGFRvIII mutant DNA in the peripheral blood of brain tumor patients. J. Neuro-Oncol. 2013, 115, 27–35. [Google Scholar] [CrossRef]
- Rao, A.M.; Quddusi, A.; Shamim, M.S. The significance of MGMT methylation in Glioblastoma Multiforme prognosis. J. Pak. Med. Assoc. 2018, 68, 1137–1139. [Google Scholar] [PubMed]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hänggi, D.; Wick, W.; et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Guo, D. EGFR mutation: Novel prognostic factor associated with immune infiltration in lower-grade glioma; an exploratory study. BMC Cancer 2019, 19, 1184. [Google Scholar] [CrossRef] [PubMed]
- Oprita, A.; Baloi, S.C.; Staicu, G.A.; Alexandru, O.; Tache, D.E.; Danoiu, S.; Micu, E.S.; Sevastre, A.S. Updated Insights on EGFR Signaling Pathways in Glioma. Int. J. Mol. Sci. 2021, 22, 587. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.-W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J.M.; Skog, J.; Akers, J.; Li, H.; Komotar, R.; Jensen, R.; Ringel, F.; Yang, I.; Kalkanis, S.; Thompson, R.; et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro-Oncology 2017, 19, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Montermini, L.; Kim, D.-K.; Meehan, B.; Roth, F.P.; Rak, J. The Impact of Oncogenic EGFRvIII on the Proteome of Extracellular Vesicles Released from Glioblastoma Cells. Mol. Cell. Proteom. 2018, 17, 1948–1964. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Li, S.F.; Wang, J.L.; Zhang, T.; Zhang, S.; Chen, H.T.; Xiao, Q.Y.; Ren, W.H.; Liu, C.; Peng, B.; et al. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020, 40, 2834–2847. [Google Scholar] [CrossRef]
- Long, J.; Wang, A.; Bai, Y.; Lin, J.; Yang, X.; Wang, D.; Yang, X.; Jiang, Y.; Zhao, H. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine 2019, 42, 363–374. [Google Scholar] [CrossRef]
- Tümen, D.; Heumann, P.; Gülow, K.; Demirci, C.N.; Cosma, L.S.; Müller, M.; Kandulski, A. Pathogenesis and Current Treatment Strategies of Hepatocellular Carcinoma. Biomedicines 2022, 10, 3202. [Google Scholar] [CrossRef]
- Marchio, A.; Amougou Atsama, M.; Béré, A.; Komas, N.-P.; Noah Noah, D.; Atangana, P.J.A.; Camengo-Police, S.-M.; Njouom, R.; Bekondi, C.; Pineau, P. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin. Exp. Med. 2018, 18, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, J.; Li, E.; Xiao, Z.; Lei, J.; Zhou, F.; Yin, X.; Hu, D.; Mao, Y.; Wu, L.; et al. TP53 mutation detected in circulating exosomal DNA is associated with prognosis of patients with hepatocellular carcinoma. Cancer Biol. Ther. 2022, 23, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, R.; Wei, Q.; Xu, X. The Landscape of Alpha Fetoprotein in Hepatocellular Carcinoma: Where Are We? Int. J. Biol. Sci. 2022, 18, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Piñero, F.; Dirchwolf, M.; Pessôa, M.G. Biomarkers in Hepatocellular Carcinoma: Diagnosis, Prognosis and Treatment Response Assessment. Cells 2020, 9, 1370. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Ye, M.; Dong, K.; Dong, R. Circulating tumor DNA in neuroblastoma. Pediatr. Blood Cancer 2020, 67, e28311. [Google Scholar] [CrossRef] [PubMed]
- Trigg, R.M.; Turner, S.D. ALK in Neuroblastoma: Biological and Therapeutic Implications. Cancers 2018, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Kurywchak, P.; Wolf-Dennen, K.; Che, S.P.Y.; Sulakhe, D.; D’Souza, M.; Xie, B.; Maltsev, N.; Gilliam, T.C.; Wu, C.-C.; et al. Unique somatic variants in DNA from urine exosomes of individuals with bladder cancer. Mol. Ther.-Methods Clin. Dev. 2021, 22, 360–376. [Google Scholar] [CrossRef]
- Xing, J.; Reynolds, J.P. Diagnostic Advances in Urine Cytology. Surg Pathol. Clin. 2018, 11, 601–610. [Google Scholar] [CrossRef]
- Feiertag, N.; Barry, E.; Abramson, M.; Park, J.Y.; Kovac, E.; Aboumohamed, A.; Watts, K.; Sankin, A. Urine Cytology Rarely Escalates Clinical Management in the Surveillance of Non-muscle-Invasive Bladder Cancer. Clin. Genitourin Cancer 2023, 21, 258–264. [Google Scholar] [CrossRef]
- Halstead, A.M.; Kapadia, C.D.; Bolzenius, J.; Chu, C.E.; Schriefer, A.; Wartman, L.D.; Bowman, G.R.; Arora, V.K. Bladder-cancer-associated mutations in RXRA activate peroxisome proliferator-activated receptors to drive urothelial proliferation. Elife 2017, 6, e30862. [Google Scholar] [CrossRef]
- Siracusano, S.; Rizzetto, R.; Porcaro, A.B. Bladder cancer genomics. Urologia 2020, 87, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Matuszczak, M.; Kiljańczyk, A.; Salagierski, M. A Liquid Biopsy in Bladder Cancer—The Current Landscape in Urinary Biomarkers. Int. J. Mol. Sci. 2022, 23, 8597. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.; Stenzl, A.; Sharma, A.; Vasdev, N. Urinary biomarkers in bladder cancer: A review of the current landscape and future directions. Urol. Oncol. Semin. Orig. Investig. 2021, 39, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Tangvarasittichai, O.; Jaiwang, W.; Tangvarasittichai, S. The Plasma DNA Concentration as a Potential Breast Cancer Screening Marker. Indian J. Clin. Biochem. 2013, 30, 55–58. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 1–10. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.-M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- Okamura, R.; Piccioni, D.E.; Boichard, A.; Lee, S.; Jimenez, R.E.; Sicklick, J.K.; Kato, S.; Kurzrock, R. High prevalence of clonal hematopoiesis-type genomic abnormalities in cell-free DNA in invasive gliomas after treatment. Int. J. Cancer 2021, 148, 2839–2847. [Google Scholar] [CrossRef]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 1–4. [Google Scholar] [CrossRef]
- Tong, L.; Ding, N.; Tong, X.; Li, J.; Zhang, Y.; Wang, X.; Xu, X.; Ye, M.; Li, C.; Wu, X.; et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics 2019, 9, 5532–5541. [Google Scholar] [CrossRef]
| Tumors/Cell Line Types | Types of exoDNA | DNA Mutations/ Expression | Tissue Origins of exoDNA | Characteristics and Applications | Diagnostic/Treatment Roles | References |
|---|---|---|---|---|---|---|
| PCa | mtDNA | RCI mutations | plasma | circulating exosomes from PCa sera carried RCI-mtDNA and mt integrity associated proteins; co-localized with the mt in the PCa cells. | the pathogenic mtDNA mutation from RCI in circulating exosomes help early PCa detection, monitoring, and surveillance. | [24,36,37,38,39] |
| Three-genes | depression | urine | EPI score > 15.6 can identify > GG2 when PSA level of 2–10 ng/mL | Assist screening for PCa, reducing unnecessary biopsy and treatment | [38,40,41,42,43] | |
| ER+ breast cancer | mtDNA | copy number | plasma of patients, the conditioned media of cancer and stromal cell cultures | The transfer of ev-mtDNA serves as a carcinogenic signal leading to endocrine therapy resistance in OXPHOS-dependent breast cancer. | [44,45,46,47,48,49] | |
| Glioblastoma | mtDNA | copy number changes and point mutations | tissue and plasma | mtDNA copy number in brain tissue and EV was lower in the glioblastoma patient group compared to the control group | mtDNA copy number changes as a biomarker for Glioblastoma | [50,51,52,53,54,55,56,57,58,59,60] |
| Ovarian cancer | mtDNA | copy number changes | whole blood/plasma | mtDNA copy number | mtDNA copy number changes can be used as a signal of ovarian cancer progression, and wb-mtDNA copy number can provide information about the appearance of early serous ovarian cancer | [61,62,63,64,65,66] |
| Tumors/Cell Lines Types | Types of exoDNA | DNA Mutations/ Expression | Tissue Origins of exoDNA | Characteristics and Applications | Diagnostic/Treatment Roles | References |
|---|---|---|---|---|---|---|
| PDAC | Nuclear DNA | KRAS, P53 | plasma | Exosomes contain dsDNA, similar to the gDNA | Early stage cancer screening and detection purposes by identifying specific mutations | [69] |
| Nuclear DNA | KRAS | plasma | GPC1+ crExos | PDAC’s biomarker | [71] | |
| Nuclear DNA | KRAS MAF | plasma | Longitudinal continuous biopsy monitoring of exoDNA | Predict the outcomes of adjuvant therapy for localized disease and the progression of metastatic disease | [6,70,72,73,74,75] | |
| Glioma | exoDNA | concentration of exoDNA | plasma | concentration of plasma exoDNA is correlated with tumor volume and mitotic index | Detecting early recurrent high-grade gliomas and asymptomatic low-grade gliomas | [76,77,78,79] |
| Nuclear DNA | IDH1G395A, ERBB2, EGFR, CDK4, AKT3, MDM2, PTEN, etc. | peripheral blood | All three types of EVs secreted by human glioblastoma cells can traverse the intact BBB. EvDNA can be used to detect IDH1G395A | The standardized use of EVs as biomarkers for brain gliomas, less invasive method of detection compared to CSF analysis | [80,81,82,83,84] | |
| Nuclear DNA | MGMT | plasma | exoDNA is a biomarker for detecting MGMT fusions | Detecting MGMT promoter hypermethylation for acquired TMZ resistance in glioblastoma patients | [85,86,87,88,89] | |
| EGFR, EGFRvIII | peripheral blood, CSF | Peripheral blood exoDNA mutations can reflect the status of tumors | EGFRvIII in exosome as a biomarker for anti-EGFRvIII targeted therapies | [20,88,90,91,92,93,94,95,96,97] | ||
| HCC | Nuclear DNA | TP53(c.747G>T) | serum | Independent risk factor for HCC prognosis | Identify the poorer RFS, provide prognosis personalized treatment | [98,99,100,101,102,103,104] |
| NB | Nuclear DNA | ALK, CHD5, SHANK2, etc. | plasma | exoDNA carries specific gene mutations that are characteristic of NB and associated with acquired drug resistance | Biomarkers for NB diagnosis and personalized drug treatments | [33,105,106] |
| BC | Nuclear DNA | RXRA, TP53, FGFR3, 30 UTR variants | urine and serum | RXRA, TP53, FGFR3, 30 UTR variants, in the DNA of tumor tissue and urine exoDNA | Combined sequencing of tumor biopsy DNA and urine exoDNA can provide a better representation of the genetic heterogeneity of tumors | [107,108,109,110,111] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, Z.; Xie, Q.; Yu, Z. Exosomal DNA: Role in Reflecting Tumor Genetic Heterogeneity, Diagnosis, and Disease Monitoring. Cancers 2024, 16, 57. https://doi.org/10.3390/cancers16010057
Xiang Z, Xie Q, Yu Z. Exosomal DNA: Role in Reflecting Tumor Genetic Heterogeneity, Diagnosis, and Disease Monitoring. Cancers. 2024; 16(1):57. https://doi.org/10.3390/cancers16010057
Chicago/Turabian StyleXiang, Ziyi, Qihui Xie, and Zili Yu. 2024. "Exosomal DNA: Role in Reflecting Tumor Genetic Heterogeneity, Diagnosis, and Disease Monitoring" Cancers 16, no. 1: 57. https://doi.org/10.3390/cancers16010057
APA StyleXiang, Z., Xie, Q., & Yu, Z. (2024). Exosomal DNA: Role in Reflecting Tumor Genetic Heterogeneity, Diagnosis, and Disease Monitoring. Cancers, 16(1), 57. https://doi.org/10.3390/cancers16010057






