Amino Acid Profiles in the Biological Fluids and Tumor Tissue of CRC Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methodologies and Biological Samples for Metabolic Profiling
2.1. Types of Biological Matrices
2.2. Common Methodologies Utilized in Metabolomics
3. Amino Acid Profiling in Colorectal Cancer Patients
3.1. Cancer Tissue Metabolomics
3.2. Serum and Plasma Metabolomics in Colorectal Cancer Patients
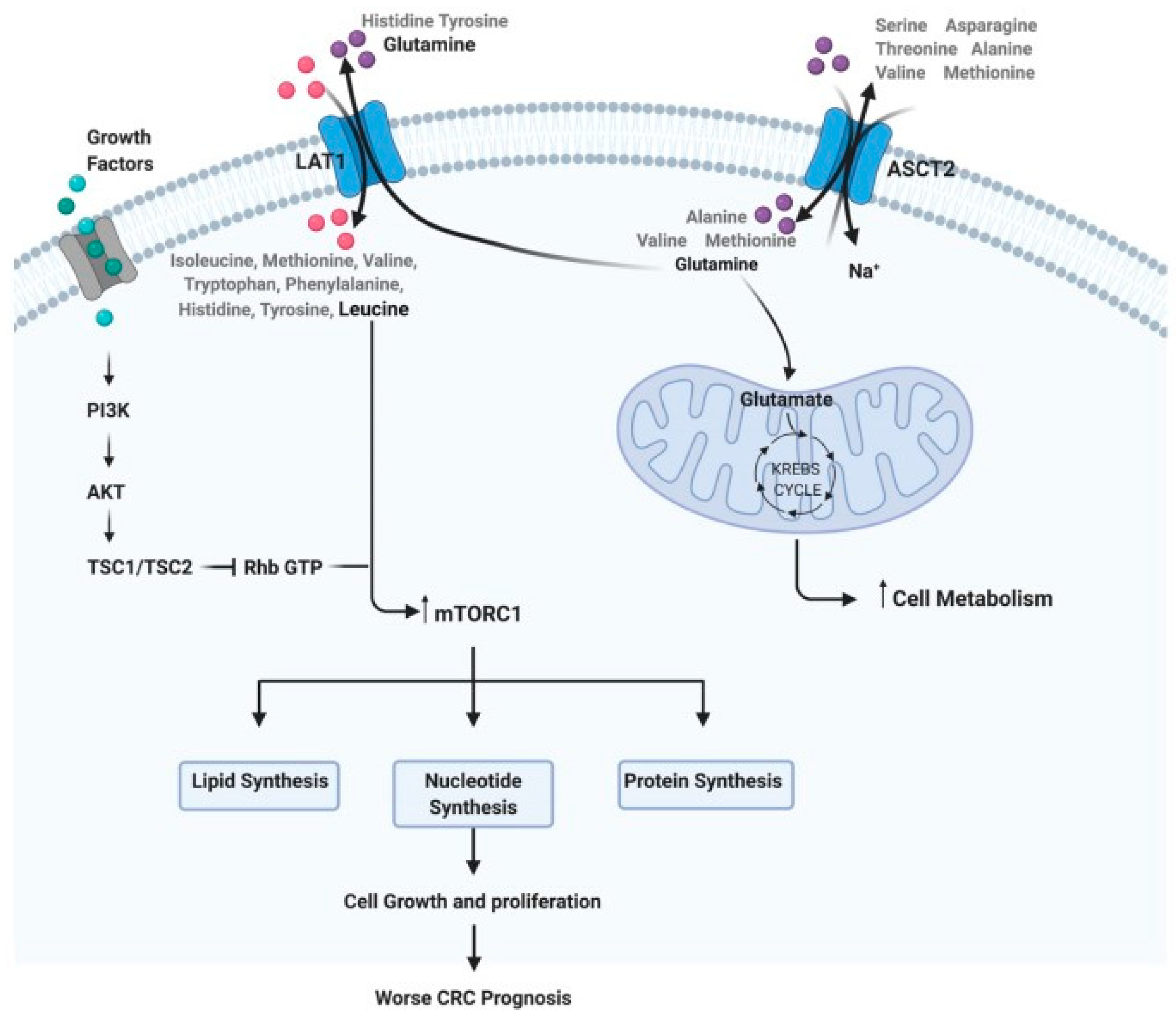
3.2.1. Glutamine and Glutamate Metabolism
3.2.2. Glycine and Serine Metabolism
3.2.3. Tryptophan Metabolism
3.2.4. Branched-Chain Amino Acid Metabolism
3.2.5. Proline Metabolism
| Reference | Year | Type of Cancer | TNM Stage | Patients (n) | Healthy Controls (n) | Biological Sample | Laboratory Techniques | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Tan et al. [6] | 2013 | CRC | I/II (68%) | 101 | 102 | Serum | GC-TOFMS/UPLC-QTOFMS | |||
| Chan et al. [20] | 2009 | CRC | Various stages | 31 | 31 | Serum | HR-MAS NMR/GC-MS | |||
| Barberini et al. [29] | 2019 | CRC | Various stages | 15 | 9 | Plasma | GC-MS | |||
| Leichtle et al. [30] | 2012 | CRC | Various stages | 59 | 58 | Serum | ET-MS | |||
| Troisi et al. [34] | 2022 | Adenomas, CRC | I/II | 150 | 50 | Serum | GC-MS | |||
| Gu et al. [35] | 2019 | Adenomas, CRC | Various stages | 62 | 38 | Serum | GC-MS | |||
| Zhu et al. [37] | 2014 | Adenomas, CRC | Various stages | 142 | 92 | Serum | LC-TMS | |||
| Nishiumi et al. [42] | 2017 | Adenomas, CRC | I/II | 282 | 291 | Plasma | GC-T/QMS | |||
| Nishiumi et al. [43] | 2012 | CRC | I/II | 59 | 63 | Serum | GC-MS | |||
| Geijsen et al. [45] | 2019 | CRC | Various stages | 268 | 353 | Plasma | UHPLC-QTOF-MS | |||
| Farshidfar et al. [46] | 2016 | Adenomas, CRC | Various stages | 320 | 254 | Serum | GC-MS | |||
| Ma et al. [47] | 2012 | CRC | Various stages | 30 | 20 | Serum | GC-MS | |||
| Wang et al. [48] | 2017 | CRC | I/II | 55 | 40 | Urine | H-NMR | |||
| Reference | Glutamate | Glutamate | Glycine | Serine | Threonine | Tryptophane | Proline | Valine | Leucine | Isoleucine |
| Tan et al. [6] | D | D | I | D | ||||||
| Chan et al. [20] | I | |||||||||
| Barberini et al. [29] | D | D | D | |||||||
| Leichtle et al. [30] | I | D | ||||||||
| Troisi et al. [34] | D | |||||||||
| Gu et al. [35] | D | I | D | D | D | D | D | D | ||
| Zhu et al. [37] | D | I | D | I | ||||||
| Nishiumi et al. [42] | D | |||||||||
| Nishiumi et al. [43] | D | I | ||||||||
| Geijsen et al. [45] | D | D | ||||||||
| Farshidfar et al. [46] | D | D | D | |||||||
| Ma et al. [47] | D | D | D | |||||||
| Wang et al. [48] | D | D | D | D | ||||||
4. Amino Acids as Discriminators of Colorectal Cancer Stage
5. Amino Acids as Prognostic Biomarkers in Colorectal Cancer
6. Amino Acids as Predictive Biomarkers of Response to Neoadjuvant Therapy
7. Usefulness, Applicability, and Limitations of Metabolic Abnormalities Analysis in Cancer in General and CRC in Particular
Highlights
- Amino acid abnormalities can be detected in serum, plasma, and tumor tissue.
- Amino acid abnormalities in tumor tissue are more pronounced than in circulation; however, the methodology is invasive and more complex.
- To study amino acids as biomarkers in CRC, plasma or serum collection is less invasive and preferred.
- Ion exchange chromatography with ninhydrin post-column derivatization is a simple and effective method to analyze plasma-free amino acids. Standardized protocols used to diagnose metabolic diseases can be employed to study amino acids in cancer patients’ circulation.
- Circulating levels of glutamine, branched-chain amino acids, and tryptophan have the potential to aid in CRC diagnosis, as they are potential hallmarks of CRC patients.
- Some studies also indicate pre-operative serum tryptophan as a possible discriminator between colon and rectal cancer.
- Amino acid plasma levels can be used as prognostic factors in colorectal cancer. Notably, histidine and citrulline levels differ in stage IV CRC. High-tumor glycine and low-serum glutamine are adverse prognostic factors in CRC.
- In LARC patients treated with neoadjuvant therapy, serum tryptophan and branched-chain amino acid levels predict the response to CRT.
8. Future Perspectives
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Weitz, J.; Koch, M.; Debus, J.; Hohler, T.; Galle, P.R.; Buchler, M.W. Colorectal cancer. Lancet 2005, 365, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Haggar, F.A.; Boushey, R.P. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009, 22, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pons, M.; Cruz-Correa, M. Colorectal Cancer Biomarkers: Where Are We Now? BioMed Res. Int. 2015, 2015, 149014. [Google Scholar] [CrossRef] [PubMed]
- Bedin, C.; Crotti, S.; D’Angelo, E.; D’Aronco, S.; Pucciarelli, S.; Agostini, M. Circulating Biomarkers for Response Prediction of Rectal Cancer to Neoadjuvant Chemoradiotherapy. Curr. Med. Chem. 2020, 27, 4274–4294. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Dong, T.; Zhao, L.; Feng, B.; Hu, X.; et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Pucciarelli, S.; Enzo, M.V.; Del Bianco, P.; Briarava, M.; Bedin, C.; Maretto, I.; Friso, M.L.; Lonardi, S.; Mescoli, C.; et al. Circulating cell-free DNA: A promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Ann. Surg. Oncol. 2011, 18, 2461–2468. [Google Scholar] [CrossRef]
- Brown, R.E.; Short, S.P.; Williams, C.S. Colorectal Cancer and Metabolism. Curr. Color. Cancer Rep. 2018, 14, 226–241. [Google Scholar] [CrossRef]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef]
- Gerner, E.W.; Meyskens, F.L., Jr. Polyamines and cancer: Old molecules, new understanding. Nat. Rev. Cancer 2004, 4, 781–792. [Google Scholar] [CrossRef]
- Dias, F.; Almeida, C.; Teixeira, A.L.; Morais, M.; Medeiros, R. LAT1 and ASCT2 Related microRNAs as Potential New Therapeutic Agents against Colorectal Cancer Progression. Biomedicines 2021, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Sotelo-Orozco, J.; Chen, S.Y.; Hertz-Picciotto, I.; Slupsky, C.M. A Comparison of Serum and Plasma Blood Collection Tubes for the Integration of Epidemiological and Metabolomics Data. Front. Mol. Biosci. 2021, 8, 682134. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Kastenmüller, G.; He, Y.; Belcredi, P.; Möller, G.; Prehn, C.; Mendes, J.; Wahl, S.; Roemisch-Margl, W.; Ceglarek, U.; et al. Differences between human plasma and serum metabolite profiles. PLoS ONE 2011, 6, e21230. [Google Scholar] [CrossRef] [PubMed]
- Segers, K.; Declerck, S.; Mangelings, D.; Heyden, Y.V.; Eeckhaut, A.V. Analytical techniques for metabolomic studies: A review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef] [PubMed]
- Rigas, P.G. Post-column labeling techniques in amino acid analysis by liquid chromatography. Anal. Bioanal. Chem. 2013, 405, 7957–7992. [Google Scholar] [CrossRef] [PubMed]
- Piraud, M.; Ruet, S.; Boyer, S.; Acquaviva, C.; Clerc-Renaud, P.; Cheillan, D.; Vianey-Saban, C. Amino acid profiling for the diagnosis of inborn errors of metabolism. Methods Mol. Biol. 2011, 708, 25–53. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef]
- Gao, P.; Zhou, C.; Zhao, L.; Zhang, G.; Zhang, Y. Tissue amino acid profile could be used to differentiate advanced adenoma from colorectal cancer. J. Pharm. Biomed. Anal. 2016, 118, 349–355. [Google Scholar] [CrossRef]
- Denkert, C.; Budczies, J.; Weichert, W.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Niesporek, S.; Noske, A.; Buckendahl, A.; Dietel, M.; et al. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol. Cancer 2008, 7, 72. [Google Scholar] [CrossRef]
- Chan, E.C.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361. [Google Scholar] [CrossRef]
- Chang, L.C.; Fan, C.W.; Tseng, W.K.; Chen, J.R.; Chein, H.P.; Hwang, C.C.; Hua, C.C. Immunohistochemical Study of the Nrf2 Pathway in Colorectal Cancer: Nrf2 Expression is Closely Correlated to Keap1 in the Tumor and Bach1 in the Normal Tissue. Appl. Immunohistochem. Mol. Morphol. AIMM Off. Publ. Soc. Appl. Immunohistochem. 2013, 21, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Piotto, M.; Moussallieh, F.-M.; Dillmann, B.; Imperiale, A.; Neuville, A.; Brigand, C.; Bellocq, J.-P.; Elbayed, K.; Namer, I. Metabolic characterization of primary human colorectal cancers using high resolution magic angle spinning 1 h magnetic resonance spectroscopy. Metabolomics 2009, 5, 292–301. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, F.; Gao, S.; Wang, Z.; Li, M.; Wei, H.; Zhong, R.; Chen, W. Simultaneous Determination of 34 Amino Acids in Tumor Tissues from Colorectal Cancer Patients Based on the Targeted UHPLC-MS/MS Method. J. Anal. Methods Chem. 2020, 2020, 4641709. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Cai, G.; Zhou, B.; Li, D.; Zhao, A.; Xie, G.; Li, H.; Cai, S.; Xie, D.; Huang, C.; et al. A distinct metabolic signature of human colorectal cancer with prognostic potential. Clin. Cancer Res. An. Off. J. Am. Assoc. Cancer Res. 2014, 20, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Ong, E.S.; Zou, L.; Li, S.; Cheah, P.Y.; Eu, K.W.; Ong, C.N. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol. Cell. Proteom. MCP 2010. [Google Scholar] [CrossRef]
- Okada, A.; Takehara, H.; Yoshida, K.; Nishi, M.; Miyake, H.; Kita, Y.; Komi, N. Increased aspartate and glutamate levels in both gastric and colon cancer tissues. Tokushima J. Exp. Med. 1993, 40, 19–25. [Google Scholar]
- Lin, Y.; Ma, C.; Bezabeh, T.; Wang, Z.; Liang, J.; Huang, Y.; Zhao, J.; Liu, X.; Ye, W.; Tang, W.; et al. (1) H NMR-based metabolomics reveal overlapping discriminatory metabolites and metabolic pathway disturbances between colorectal tumor tissues and fecal samples. Int. J. Cancer. 2019, 145, 1679–1689. [Google Scholar] [CrossRef]
- Rodriguez-Tomas, E.; Arenas, M.; Gomez, J.; Acosta, J.; Trilla, J.; Lopez, Y.; Arquez, M.; Torres, L.; Araguas, P.; Hernandez-Aguilera, A.; et al. Identification of potential metabolic biomarkers of rectal cancer and of the effect of neoadjuvant radiochemotherapy. PLoS ONE 2021, 16, e0250453. [Google Scholar] [CrossRef]
- Barberini, L.; Restivo, A.; Noto, A.; Deidda, S.; Fattuoni, C.; Fanos, V.; Saba, L.; Zorcolo, L.; Mussap, M. A gas chromatography-mass spectrometry (GC-MS) metabolomic approach in human colorectal cancer (CRC): The emerging role of monosaccharides and amino acids. Ann. Transl. Med. 2019, 7, 727. [Google Scholar] [CrossRef]
- Leichtle, A.B.; Nuoffer, J.M.; Ceglarek, U.; Kase, J.; Conrad, T.; Witzigmann, H.; Thiery, J.; Fiedler, G.M. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 2012, 8, 643–653. [Google Scholar] [CrossRef]
- Miyagi, Y.; Higashiyama, M.; Gochi, A.; Akaike, M.; Ishikawa, T.; Miura, T.; Saruki, N.; Bando, E.; Kimura, H.; Imamura, F.; et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS ONE 2011, 6, e24143. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Chen, M.J.; Chang, C.H.; Tiai, Y.F.; Lin, P.W.; Lai, H.S.; Wang, S.T. Plasma amino acid levels in patients with colorectal cancers and liver cirrhosis with hepatocellular carcinoma. Hepato-Gastroenterol. 2003, 50, 1269–1273. [Google Scholar]
- Wise, D.R.; Thompson, C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010, 35, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Tafuro, M.; Lombardi, M.; Scala, G.; Richards, S.M.; Symes, S.J.K.; Ascierto, P.A.; Delrio, P.; Tatangelo, F.; Buonerba, C.; et al. A Metabolomics-Based Screening Proposal for Colorectal Cancer. Metabolites 2022, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics Analysis in Serum from Patients with Colorectal Polyp and Colorectal Cancer by 1H-NMR Spectrometry. Dis. Markers 2019, 2019, 3491852. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef]
- Qiu, Y.; Cai, G.; Su, M.; Chen, T.; Zheng, X.; Xu, Y.; Ni, Y.; Zhao, A.; Xu, L.X.; Cai, S.; et al. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 2009, 8, 4844–4850. [Google Scholar] [CrossRef]
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503. [Google Scholar] [CrossRef]
- Uyttenhove, C.; Pilotte, L.; Theate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef]
- Huang, A.; Fuchs, D.; Widner, B.; Glover, C.; Henderson, D.C.; Allen-Mersh, T.G. Serum tryptophan decrease correlates with immune activation and impaired quality of life in colorectal cancer. Br. J. Cancer 2002, 86, 1691–1696. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Shi, L.; Yu, X.; Gu, Y.; Sun, X. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle 2018, 17, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Kawana, S.; Unno, Y.; Sakai, T.; Okamoto, K.; Yamada, Y.; Sudo, K.; Yamaji, T.; Saito, Y.; et al. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 2017, 8, 17115–17126. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, M.; Wu, Q. Identification of potential metabolite markers for colon cancer and rectal cancer using serum metabolomics. J. Clin. Lab. Anal. 2020, 34, e23333. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, A.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer. 2019, 145, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.A.; Hilsden, R.; McGregor, S.E.; Buie, W.D.; MacLean, A.; Vogel, H.J.; Bathe, O.F. A validated metabolomic signature for colorectal cancer: Exploration of the clinical value of metabolomics. Br. J. Cancer 2016, 115, 848–857. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, P.; Wang, F.; Liu, W.; Yang, J.; Qin, H. An integrated proteomics and metabolomics approach for defining oncofetal biomarkers in the colorectal cancer. Ann. Surg. 2012, 255, 720–730. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, Y.; Liang, J.; Huang, Y.; Ma, C.; Liu, X.; Yang, J. NMR-based metabolomic techniques identify potential urinary biomarkers for early colorectal cancer detection. Oncotarget 2017, 8, 105819–105831. [Google Scholar] [CrossRef]
- Geijsen, A.; van Roekel, E.H.; van Duijnhoven, F.J.B.; Achaintre, D.; Bachleitner-Hofmann, T.; Baierl, A.; Bergmann, M.M.; Boehm, J.; Bours, M.J.L.; Brenner, H.; et al. Plasma metabolites associated with colorectal cancer stage: Findings from an international consortium. Int. J. Cancer. 2020, 146, 3256–3266. [Google Scholar] [CrossRef]
- Di Donato, S.; Vignoli, A.; Biagioni, C.; Malorni, L.; Mori, E.; Tenori, L.; Calamai, V.; Parnofiello, A.; Di Pierro, G.; Migliaccio, I.; et al. A Serum Metabolomics Classifier Derived from Elderly Patients with Metastatic Colorectal Cancer Predicts Relapse in the Adjuvant Setting. Cancers 2021, 13, 2762. [Google Scholar] [CrossRef]
- Vahabi, F.; Sadeghi, S.; Arjmand, M.; Mirkhani, F.; Hosseini, E.; Mehrabanfar, M.; Hajhosseini, R.; Iravani, A.; Bayat, P.; Zamani, Z. Staging of colorectal cancer using serum metabolomics with 1HNMR Spectroscopy. Iran. J. Basic Med. Sci. 2017, 20, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Farshidfar, F.; Weljie, A.M.; Kopciuk, K.; Buie, W.D.; Maclean, A.; Dixon, E.; Sutherland, F.R.; Molckovsky, A.; Vogel, H.J.; Bathe, O.F. Serum metabolomic profile as a means to distinguish stage of colorectal cancer. Genome Med. 2012, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Redalen, K.R.; Sitter, B.; Bathen, T.F.; Groholt, K.K.; Hole, K.H.; Dueland, S.; Flatmark, K.; Ree, A.H.; Seierstad, T. High tumor glycine concentration is an adverse prognostic factor in locally advanced rectal cancer. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2016, 118, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.H.; Pan, Y.P.; Fan, C.W.; Tseng, W.K.; Huang, J.S.; Wu, T.H.; Chou, W.C.; Wang, C.H.; Yeh, K.Y.; Chang, P.H. Clinical Significance of Serum Glutamine Level in Patients with Colorectal Cancer. Nutrients 2019, 11, 898. [Google Scholar] [CrossRef]
- Sirnio, P.; Vayrynen, J.P.; Klintrup, K.; Makela, J.; Karhu, T.; Herzig, K.H.; Minkkinen, I.; Makinen, M.J.; Karttunen, T.J.; Tuomisto, A. Alterations in serum amino-acid profile in the progression of colorectal cancer: Associations with systemic inflammation, tumour stage and patient survival. Br. J. Cancer 2019, 120, 238–246. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, C.; Zheng, Y.; Liu, C.; Wang, X.; Cong, X. Glutamine deficiency promotes recurrence and metastasis in colorectal cancer through enhancing epithelial-mesenchymal transition. J. Transl. Med. 2022, 20, 330. [Google Scholar] [CrossRef]
- Bertini, I.; Cacciatore, S.; Jensen, B.V.; Schou, J.V.; Johansen, J.S.; Kruhoffer, M.; Luchinat, C.; Nielsen, D.L.; Turano, P. Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res. 2012, 72, 356–364. [Google Scholar] [CrossRef]
- Santos, M.D.; Silva, C.; Rocha, A.; Matos, E.; Nogueira, C.; Lopes, C. Tumor regression grades: Can they influence rectal cancer therapy decision tree? Int. J. Surg. Oncol. 2013, 2013, 572149. [Google Scholar] [CrossRef]
- Santos, M.D.; Silva, C.; Rocha, A.; Matos, E.; Nogueira, C.; Lopes, C. Prognostic value of mandard and dworak tumor regression grading in rectal cancer: Study of a single tertiary center. ISRN Surg. 2014, 2014, 310542. [Google Scholar] [CrossRef]
- Yang, K.; Zhang, F.; Han, P.; Wang, Z.Z.; Deng, K.; Zhang, Y.Y.; Zhao, W.W.; Song, W.; Cai, Y.Q.; Li, K.; et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for colorectal cancer. Metabolomics 2018, 14, 110. [Google Scholar] [CrossRef]
- Jia, H.; Shen, X.; Guan, Y.; Xu, M.; Tu, J.; Mo, M.; Xie, L.; Yuan, J.; Zhang, Z.; Cai, S.; et al. Predicting the pathological response to neoadjuvant chemoradiation using untargeted metabolomics in locally advanced rectal cancer. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2018, 128, 548–556. [Google Scholar] [CrossRef]
- Crotti, S.; Fraccaro, A.; Bedin, C.; Bertazzo, A.; Di Marco, V.; Pucciarelli, S.; Agostini, M. Tryptophan Catabolism and Response to Therapy in Locally Advanced Rectal Cancer (LARC) Patients. Front. Oncol. 2020, 10, 583228. [Google Scholar] [CrossRef]
- Eagle, H. Nutrition needs of mammalian cells in tissue culture. Science 1955, 122, 501–514. [Google Scholar] [CrossRef]
- Kim, H.J.; Jang, S.H.; Ryu, J.S.; Lee, J.E.; Kim, Y.C.; Lee, M.K.; Jang, T.W.; Lee, S.Y.; Nakamura, H.; Nishikata, N.; et al. The performance of a novel amino acid multivariate index for detecting lung cancer: A case control study in Korea. Lung Cancer 2015, 90, 522–527. [Google Scholar] [CrossRef]
- Eniu, D.T.; Romanciuc, F.; Moraru, C.; Goidescu, I.; Eniu, D.; Staicu, A.; Rachieriu, C.; Buiga, R.; Socaciu, C. The decrease of some serum free amino acids can predict breast cancer diagnosis and progression. Scand. J. Clin. Lab. Investig. 2019, 79, 17–24. [Google Scholar] [CrossRef]
- Cadoni, G.; Giraldi, L.; Chiarla, C.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Settimi, S.; Galli, J.; Paludetti, G.; Arzani, D.; et al. Prognostic Role of Serum Amino Acids in Head and Neck Cancer. Dis. Markers 2020, 2020, 2291759. [Google Scholar] [CrossRef]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Wilkinson, A.C. Branched-chain amino acid metabolism in cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64–70. [Google Scholar] [CrossRef]
- Greene, L.I.; Bruno, T.C.; Christenson, J.L.; D’Alessandro, A.; Culp-Hill, R.; Torkko, K.; Borges, V.F.; Slansky, J.E.; Richer, J.K. A Role for Tryptophan-2,3-dioxygenase in CD8 T-cell Suppression and Evidence of Tryptophan Catabolism in Breast Cancer Patient Plasma. Mol. Cancer Res. MCR 2019, 17, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, B.; Hu, Y.; Zhao, Y. New Insights Into Gut-Bacteria-Derived Indole and Its Derivatives in Intestinal and Liver Diseases. Front. Pharmacol. 2021, 12, 769501. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Yin, M.; Sun, F.; Zhang, T.; Zhou, X.; Li, H.; Zheng, J.; Chen, X.; Li, C.; Ning, X.; et al. A metabolomics approach for predicting the response to neoadjuvant chemotherapy in cervical cancer patients. Mol. Biosyst. 2014, 10, 2126–2133. [Google Scholar] [CrossRef]
- Wei, S.; Liu, L.; Zhang, J.; Bowers, J.; Gowda, G.A.; Seeger, H.; Fehm, T.; Neubauer, H.J.; Vogel, U.; Clare, S.E.; et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol. Oncol. 2013, 7, 297–307. [Google Scholar] [CrossRef]
- He, X.; Gu, J.; Zou, D.; Yang, H.; Zhang, Y.; Ding, Y.; Teng, L. NMR-Based Metabolomics Analysis Predicts Response to Neoadjuvant Chemotherapy for Triple-Negative Breast Cancer. Front. Mol. Biosci. 2021, 8, 708052. [Google Scholar] [CrossRef]
- Ramspek, C.L.; Evans, M.; Wanner, C.; Drechsler, C.; Chesnaye, N.C.; Szymczak, M.; Krajewska, M.; Torino, C.; Porto, G.; Hayward, S.; et al. Kidney Failure Prediction Models: A Comprehensive External Validation Study in Patients with Advanced CKD. J. Am. Soc. Nephrol. JASN 2021, 32, 1174–1186. [Google Scholar] [CrossRef]
- Pomyen, Y.; Wanichthanarak, K.; Poungsombat, P.; Fahrmann, J.; Grapov, D.; Khoomrung, S. Deep metabolome: Applications of deep learning in metabolomics. Comput. Struct. Biotechnol. J. 2020, 18, 2818–2825. [Google Scholar] [CrossRef]
- Huang, L.; Wang, L.; Hu, X.; Chen, S.; Tao, Y.; Su, H.; Yang, J.; Xu, W.; Vedarethinam, V.; Wu, S.; et al. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 2020, 11, 3556. [Google Scholar] [CrossRef]
- Kim, J.Y.; Jeon, E.; Kwon, S.; Jung, H.; Joo, S.; Park, Y.; Lee, S.K.; Lee, J.E.; Nam, S.J.; Cho, E.Y.; et al. Prediction of pathologic complete response to neoadjuvant chemotherapy using machine learning models in patients with breast cancer. Breast Cancer Res. Treat. 2021, 189, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Sun, N.; Zens, P.; Kunzke, T.; Buck, A.; Prade, V.M.; Wang, J.; Wang, Q.; Hu, R.; Feuchtinger, A.; et al. Spatial metabolomics for evaluating response to neoadjuvant therapy in non-small cell lung cancer patients. Cancer Commun 2022, 42, 517–535. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Zhou, N.; Feng, L.; Li, Z.; Fang, Y.; Fan, X.; Ling, Y.; Liu, H.; Zou, X.; Wang, J.; et al. Deep Learning Model for Predicting the Pathological Complete Response to Neoadjuvant Chemoradiotherapy of Locally Advanced Rectal Cancer. Front. Oncol. 2022, 12, 807264. [Google Scholar] [CrossRef] [PubMed]
| Reference | Year | Disease | Stage | Early Stage | Patients (n) | Healthy Controls (n) | Biological Sample | Techniques | Findings |
|---|---|---|---|---|---|---|---|---|---|
| Qiu et al. [37] | 2014 | CRC | TNM 0–IV | 46% | 227 | Na | Tumor biopsies | Gas chromatography–time-of-flight mass spectrometry | Glutamate, aspartate, cysteine, and beta-alanine can distinguish patients with a longer time to recurrence and better 5-year recurrence. |
| Farshidfar et al. [52] | 2016 | CRC | TNM I–IV | 33% | 320 | 254 | Serum | Gas chromatography–mass spectrometry | A model with 35 metabolites, including ornithine, proline, and aspartate, is able to discriminate recurring from non-recurring stage II patients. |
| Redalen et al. [53] | 2016 | LARC | T3–4/N0/M0 or T2-4/N1-2M0- | na | 54 | Na | Tumor biopsies | High-resolution magic angle spinning magnetic resonance spectroscopy | High tumor glycine (1.77 μmol/g) is associated with worse progression-free survival. |
| Ling et al. [54] | 2019 | CRC | TNM I–IV | 49% | 123 | Na | Serum | Enzy ChromTM Glutamine Assay Kit | Lower plasma glutamine (<52 ng/μL) is associated with worse overall survival and progression-free survival. |
| Sirnio et al. [55] | 2018 | CRC | TNM I–IV | 51% | 357 | Na | Serum | Nuclear magnetic resonance | Lower glutamine (<410 μmol/L) and histidine (55 μmol/L) and higher phenylalanine (>93 μmol/L) and glycine (>263 μmol/L) are associated with decreased cancer-specific survival. |
| Bertini et al. [57] | 2012 | mCRC | IV | na | 181 | 139 | Serum | Nuclear magnetic resonance | Lower serum creatine and valine are associated with shorter overall survival. |
| Reference | Year | Type Ofcancer | Stages | Patients (n) | Healthy Controls (n) | Biological Sample | Techniques | Findings |
|---|---|---|---|---|---|---|---|---|
| Rodríguez-Tomàs et al. [28] | 2021 | LARC- | TNM T3–4 and/or N+ | 32 | 48 | Plasma | GC-EI-QTOF-MS | Significantly lower plasma valine in pre-nCRT samples from pathological complete response patients. |
| Yang et al. [60] | 2018 | Colon and Rectum | TNM II–III | 47 Rectum; 10 colon | na | Plasma before nCRT | UHPLC—quadruple time-of-flight)/mass spectrometry analyses | Leucine increased in non-responsive patients with superior sensitivity to CAE and CA 199Jia. |
| Jia et al. [61] | 2018 | LARC | TNM T3–4 and/or N+ | 105 | na | Serum | Liquid chromatography–mass spectrometry | 3-methylhistidine, 4-imidazoleacetic acid, and dimethylglycine downregulated in nCRT-resistant patientsRodrı. |
| Crotti et al. [62] | 2020 | LARC | TNM T3–4 and/or N+ | 45 | na | Plasma | UHPLC-UV-VIS/FLD and LC-MS/MS | Significantly increased tryptophan in non-responsive patients. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, M.D.; Barros, I.; Brandão, P.; Lacerda, L. Amino Acid Profiles in the Biological Fluids and Tumor Tissue of CRC Patients. Cancers 2024, 16, 69. https://doi.org/10.3390/cancers16010069
Santos MD, Barros I, Brandão P, Lacerda L. Amino Acid Profiles in the Biological Fluids and Tumor Tissue of CRC Patients. Cancers. 2024; 16(1):69. https://doi.org/10.3390/cancers16010069
Chicago/Turabian StyleSantos, Marisa Domingues, Ivo Barros, Pedro Brandão, and Lúcia Lacerda. 2024. "Amino Acid Profiles in the Biological Fluids and Tumor Tissue of CRC Patients" Cancers 16, no. 1: 69. https://doi.org/10.3390/cancers16010069
APA StyleSantos, M. D., Barros, I., Brandão, P., & Lacerda, L. (2024). Amino Acid Profiles in the Biological Fluids and Tumor Tissue of CRC Patients. Cancers, 16(1), 69. https://doi.org/10.3390/cancers16010069






