Multilevel Heterogeneity of Colorectal Cancer Liver Metastasis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Genetic Heterogeneity
2.1. Key Driver Genes
2.2. Chromosomal Instability (CIN)
2.3. Microsatellite Instability (MSI) Status
3. Transcriptomic Heterogeneity
3.1. MicroRNAs (miRNAs)
3.2. circRNAs
3.3. LncRNAs
3.4. Epigenetic Heterogeneity
4. Protein Heterogeneity
4.1. EMT-Related Proteins
4.2. Transcription Factors
4.3. Other Proteins
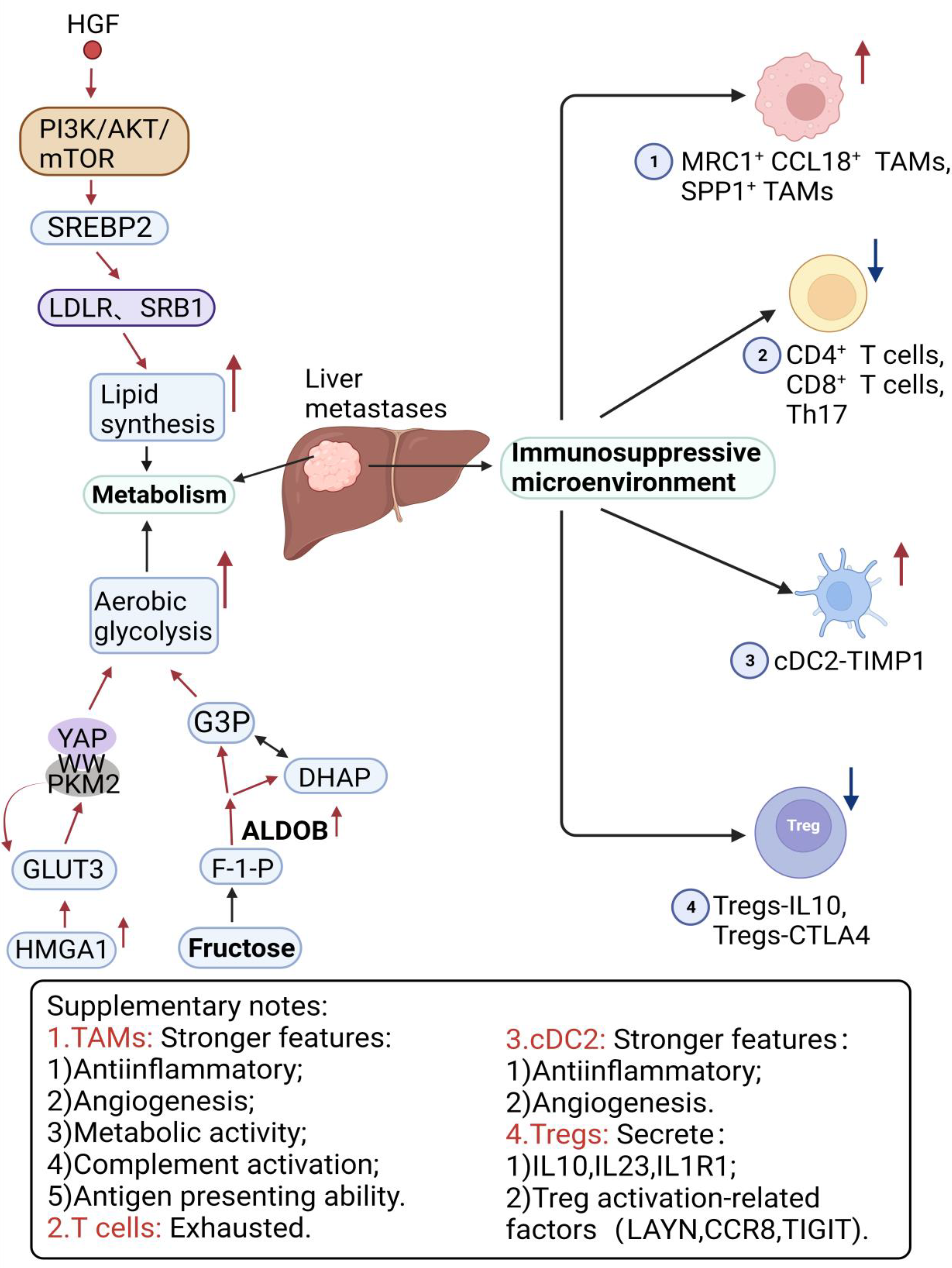
5. Metabolic Heterogeneity
6. Immune Heterogeneity
6.1. Macrophages
6.2. T Cells
6.3. Dendritic Cells
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Zhang, S.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer incidence and mortality in China, 2016. J. Natl. Cancer Cent. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G.S.; Barzi, A.; Jemal, A. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Van der Geest, L.G.M.; Lam-Boer, J.T.; Koopman, M.; Verhoef, C.; Elferink, M.A.G.; de Wilt, J.H.W. Nationwide trends in incidence, treatment and survival of colorectal cancer patients with synchronous metastases. Clin. Exp. Metastasis 2015, 32, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Brocks, D.; Gerhauser, C. Intratumor heterogeneity in epigenetic patterns. Semin. Cancer Biol. 2018, 51, 12–21. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, S.; Ma, J.; Chen, Z.; Song, G.; Rao, D.; Cheng, Y.; Huang, S.; Liu, Y.; Jiang, S.; et al. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov. 2022, 12, 134–153. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhu, L.; Liu, J.; Xie, M.; Chen, J.; Li, J. Emerging Role of Immunotherapy for Colorectal Cancer with Liver Metastasis. OncoTargets Ther. 2020, 13, 11645–11658. [Google Scholar] [CrossRef]
- Xu, J.; Fan, J.; Qin, X.; Cai, J.; Gu, J.; Wang, S.; Wang, X.; Zhang, S.; Zhang, Z. Chinese guidelines for the diagnosis and comprehensive treatment of colorectal liver metastases (version 2018). J. Cancer Res. Clin. Oncol. 2019, 145, 725–736. [Google Scholar] [CrossRef]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef]
- Gelsomino, F.; Barbolini, M.; Spallanzani, A.; Pugliese, G.; Cascinu, S. The evolving role of microsatellite instability in colorectal cancer: A review. Cancer Treat. Rev. 2016, 51, 19–26. [Google Scholar] [CrossRef]
- Willis, J.A.; Reyes-Uribe, L.; Chang, K.; Lipkin, S.M.; Vilar, E. Immune Activation in Mismatch Repair-Deficient Carcinogenesis: More Than Just Mutational Rate. Clin. Cancer Res. 2020, 26, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Tamiya, M.; Tamiya, A.; Inoue, T.; Kimura, M.; Kunimasa, K.; Nakahama, K.; Taniguchi, Y.; Shiroyama, T.; Isa, S.-I.; Nishino, K.; et al. Metastatic site as a predictor of nivolumab efficacy in patients with advanced non-small cell lung cancer: A retrospective multicenter trial. PLoS ONE 2018, 13, e0192227. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Guo, H.; Cao, C.; Li, X.; Hu, B.; Zhu, P.; Wu, X.; Wen, L.; Tang, F.; Huang, Y.; et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016, 26, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Van der Jeught, K.; Xu, H.-C.; Li, Y.-J.; Lu, X.-B.; Ji, G. Drug resistance and new therapies in colorectal cancer. World J. Gastroenterol. 2018, 24, 3834–3848. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef] [PubMed]
- Sagaert, X.; Vanstapel, A.; Verbeek, S. Tumor Heterogeneity in Colorectal Cancer: What Do We Know So Far? Pathobiology 2018, 85, 72–84. [Google Scholar] [CrossRef]
- Koulouridi, A.; Karagianni, M.; Messaritakis, I.; Sfakianaki, M.; Voutsina, A.; Trypaki, M.; Bachlitzanaki, M.; Koustas, E.; Karamouzis, M.V.; Ntavatzikos, A.; et al. Prognostic Value of Mutations in Colorectal Cancer Patients. Cancers 2022, 14, 3320. [Google Scholar] [CrossRef]
- Santos, C.; Azuara, D.; Viéitez, J.M.; Páez, D.; Falcó, E.; Élez, E.; López-López, C.; Valladares, M.; Robles-Díaz, L.; García-Alfonso, P.; et al. Phase II study of high-sensitivity genotyping of KRAS, NRAS, BRAF and PIK3CA to ultra-select metastatic colorectal cancer patients for panitumumab plus FOLFIRI: The ULTRA trial. Ann. Oncol. 2019, 30, 796–803. [Google Scholar] [CrossRef]
- Di Nicolantonio, F.; Martini, M.; Molinari, F.; Sartore-Bianchi, A.; Arena, S.; Saletti, P.; De Dosso, S.; Mazzucchelli, L.; Frattini, M.; Siena, S.; et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 5705–5712. [Google Scholar] [CrossRef]
- Brunsell, T.H.; Sveen, A.; Bjørnbeth, B.A.; Røsok, B.I.; Danielsen, S.A.; Brudvik, K.W.; Berg, K.C.G.; Johannessen, B.; Cengija, V.; Abildgaard, A.; et al. High Concordance and Negative Prognostic Impact of RAS/BRAF/PIK3CA Mutations in Multiple Resected Colorectal Liver Metastases. Clin. Color. Cancer 2020, 19, e26–e47. [Google Scholar] [CrossRef] [PubMed]
- Baldus, S.E.; Schaefer, K.-L.; Engers, R.; Hartleb, D.; Stoecklein, N.H.; Gabbert, H.E. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin. Cancer Res. 2010, 16, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Knijn, N.; Mekenkamp, L.J.M.; Klomp, M.; Vink-Börger, M.E.; Tol, J.; Teerenstra, S.; Meijer, J.W.R.; Tebar, M.; Riemersma, S.; van Krieken, J.H.J.M.; et al. KRAS mutation analysis: A comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br. J. Cancer 2011, 104, 1020–1026. [Google Scholar] [CrossRef] [PubMed]
- Brannon, A.R.; Vakiani, E.; Sylvester, B.E.; Scott, S.N.; McDermott, G.; Shah, R.H.; Kania, K.; Viale, A.; Oschwald, D.M.; Vacic, V.; et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014, 15, 454. [Google Scholar] [CrossRef] [PubMed]
- Vakiani, E.; Janakiraman, M.; Shen, R.; Sinha, R.; Zeng, Z.; Shia, J.; Cercek, A.; Kemeny, N.; D’Angelica, M.; Viale, A.; et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J. Clin. Oncol. 2012, 30, 2956–2962. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhang, Y.; Zhu, Z. Gene heterogeneity in metastasis of colorectal cancer to the lung. Semin. Cell Dev. Biol. 2017, 64, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Lipton, L.; Desai, J.; Gibbs, P.; Jorissen, R.N.; Christie, M.; Drummond, K.J.; Thomson, B.N.J.; Usatoff, V.; Evans, P.M.; et al. KRAS mutation is associated with lung metastasis in patients with curatively resected colorectal cancer. Clin. Cancer Res. 2011, 17, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- David, C.J.; Huang, Y.-H.; Chen, M.; Su, J.; Zou, Y.; Bardeesy, N.; Iacobuzio-Donahue, C.A.; Massagué, J. TGF-β Tumor Suppression through a Lethal EMT. Cell 2016, 164, 1015–1030. [Google Scholar] [CrossRef]
- Itatani, Y.; Kawada, K.; Fujishita, T.; Kakizaki, F.; Hirai, H.; Matsumoto, T.; Iwamoto, M.; Inamoto, S.; Hatano, E.; Hasegawa, S.; et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology 2013, 145, 1064–1075. [Google Scholar] [CrossRef]
- Wang, H.-W.; Yan, X.-L.; Wang, L.-J.; Zhang, M.-H.; Yang, C.-H.; Wei, L.; Jin, K.-M.; Bao, Q.; Li, J.; Wang, K.; et al. Characterization of genomic alterations in Chinese colorectal cancer patients with liver metastases. J. Transl. Med. 2021, 19, 313. [Google Scholar] [CrossRef]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150. [Google Scholar] [CrossRef]
- Vargas-Rondón, N.; Villegas, V.E.; Rondón-Lagos, M. The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers 2017, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.-A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Løes, I.M.; Alagaratnam, S.; Nilsen, G.; Høland, M.; Lingjærde, O.C.; Sorbye, H.; Berg, K.C.G.; Horn, A.; Angelsen, J.-H.; et al. Intra-patient Inter-metastatic Genetic Heterogeneity in Colorectal Cancer as a Key Determinant of Survival after Curative Liver Resection. PLoS Genet. 2016, 12, e1006225. [Google Scholar] [CrossRef] [PubMed]
- Gambaro, K.; Marques, M.; McNamara, S.; Couetoux du Tertre, M.; Diaz, Z.; Hoffert, C.; Srivastava, A.; Hebert, S.; Samson, B.; Lesperance, B.; et al. Copy number and transcriptome alterations associated with metastatic lesion response to treatment in colorectal cancer. Clin. Transl. Med. 2021, 11, e401. [Google Scholar] [CrossRef]
- Mogensen, M.B.; Rossing, M.; Østrup, O.; Larsen, P.N.; Heiberg Engel, P.J.; Jørgensen, L.N.; Hogdall, E.V.; Eriksen, J.; Ibsen, P.; Jess, P.; et al. Genomic alterations accompanying tumour evolution in colorectal cancer: Tracking the differences between primary tumours and synchronous liver metastases by whole-exome sequencing. BMC Cancer 2018, 18, 752. [Google Scholar] [CrossRef] [PubMed]
- Mamlouk, S.; Childs, L.H.; Aust, D.; Heim, D.; Melching, F.; Oliveira, C.; Wolf, T.; Durek, P.; Schumacher, D.; Bläker, H.; et al. DNA copy number changes define spatial patterns of heterogeneity in colorectal cancer. Nat. Commun. 2017, 8, 14093. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Saad, R.S.; Ghorab, Z.; Khalifa, M.A.; Xu, M. CDX2 as a marker for intestinal differentiation: Its utility and limitations. World J. Gastrointest. Surg. 2011, 3, 159–166. [Google Scholar] [CrossRef]
- Mekenkamp, L.J.M.; Haan, J.C.; Israeli, D.; van Essen, H.F.B.; Dijkstra, J.R.; van Cleef, P.; Punt, C.J.A.; Meijer, G.A.; Nagtegaal, I.D.; Ylstra, B. Chromosomal copy number aberrations in colorectal metastases resemble their primary counterparts and differences are typically non-recurrent. PLoS ONE 2014, 9, e86833. [Google Scholar] [CrossRef]
- Yamamoto, S.; Midorikawa, Y.; Morikawa, T.; Nishimura, Y.; Sakamoto, H.; Ishikawa, S.; Akagi, K.; Aburatani, H. Identification of chromosomal aberrations of metastatic potential in colorectal carcinoma. Genes Chromosomes Cancer 2010, 49, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Chen, W.-D.; Parmigiani, G.; Diehl, F.; Beerenwinkel, N.; Antal, T.; Traulsen, A.; Nowak, M.A.; Siegel, C.; Velculescu, V.E.; et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 4283–4288. [Google Scholar] [CrossRef] [PubMed]
- He, W.-Z.; Hu, W.-M.; Wang, F.; Rong, Y.-M.; Yang, L.; Xie, Q.-K.; Yang, Y.-Z.; Jiang, C.; Qiu, H.-J.; Lu, J.-B.; et al. Comparison of Mismatch Repair Status Between Primary and Matched Metastatic Sites in Patients with Colorectal Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 1174–1183. [Google Scholar] [CrossRef]
- Evrard, C.; Messina, S.; Sefrioui, D.; Frouin, É.; Auriault, M.-L.; Chautard, R.; Zaanan, A.; Jaffrelot, M.; De La Fouchardière, C.; Aparicio, T.; et al. Heterogeneity of Mismatch Repair Status and Microsatellite Instability between Primary Tumour and Metastasis and Its Implications for Immunotherapy in Colorectal Cancers. Int. J. Mol. Sci. 2022, 23, 4427. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Kang, Y.; Lee, Y.J.; Kim, E.; Ahn, B.; Lee, E.; Kim, J.Y.; Lee, J.H.; Lee, Y.; Kim, C.H.; et al. Comparison of the Mismatch Repair System between Primary and Metastatic Colorectal Cancers Using Immunohistochemistry. J. Pathol. Transl. Med. 2017, 51, 129–136. [Google Scholar] [CrossRef]
- Haraldsdottir, S.; Roth, R.; Pearlman, R.; Hampel, H.; Arnold, C.A.; Frankel, W.L. Mismatch repair deficiency concordance between primary colorectal cancer and corresponding metastasis. Fam. Cancer 2016, 15, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Imaoka, H.; Toiyama, Y.; Fujikawa, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; Kato, T.; et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. 2016, 27, 1879–1886. [Google Scholar] [CrossRef]
- Nassar, F.J.; Msheik, Z.S.; Itani, M.M.; Helou, R.E.; Hadla, R.; Kreidieh, F.; Bejjany, R.; Mukherji, D.; Shamseddine, A.; Nasr, R.R.; et al. Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis. Diagnostics 2021, 11, 341. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Pesta, M.; Radova, L.; Liska, V.; Daum, O.; Kala, Z.; Svoboda, M.; Kiss, I.; Slaby, O. Genome-wide microRNA Expression Profiling in Primary Tumors and Matched Liver Metastasis of Patients with Colorectal Cancer. Cancer Genom. Proteom. 2016, 13, 311–316. [Google Scholar]
- Hur, K.; Toiyama, Y.; Schetter, A.J.; Okugawa, Y.; Harris, C.C.; Boland, C.R.; Goel, A. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J. Natl. Cancer Inst. 2015, 107, dju492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Guo, J.; Gu, J.; Wang, Z.; Wang, G.; Li, H.; Wang, J. Identifying the key genes and microRNAs in colorectal cancer liver metastasis by bioinformatics analysis and in vitro experiments. Oncol. Rep. 2019, 41, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-C.; Hsu, P.W.-C.; Lai, T.-C.; Chau, G.-Y.; Lin, C.-W.; Chen, C.-M.; Lin, C.-D.; Liao, Y.-L.; Wang, J.-L.; Chau, Y.-P.; et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 2009, 49, 1571–1582. [Google Scholar] [CrossRef] [PubMed]
- Thakral, S.; Ghoshal, K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene Ther. 2015, 15, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.S.-C.; Ng, L.; Chow, A.K.-M.; Wan, T.M.-H.; Yau, S.; Cheng, N.S.-M.; Wong, S.K.-M.; Man, J.H.-W.; Lo, O.S.-H.; Foo, D.C.-C.; et al. Identification of microRNA 885-5p as a novel regulator of tumor metastasis by targeting CPEB2 in colorectal cancer. Oncotarget 2017, 8, 26858–26870. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Qin, B.; Liu, F.; Chen, Y.; Zhang, R. miR-885-5p upregulation promotes colorectal cancer cell proliferation and migration by targeting suppressor of cytokine signaling. Oncol. Lett. 2018, 16, 65–72. [Google Scholar] [CrossRef]
- Pizzini, S.; Bisognin, A.; Mandruzzato, S.; Biasiolo, M.; Facciolli, A.; Perilli, L.; Rossi, E.; Esposito, G.; Rugge, M.; Pilati, P.; et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genom. 2013, 14, 589. [Google Scholar] [CrossRef]
- Song, J.J.; Li, W. MiR-10b suppresses the growth and metastasis of colorectal cancer cell by targeting FGF13. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 576–587. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Okugawa, Y.; Ide, S.; Imaoka, H.; Boland, C.R.; Goel, A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017, 66, 654–665. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Takahashi, M.; Balaguer, F.; Nagasaka, T.; Koike, J.; Hemmi, H.; Koi, M.; Boland, C.R.; Goel, A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013, 62, 1315–1326. [Google Scholar] [CrossRef]
- Diao, Y.; Guo, X.; Jiang, L.; Wang, G.; Zhang, C.; Wan, J.; Jin, Y.; Wu, Z. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in rhabdomyosarcoma. J. Biol. Chem. 2014, 289, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Jiang, N.; Zhang, G.-X.; Sun, Q. MiR-203 inhibits tumor invasion and metastasis in gastric cancer by ATM. Acta Biochim. Biophys. Sin. 2016, 48, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, B.; Cui, J.; Liang, Z.; Liu, K. miR-203 Inhibits the Invasion and EMT of Gastric Cancer Cells by Directly Targeting Annexin A4. Oncol. Res. 2019, 27, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Torres, S.; Garcia-Palmero, I.; Bartolomé, R.A.; Fernandez-Aceñero, M.J.; Molina, E.; Calviño, E.; Segura, M.F.; Casal, J.I. Combined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasis. J. Pathol. 2017, 242, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.; Baird, A.-M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon Skipping Is Correlated with Exon Circularization. J. Mol. Biol. 2015, 427, 2414–2417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-O.; Wang, H.-B.; Zhang, Y.; Lu, X.; Chen, L.-L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Guo, Y.; Chen, C.; Fan, D.; Wu, X.; Zhao, L.; Shao, B.; Sun, Z.; Ji, Z. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: Involvement of miR-30c-5p/TCF7 axis. Mol. Cancer 2021, 20, 93. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Huang, Z.; Mo, X.; Song, Y.; Li, X.; Li, X.; Zhang, M. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J. Exp. Clin. Cancer Res. 2020, 39, 91. [Google Scholar] [CrossRef]
- Long, F.; Lin, Z.; Li, L.; Ma, M.; Lu, Z.; Jing, L.; Li, X.; Lin, C. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol. Cancer 2021, 20, 26. [Google Scholar] [CrossRef]
- Xu, H.; Wang, C.; Song, H.; Xu, Y.; Ji, G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol. Cancer 2019, 18, 8. [Google Scholar] [CrossRef]
- Chen, R.-X.; Chen, X.; Xia, L.-P.; Zhang, J.-X.; Pan, Z.-Z.; Ma, X.-D.; Han, K.; Chen, J.-W.; Judde, J.-G.; Deas, O.; et al. N-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, X.; Geng, X.; Zhang, Y.; Wang, J.; Wang, Y.; Jing, J.; Zhou, X.; Pan, W. Circular RNA hsa_circ_0006401 promotes proliferation and metastasis in colorectal carcinoma. Cell Death Dis. 2021, 12, 443. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, Z.; Xu, C.; Zhou, Z.; Zhu, Z.; You, T. HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Lett. 2014, 355, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Nwakeze, C.L.; Horta, A.; Rajkumar, S.M.; Coffey, E.L.; Duffy, E.E.; Duffié, R.; Monahan, K.; O’Keeffe, S.; Simon, M.D.; et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 2019, 177, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Guan, B.; Mi, Y.; Shi, D.; Wei, P.; Gu, Y.; Cai, S.; Xu, Y.; Li, X.; Yan, D.; et al. LncRNA MIR17HG promotes colorectal cancer liver metastasis by mediating a glycolysis-associated positive feedback circuit. Oncogene 2021, 40, 4709–4724. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Kang, H.; Xiao, Y.; Du, Y.; Xiao, Y.; Song, G.; Zhang, Y.; Guo, Y.; Yang, F.; He, F.; et al. LncRNA GAL promotes colorectal cancer liver metastasis through stabilizing GLUT1. Oncogene 2022, 41, 1882–1894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S.; Kang, W.; Liu, C.; Dong, Y.; Ren, F.; Wang, Y.; Zhang, J.; Wang, G.; To, K.F.; et al. CREPT facilitates colorectal cancer growth through inducing Wnt/β-catenin pathway by enhancing p300-mediated β-catenin acetylation. Oncogene 2018, 37, 3485–3500. [Google Scholar] [CrossRef]
- Li, L.; Liu, J.; Xue, H.; Li, C.; Liu, Q.; Zhou, Y.; Wang, T.; Wang, H.; Qian, H.; Wen, T. A TGF-β-MTA1-SOX4-EZH2 signaling axis drives epithelial-mesenchymal transition in tumor metastasis. Oncogene 2020, 39, 2125–2139. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Zhao, N.; Yang, X.; Li, Y.; Zhai, F.; Zang, X.; Cui, W. Checkpoint regulator B7x is epigenetically regulated by HDAC3 and mediates resistance to HDAC inhibitors by reprogramming the tumor immune environment in colorectal cancer. Cell Death Dis. 2020, 11, 753. [Google Scholar] [CrossRef]
- Chen, X.; Xu, M.; Xu, X.; Zeng, K.; Liu, X.; Pan, B.; Li, C.; Sun, L.; Qin, J.; Xu, T.; et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol. Cancer 2020, 19, 106. [Google Scholar] [CrossRef] [PubMed]
- Teng, S.; Li, Y.E.; Yang, M.; Qi, R.; Huang, Y.; Wang, Q.; Zhang, Y.; Chen, S.; Li, S.; Lin, K.; et al. Tissue-specific transcription reprogramming promotes liver metastasis of colorectal cancer. Cell Res. 2020, 30, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Miranda, E.; Bianchi, P.; Destro, A.; Morenghi, E.; Malesci, A.; Santoro, A.; Laghi, L.; Roncalli, M. Genetic and epigenetic alterations in primary colorectal cancers and related lymph node and liver metastases. Cancer 2013, 119, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Hannon, G.J.; Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, P.-S.; Zuo, Z.; Lin, J.-F.; Li, X.; Wu, Q.-N.; Chen, Z.-H.; Zeng, Z.-L.; Wang, F.; Zheng, J.; et al. METTL3 facilitates tumor progression via an mA-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer 2019, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Wang, B.; Luo, J.; Jiang, K.; Zhang, H.; Mustonen, H.; Puolakkainen, P.; Zhu, J.; Ye, Y.; Wang, S. Global-scale profiling of differential expressed lysine acetylated proteins in colorectal cancer tumors and paired liver metastases. J. Proteom. 2016, 142, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Ye, Y.; Yang, X.; Liu, B.; Wang, Z.; Chen, S.; Jiang, K.; Zhang, W.; Jiang, H.; Mustonen, H.; et al. SIRT2-dependent IDH1 deacetylation inhibits colorectal cancer and liver metastases. EMBO Rep. 2020, 21, e48183. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, Y.; Guo, S.; Jin, H.; Wang, W.; Yang, P. Large scale systematic proteomic quantification from non-metastatic to metastatic colorectal cancer. Sci. Rep. 2015, 5, 12120. [Google Scholar] [CrossRef]
- Liu, X.; Xu, D.; Liu, Z.; Li, Y.; Zhang, C.; Gong, Y.; Jiang, Y.; Xing, B. THBS1 facilitates colorectal liver metastasis through enhancing epithelial-mesenchymal transition. Clin. Transl. Oncol. 2020, 22, 1730–1740. [Google Scholar] [CrossRef]
- Bae, Y.K.; Kim, A.; Kim, M.K.; Choi, J.E.; Kang, S.H.; Lee, S.J. Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum. Pathol. 2013, 44, 2028–2037. [Google Scholar] [CrossRef] [PubMed]
- Hope, C.; Emmerich, P.B.; Papadas, A.; Pagenkopf, A.; Matkowskyj, K.A.; Van De Hey, D.R.; Payne, S.N.; Clipson, L.; Callander, N.S.; Hematti, P.; et al. Versican-Derived Matrikines Regulate Batf3-Dendritic Cell Differentiation and Promote T Cell Infiltration in Colorectal Cancer. J. Immunol. 2017, 199, 1933–1941. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Xu, S.; Zhang, H.; Wang, Y.; Xiao, C.; Jiang, T.; Wu, L.; Zhang, T.; Sun, X.; Zhong, L.; et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J. Exp. Clin. Cancer Res. 2016, 35, 148. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, X.; Yang, Y.; Yang, S.; Dong, Z.; Du, L.; Wang, L.; Wang, C. Periostin expression and its prognostic value for colorectal cancer. Int. J. Mol. Sci. 2015, 16, 12108–12118. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wu, W.K.K.; Li, X.; He, J.; Li, X.-X.; Ng, S.S.M.; Yu, C.; Gao, Z.; Yang, J.; Li, M.; et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 2015, 64, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xi, Y.; Zhu, G.; Jia, J.; Huang, H.; Liu, Y.; Guo, Y.; Liu, L. Downregulated IGFBP7 facilitates liver metastasis by modulating epithelial-mesenchymal transition in colon cancer. Oncol. Rep. 2019, 42, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, M.B.; Dhawan, P.; Baumert, T.F. Tight junction proteins in gastrointestinal and liver disease. Gut 2019, 68, 547–561. [Google Scholar] [CrossRef]
- Wang, K.; Li, T.; Xu, C.; Ding, Y.; Li, W.; Ding, L. Claudin-7 downregulation induces metastasis and invasion in colorectal cancer via the promotion of epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2019, 508, 797–804. [Google Scholar] [CrossRef]
- Georges, R.; Bergmann, F.; Hamdi, H.; Zepp, M.; Eyol, E.; Hielscher, T.; Berger, M.R.; Adwan, H. Sequential biphasic changes in claudin1 and claudin4 expression are correlated to colorectal cancer progression and liver metastasis. J. Cell. Mol. Med. 2012, 16, 260–272. [Google Scholar] [CrossRef]
- Tian, T.; Chen, Z.-H.; Zheng, Z.; Liu, Y.; Zhao, Q.; Liu, Y.; Qiu, H.; Long, Q.; Chen, M.; Li, L.; et al. Investigation of the role and mechanism of ARHGAP5-mediated colorectal cancer metastasis. Theranostics 2020, 10, 5998–6010. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Qi, X.; Liu, J.; Zhou, R.; Lin, C.; Shangguan, J.; Zhang, Z.; Zhao, L.; Li, G. MYH9 Promotes Growth and Metastasis via Activation of MAPK/AKT Signaling in Colorectal Cancer. J. Cancer 2019, 10, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lai, Q.; He, C.; Fang, Y.; Yan, Q.; Zhang, Y.; Wang, X.; Gu, C.; Wang, Y.; Ye, L.; et al. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. J. Exp. Clin. Cancer Res. 2019, 38, 334. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Liang, W.; Liao, Y.; Li, Z.; Wang, Y.; Yan, C. PEA15 promotes liver metastasis of colorectal cancer by upregulating the ERK/MAPK signaling pathway. Oncol. Rep. 2019, 41, 43–56. [Google Scholar] [CrossRef]
- Cotter, K.; Capecci, J.; Sennoune, S.; Huss, M.; Maier, M.; Martinez-Zaguilan, R.; Forgac, M. Activity of plasma membrane V-ATPases is critical for the invasion of MDA-MB231 breast cancer cells. J. Biol. Chem. 2015, 290, 3680–3692. [Google Scholar] [CrossRef]
- Wang, J.; Chen, D.; Song, W.; Liu, Z.; Ma, W.; Li, X.; Zhang, C.; Wang, X.; Wang, Y.; Yang, Y.; et al. ATP6L promotes metastasis of colorectal cancer by inducing epithelial-mesenchymal transition. Cancer Sci. 2020, 111, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Ku, X.; Xu, Y.; Cai, C.; Yang, Y.; Cui, L.; Yan, W. In-Depth Characterization of Mass Spectrometry-Based Proteomic Profiles Revealed Novel Signature Proteins Associated with Liver Metastatic Colorectal Cancers. Anal. Cell. Pathol. 2019, 2019, 7653230. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Bushweller, J.H. Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef]
- Wang, X.; Lin, Q.; Lv, F.; Liu, N.; Xu, Y.; Liu, M.; Chen, Y.; Yi, Z. LG-362B targets PML-RARα and blocks ATRA resistance of acute promyelocytic leukemia. Leukemia 2016, 30, 1465–1474. [Google Scholar] [CrossRef]
- Wu, S.; Wang, H.; Li, Y.; Xie, Y.; Huang, C.; Zhao, H.; Miyagishi, M.; Kasim, V. Transcription Factor YY1 Promotes Cell Proliferation by Directly Activating the Pentose Phosphate Pathway. Cancer Res. 2018, 78, 4549–4562. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, F.; Zhu, X.; Guo, W.; Fu, T.; Wang, W. Death Domain-Associated Protein Promotes Colon Cancer Metastasis through Direct Interaction with ZEB1. J. Cancer 2020, 11, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Kamal, Y.; Schmit, S.L.; Hoehn, H.J.; Amos, C.I.; Frost, H.R. Transcriptomic Differences between Primary Colorectal Adenocarcinomas and Distant Metastases Reveal Metastatic Colorectal Cancer Subtypes. Cancer Res. 2019, 79, 4227–4241. [Google Scholar] [CrossRef] [PubMed]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 2015, 5, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-K.; Song, M.-J.; Jung, Y.; Lee, W.-S.; Jang, H.H. Proteomic Analysis of Primary Colon Cancer and Synchronous Solitary Liver Metastasis. Cancer Genom. Proteom. 2019, 16, 583–592. [Google Scholar] [CrossRef]
- Kwon, C.H.; Park, H.J.; Lee, J.R.; Kim, H.K.; Jeon, T.Y.; Jo, H.J.; Kim, D.H.; Kim, G.H.; Park, D.Y. Serpin peptidase inhibitor clade A member 1 is a biomarker of poor prognosis in gastric cancer. Br. J. Cancer 2014, 111, 1993–2002. [Google Scholar] [CrossRef]
- Ercetin, E.; Richtmann, S.; Delgado, B.M.; Gomez-Mariano, G.; Wrenger, S.; Korenbaum, E.; Liu, B.; DeLuca, D.; Kühnel, M.P.; Jonigk, D.; et al. Clinical Significance of SERPINA1 Gene and Its Encoded Alpha1-antitrypsin Protein in NSCLC. Cancers 2019, 11, 1306. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.H.; Jung, W.-H.; Koo, J.S. Succinate dehydrogenase expression in breast cancer. Springerplus 2013, 2, 299. [Google Scholar] [CrossRef]
- Ricketts, C.; Woodward, E.R.; Killick, P.; Morris, M.R.; Astuti, D.; Latif, F.; Maher, E.R. Germline SDHB mutations and familial renal cell carcinoma. J. Natl. Cancer Inst. 2008, 100, 1260–1262. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Kuo, C.-C.; Ling, H.-H.; Chiang, M.-C.; Chung, C.-H.; Lee, W.-Y.; Chu, C.-Y.; Wu, Y.-C.; Chen, C.-H.; Lai, Y.-W.; Tsai, I.L.; et al. Metastatic Colorectal Cancer Rewrites Metabolic Program Through a Glut3-YAP-dependent Signaling Circuit. Theranostics 2019, 9, 2526–2540. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.-H.; Chang, W.-W.; Yeh, B.-W.; Chu, Y.-S.; Lee, Y.-C.; Lee, H.-T. Glucose Transporter 3 is Essential for the Survival of Breast Cancer Cells in the Brain. Cells 2019, 8, 1568. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Hao, Y.; Zhou, X.; Li, H.; Liu, F.; Zhu, H.; Song, X.; Niu, Z.; Ni, Q.; Chen, M.-S.; et al. Prognostic value of glucose transporter 3 expression in hepatocellular carcinoma. Oncol. Lett. 2020, 19, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Levantini, E.; Fhu, C.W.; Teo, J.T.; Clohessy, J.G.; Goggi, J.L.; Wu, C.-S.; Chen, L.; Chin, T.M.; Tenen, D.G. CAV1–GLUT3 signaling is important for cellular energy and can be targeted by Atorvastatin in Non-Small Cell Lung Cancer. Theranostics 2019, 9, 6157–6174. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Guo, Y.; Liu, X.; Liu, N. HMGA1 Promotes Hepatic Metastasis of Colorectal Cancer by Inducing Expression of Glucose Transporter 3 (GLUT3). Med. Sci. Monit. 2020, 26, e924975. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-J.; Hsieh, Y.-J.; Cheng, W.-C.; Lin, C.-P.; Lin, Y.-s.; Yang, S.-F.; Chen, C.-C.; Izumiya, Y.; Yu, J.-S.; Kung, H.-J.; et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α-mediated glucose metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Zhou, R.; Lin, C.; Yang, S.; Wang, H.; Li, W.; Zheng, K.; Lin, W.; Li, X.; Yao, X.; et al. Tumor-secreted dickkopf2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics 2019, 9, 1001–1014. [Google Scholar] [CrossRef]
- Fahrner, M.; Bronsert, P.; Fichtner-Feigl, S.; Jud, A.; Schilling, O. Proteome biology of primary colorectal carcinoma and corresponding liver metastases. Neoplasia 2021, 23, 1240–1251. [Google Scholar] [CrossRef]
- Bu, P.; Chen, K.-Y.; Xiang, K.; Johnson, C.; Crown, S.B.; Rakhilin, N.; Ai, Y.; Wang, L.; Xi, R.; Astapova, I.; et al. Aldolase B-Mediated Fructose Metabolism Drives Metabolic Reprogramming of Colon Cancer Liver Metastasis. Cell Metab. 2018, 27, 1249–1262. [Google Scholar] [CrossRef]
- Zhang, K.-L.; Zhu, W.-W.; Wang, S.-H.; Gao, C.; Pan, J.-J.; Du, Z.-G.; Lu, L.; Jia, H.-L.; Dong, Q.-Z.; Chen, J.-H.; et al. Organ-specific cholesterol metabolic aberration fuels liver metastasis of colorectal cancer. Theranostics 2021, 11, 6560–6572. [Google Scholar] [CrossRef]
- Williams, M.D.; Zhang, X.; Park, J.-J.; Siems, W.F.; Gang, D.R.; Resar, L.M.S.; Reeves, R.; Hill, H.H. Characterizing metabolic changes in human colorectal cancer. Anal. Bioanal. Chem. 2015, 407, 4581–4595. [Google Scholar] [CrossRef] [PubMed]
- Halama, N.; Michel, S.; Kloor, M.; Zoernig, I.; Benner, A.; Spille, A.; Pommerencke, T.; von Knebel, D.M.; Folprecht, G.; Luber, B.; et al. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. 2011, 71, 5670–5677. [Google Scholar] [CrossRef] [PubMed]
- Wculek, S.K.; Malanchi, I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 2015, 528, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Shema, E.; Loi, S.; Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021, 27, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Sun, H.; Zhang, Y.; Wang, Z.; Xun, Z.; Li, Z.; Ding, X.; Bao, R.; Hong, L.; Jia, W.; et al. Single-cell and spatial analysis reveal interaction of FAP fibroblasts and SPP1 macrophages in colorectal cancer. Nat. Commun. 2022, 13, 1742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, J.; Zhao, Z.; Yang, M.; Chen, M.; Liu, C.; Ji, J.; Zhu, D. Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett. 2020, 470, 84–94. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Q.; Xing, B.; Luo, N.; Gao, R.; Yu, K.; Hu, X.; Bu, Z.; Peng, J.; Ren, X.; et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell 2022, 40, 424–437. [Google Scholar] [CrossRef]
- Zilionis, R.; Engblom, C.; Pfirschke, C.; Savova, V.; Zemmour, D.; Saatcioglu, H.D.; Krishnan, I.; Maroni, G.; Meyerovitz, C.V.; Kerwin, C.M.; et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity 2019, 50, 1317–1334. [Google Scholar] [CrossRef]
- Tu, W.; Gong, J.; Zhou, Z.; Tian, D.; Wang, Z. TCF4 enhances hepatic metastasis of colorectal cancer by regulating tumor-associated macrophage via CCL2/CCR2 signaling. Cell Death Dis. 2021, 12, 882. [Google Scholar] [CrossRef]
- Ballotta, V.; Driessen-Mol, A.; Bouten, C.V.C.; Baaijens, F.P.T. Strain-dependent modulation of macrophage polarization within scaffolds. Biomaterials 2014, 35, 4919–4928. [Google Scholar] [CrossRef]
- Waldo, S.W.; Li, Y.; Buono, C.; Zhao, B.; Billings, E.M.; Chang, J.; Kruth, H.S. Heterogeneity of human macrophages in culture and in atherosclerotic plaques. Am. J. Pathol. 2008, 172, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Gordon, S.; Hume, D.A.; Mowat, A.M.; Randolph, G.J. Unravelling mononuclear phagocyte heterogeneity. Nat. Rev. Immunol. 2010, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Yona, S.; Gordon, S. From the Reticuloendothelial to Mononuclear Phagocyte System—The Unaccounted Years. Front. Immunol. 2015, 6, 328. [Google Scholar] [CrossRef]
- Donadon, M.; Torzilli, G.; Cortese, N.; Soldani, C.; Di Tommaso, L.; Franceschini, B.; Carriero, R.; Barbagallo, M.; Rigamonti, A.; Anselmo, A.; et al. Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis. J. Exp. Med. 2020, 217, e20191847. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Nishikawa, H. Roles of regulatory T cells in cancer immunity. Int. Immunol. 2016, 28, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zou, Y.; Lian, L.; Wu, X.; He, X.; He, X.; Wu, X.; Huang, Y.; Lan, P. Changes of T cells and cytokines TGF-β1 and IL-10 in mice during liver metastasis of colon carcinoma: Implications for liver anti-tumor immunity. J. Gastrointest. Surg. 2013, 17, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, Z.; Zhang, N.; Zhu, C.; Lin, X.; Yu, J.; Chen, Z.; Lan, P.; Wan, Y. Increase in CD4FOXP3 regulatory T cell number and upregulation of the HGF/c-Met signaling pathway during the liver metastasis of colorectal cancer. Oncol. Lett. 2020, 20, 2113–2118. [Google Scholar] [CrossRef]
- Oft, M. IL-10: Master switch from tumor-promoting inflammation to antitumor immunity. Cancer Immunol. Res. 2014, 2, 194–199. [Google Scholar] [CrossRef]
- Mannino, M.H.; Zhu, Z.; Xiao, H.; Bai, Q.; Wakefield, M.R.; Fang, Y. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 2015, 367, 103–107. [Google Scholar] [CrossRef]
- Neurath, M.F. IL-23 in inflammatory bowel diseases and colon cancer. Cytokine Growth Factor Rev. 2019, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Guo, X.; Wu, T.; Fei, K.; Wu, L. Identification of a novel cDC2-committed progenitor within mouse common dendritic cell progenitor population. Protein Cell 2022, 13, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; O’Brien, L.J.; Walpole, C.M.; Pearson, F.E.; Leal-Rojas, I.M.; Masterman, K.-A.; Atkinson, V.; Barbour, A.; Radford, K.J. Human CD141 dendritic cells (cDC1) are impaired in patients with advanced melanoma but can be targeted to enhance anti-PD-1 in a humanized mouse model. J. Immunother. Cancer 2021, 9, e001963. [Google Scholar] [CrossRef] [PubMed]
- Cytlak, U.; Resteu, A.; Pagan, S.; Green, K.; Milne, P.; Maisuria, S.; McDonald, D.; Hulme, G.; Filby, A.; Carpenter, B.; et al. Differential IRF8 Transcription Factor Requirement Defines Two Pathways of Dendritic Cell Development in Humans. Immunity 2020, 53, 353–370. [Google Scholar] [CrossRef] [PubMed]
- Bourdely, P.; Anselmi, G.; Vaivode, K.; Ramos, R.N.; Missolo-Koussou, Y.; Hidalgo, S.; Tosselo, J.; Nuñez, N.; Richer, W.; Vincent-Salomon, A.; et al. Transcriptional and Functional Analysis of CD1c Human Dendritic Cells Identifies a CD163 Subset Priming CD8CD103 T Cells. Immunity 2020, 53, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Nassar, D.; Blanpain, C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu. Rev. Pathol. 2016, 11, 47–76. [Google Scholar] [CrossRef]
- Ju, H.-Q.; Lu, Y.-X.; Chen, D.-L.; Tian, T.; Mo, H.-Y.; Wei, X.-L.; Liao, J.-W.; Wang, F.; Zeng, Z.-L.; Pelicano, H.; et al. Redox Regulation of Stem-like Cells Though the CD44v-xCT Axis in Colorectal Cancer: Mechanisms and Therapeutic Implications. Theranostics 2016, 6, 1160–1175. [Google Scholar] [CrossRef]
- De Sousa e Melo, F.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef]
- Zhang, N.; Ng, A.S.; Cai, S.; Li, Q.; Yang, L.; Kerr, D. Novel therapeutic strategies: Targeting epithelial-mesenchymal transition in colorectal cancer. Lancet Oncol. 2021, 22, e358–e368. [Google Scholar] [CrossRef]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Loupakis, F.; Cremolini, C.; Masi, G.; Lonardi, S.; Zagonel, V.; Salvatore, L.; Cortesi, E.; Tomasello, G.; Ronzoni, M.; Spadi, R.; et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N. Engl. J. Med. 2014, 371, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell. Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Liao, K.; Zhou, Y.; Wen, T.; Quan, G.; Pan, X.; Wu, C. Application of glutathione depletion in cancer therapy: Enhanced ROS-based therapy, ferroptosis, and chemotherapy. Biomaterials 2021, 277, 121110. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ramadori, P.; Pfister, D.; Seehawer, M.; Zender, L.; Heikenwalder, M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer 2021, 21, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Mehdizadeh, S.; Smith, J.; Young, A.; Mufazalov, I.A.; Mowery, C.T.; Daud, A.; Bluestone, J.A. Regulatory T cell control of systemic immunity and immunotherapy response in liver metastasis. Sci. Immunol. 2020, 5, eaba0759. [Google Scholar] [CrossRef]

| Items | Factors | Change | References |
|---|---|---|---|
| APC, RAS, BRAF, PIK3CA, TP53, SMAD4, MSI status | - | [21,22,23,24,25,26,27,30,43,44,45,46] | |
| DNA copy numbers | -/↑ | [34,35,36,37,40,41] | |
| Items | Factors | Change | References |
|---|---|---|---|
| MiRNAs | MiR-122, MiR-122*, MiR-885-5p, MiR-203, MiR-200c, MiR-424-3p, MiR-503, MiR-1292 | ↑ | [51,52,53,60,61,65] |
| MiR-143, MiR-10b, MiR-28-5p | ↓ | [51,52,53] | |
| CircRNAs | Circ0001178, Circ0000826, CircNSUN2, Circ0006401 | ↑ | [71,72,73,74,75,76,77,78,79,80,81,82,83] |
| LncRNAs | LncRNA GAL, LncRNA MIR17HG | ↑ | [86,87] |
| DNA methylation | RASSF1a, CircNSUN2, METTL3 | ↑ | [81,94,95] |
| p16INK4a | ↓ | [94] | |
| Protein acetylation | H2BLK121Ac, ADH1B K331Ac, IDH1 | ↑ | [96,97] |
| HIST2H3AK19Ac, TPM2 K152Ac | ↓ | [96] |
| Items | Factors | Change | References |
|---|---|---|---|
| EMT-related proteins | Migration-associated proteins: VTN, ARP3, FN1, TIMP1, VCAN, POSTN, THBS1 | ↑ | [99,100] |
| IGFBP7, Adhesion protein: claudins, ITA5 | ↓ | [99,106,107,108,109,110] | |
| ARHGAP5, PEA15, ATP6L, FILIP1L | ↑ | [111,114,117] | |
| Transcription factors | MYC, HIF1α | ↑ | [123] |
| DAXX | ↓ | [122] | |
| Other proteins | PLG, Serpin A1, APOA1A, CA1, SDHA | ↑ | [114,125] |
| the mitochondrial matrix, the mitochondrial intermembrane space, the proteasome complex, and the actin cytoskeleton | ↓ | [125] |
| Items | Factors | Change | References |
|---|---|---|---|
| Aerobic glycolysis | GLUT3, HMGA1, PKM2, DKK2, Pyruvate carboxylase, Fructose-bisphosphate aldolase B, Fructose-1,6-bisphosphatase 1 | ↑ | [131,135,136,137,138] |
| Fructose metabolism | ALDOB | ↑ | [139] |
| Cholesterol metabolism | SREBP2, LDLR, SRB1 | ↑ | [140] |
| Fatty acids, acylcarnitines, oxidative compounds, polyamines | GSH, putrescine | ↑ | [141] |
| Items | Factors | Change | References |
|---|---|---|---|
| TAMs | MRC1+ CCL18+ TAMs, SPP1+ TAMs | ↑ | [6,148,150] |
| T cells | CD4+ FOXP3+ Tregs | ↑ | [157,158] |
| Th17, CD4+ T cells, CD8+ T cells, Treg-IL10, Treg-CTLA4 | ↓ | [148,157,158] | |
| DCs | cDC2-TIMP1 | ↑ | [148] |
| cDC2-C1QC(DC3s) | ↓ | [148] | |
| Neutrophils | ↑ | [6] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Zhai, C.; Xu, X.; Wang, H.; Han, W.; Shen, J. Multilevel Heterogeneity of Colorectal Cancer Liver Metastasis. Cancers 2024, 16, 59. https://doi.org/10.3390/cancers16010059
Chen H, Zhai C, Xu X, Wang H, Han W, Shen J. Multilevel Heterogeneity of Colorectal Cancer Liver Metastasis. Cancers. 2024; 16(1):59. https://doi.org/10.3390/cancers16010059
Chicago/Turabian StyleChen, Hao, Chongya Zhai, Xian Xu, Haidong Wang, Weidong Han, and Jiaying Shen. 2024. "Multilevel Heterogeneity of Colorectal Cancer Liver Metastasis" Cancers 16, no. 1: 59. https://doi.org/10.3390/cancers16010059
APA StyleChen, H., Zhai, C., Xu, X., Wang, H., Han, W., & Shen, J. (2024). Multilevel Heterogeneity of Colorectal Cancer Liver Metastasis. Cancers, 16(1), 59. https://doi.org/10.3390/cancers16010059







