Simple Summary
The extracellular matrix acts as scaffolding to support the structure and function of the cells in our body. In cancer, this matrix is altered in a way that helps cancer cells to grow, spread and avoid the immune system. The altered matrix can also prevent cancer treatments like chemotherapy and immunotherapy from working. Targeting the matrix with drugs is emerging as an exciting way to modify the scaffold in a precise way to improve the effectiveness of cancer treatments in patients. In this review, we will examine strategies to target the matrix with drugs, which can help the immune system fight cancer and improve the response to existing cancer therapies.
Abstract
The extracellular matrix (ECM) is composed of complex fibrillar proteins, proteoglycans, and macromolecules, generated by stromal, immune, and cancer cells. The components and organisation of the matrix evolves as tumours progress to invasive disease and metastasis. In many solid tumours, dense fibrotic ECM has been hypothesised to impede therapy response by limiting drug and immune cell access. Interventions to target individual components of the ECM, collectively termed the matrisome, have, however, revealed complex tumour-suppressor, tumour-promoter, and immune-modulatory functions, which have complicated clinical translation. The degree to which distinct components of the matrisome can dictate tumour phenotypes and response to therapy is the subject of intense study. A primary aim is to identify therapeutic opportunities within the matrisome, which might support a better response to existing therapies. Many matrix signatures have been developed which can predict prognosis, immune cell content, and immunotherapy responses. In this review, we will examine key components of the matrisome which have been associated with advanced tumours and therapy resistance. We have primarily focussed here on targeting matrisome components, rather than specific cell types, although several examples are described where cells of origin can dramatically affect tumour roles for matrix components. As we unravel the complex biochemical, biophysical, and intracellular transduction mechanisms associated with the ECM, numerous therapeutic opportunities will be identified to modify tumour progression and therapy response.
1. Matrix Signatures Are Prognostic and Predict Immune Content
Deregulation of the tumour matrisome is a characteristic of all solid tumours, where an aberrant matrix can dictate the hallmarks of cancer [1]. The matrix is composed of a complex network or fibrillar proteins, proteoglycans, and other macromolecules, which are intricately crosslinked and subjected to post-translational modification. The term matrisome is used to describe an inventory of genes or proteins, which form the matrix or regulate matrix composition, structure, or function. In mammals, there are approximately ~300 core matrisome components (e.g., NABA_CORE_MATRISOME gene set [2]) and a large number of matrix-associated proteins and enzymatic regulators, which are variously included within the broad definition of the matrisome [3]. Approximately 1000 genes encoding matrix or matrix-associated proteins are commonly included in matrisome gene ontology lists [4]. The matrix provides the scaffold in which tissues are embedded and also directs cellular behaviour through direct interaction with numerous cellular receptors, tissue biomechanics, the regulation of growth factor availability, and physical restriction [5]. Tissue organisation governed by the matrix also critically regulates how and when distinct cell types interact with each other. In cancer, this is particularly pertinent to stromal interactions, tumour vascularisation, and immune landscape [6].
Matrix deregulation in solid cancers is perhaps most apparent as tumour desmoplasia, where excessive fibrotic deposition of the ECM leads to tissue stiffening. In many aggressive tumours, including PDAC, breast cancers, and ovarian cancer, desmoplasia is a defining feature, which often correlates with poor outcome and treatment failure [7,8,9,10,11,12]. However, unravelling the correlation of desmoplasia with causative roles in aggressive disease has proven more difficult. Supporting a causative role, tissue stiffness and fibrosis are well-known risk factors for several cancers. Mammographic density identified through population screening is among the highest risk indicators for developing breast cancer [13,14]. Furthermore, fibrotic conditions, such as liver cirrhosis and pancreatitis, are associated with significant elevated cancer risk [15,16,17]. Nonetheless, interventions to modify tumour fibrosis have had mixed outcomes in pre-clinical models and clinical translation remains elusive [7,9,18,19,20,21]. In this review, we have focussed on the specific targeting of matrisome components to try to unravel causative and correlative relationships. Examining individual matrix components often reveals strikingly complex extracellular signalling roles, which challenges us to reconsider the importance of the ECM in the processing of information. The most promising strategies for cancer treatment appear to work through modulation of the immune landscape, and successful targeting of the stroma to improve outcome and therapy response remains an exciting prospect.
2. Common Matrix Signatures Evolve to Support Invasion and Metastasis
While the matrices in distinct tissues each have their own characteristic composition and biomechanical properties, commonalities also exist: the primary matrix component in most tissues is fibrillar type 1 collagen; most parenchymal tissues and early stage tumours are surrounded by basement membranes, a specialised type of barrier matrix rich in collagen IV and laminins; and the matrix provides a scaffold for the parenchyma and vascular trees providing access for nutrients and immune cells and egress for waste. All solid tumours must subvert these physiological properties to flourish. The rapid advance of omics technologies has allowed us to catalogue temporal and spatial changes to the matrisome during tumour progression for diverse cancer types uncovering both commonalities and tumour-specific mechanisms.
In solid cancers, the matrix co-evolves with the malignant component to support tumour growth, evade immune surveillance, and promote disease progression [22]. Many studies have revealed conserved matrisome features and signatures associated with poor outcome and metastasis (Table 1) [7,8,23,24,25]. Moffit et al. used an in silico approach to separate stromal and epithelial signatures from bulk transcriptomic data and used this to define signatures of the activated vs. normal pancreatic cancer stroma; interestingly, the prognostic power of the activated stromal signature was dependent on the epithelial subtype, predicting worse outcome in basal-like tumours [26]. Most identified genes were matrix-related, including many collagens and FN1. Pearce et al. employed a multi-omics approach to examine features associated with metastatic progression in ovarian cancer [8]. They defined a transcriptomic Matrix Index (MI) scoring system, based on the 22 genes most significantly associated with disease severity and tissue stiffness. Importantly, the MI is highly prognostic, not only for ovarian cancer but also for many epithelial and mesenchymal solid tumours. The MI correlated robustly with regulatory T-cell content (FOXP3+), though a relationship with CD8+ T-cells was less clear. Many genes overlap between the MI and Moffitt signatures, despite distinct tissue origins, including COL1A1, FN1, VCAN, and COMP.
In breast cancer, matrisome signatures associated with metastasis have been documented, highlighting FN1, Tenascin C, and lysyl oxidases as poor prognostic indicators [23]. Collagen I again features in this prometastatic signature, alongside COLV and COLVI; importantly, COLVI has also been associated with a functional role in TNBC metastasis in a cell-derived matrix model [27], while COL1A1 or FN1 alone can also drive an aggressive breast cancer phenotype in culture models [28]. Indeed, both COL1A1 and FN1 have been identified as individual prognostic biomarkers in a variety of cancers [29,30,31,32,33,34,35,36]. Taking a proteomic approach to map the evolving matrisome in breast cancer, Papanicolaou et al. identified and characterised a role for collagen XII as a metastasis-related component, though, interestingly, this was functionally linked to regulation of collagen I fibril organisation [37].
A picture emerges where the tumour-associated matrix promotes an immune-evasive, pro-metastatic environment, often through modulation of common and abundant matrix components, including collagen I, FN1, and hyaluronic acid (HA). Organisation, alignment, crosslinking, and degradation all play important roles, which offer numerous therapeutic opportunities.
3. Matrix Signatures Associated with Immune Evasion and Immune Checkpoint Response
Historically, tumour matrix studies have focussed on invasion and metastasis, whereas recent emphasis has turned to immune evasion (Table 1). Significant effort is now being made to understand the role of the tumour microenvironment (TME) in the regulation of anti-tumour immunity and ICB response. Taking a pan-cancer approach, Chakravarthy et al. characterised ECM changes between normal and malignant tissue across multiple solid tumour TCGA datasets and defined a conserved gene expression signature associated with malignancy [25]; notably, the cancer-associated ECM signature (C-ECM) is not only associated with disease prognosis but also predicts immune evasion and immune-checkpoint blockade (ICB) failure. Many of the ECM genes identified are TGFβ-responsive targets in cancer-associated fibroblasts (CAFs), and these data concur with a broad range of studies implicating the TGFβ-activated stroma with poor ICB response [38,39,40,41]. The intimate link between the matrisome and immune content implies that the direct targeting of specific matrisome components may provide benefit to augment immunotherapy and chemotherapy response [6].
In breast cancer, an early stromal gene expression study by Finak et al. also highlighted the critical contribution of the stroma–immune relationship to breast cancer prognosis [42]. A clinically useful example is provided by the colorectal cancer (CRC) immunoscore, largely based on CD3 and CD8 T-cell content. The immunoscore is a powerful predictor of both prognosis and ICB response in MSI-positive CRC tumours [43,44,45]; immunotherapy is now the frontline therapy for the management of MSI-positive metastatic CRC, where biomarkers for response are critical [46]. Similarly in melanoma, where immunotherapy is offering huge clinical benefit, signatures associated with T-cell exclusion are powerful predictors of ICB response [47].
While T-cell content is a powerful predictor in some cancer settings such as CRC, the relationship between CD8+ T-cells and ICB response is clearly complex, as many factors can govern the ability of T-effector cells to target tumour cells, including neo-antigen load, PD-L1 expression, T-cell distribution, and the presence of immune-suppressive Tregs, γδ T cells, and myeloid suppressor cells [48,49,50,51]. To integrate these factors, the Tumour Immune Dysfunction and Exclusion study (TIDE) defined a scoring system which considers many critical factors governing anti-tumour immune responses, including T-cell exclusion, T-cell exhaustion, immune cell repertoire, and stromal signatures [52]. Examining melanoma trial results, the study concludes that high CD8+ T-cell content is an effective indicator of ICB response only when associated with a low TGFβ signature [52]. The TME risk signatures developed for lung [53] and HCC [54], as well as the immune checkpoint inhibitor scoring system (IMS) for bladder cancer [55], provide additional informative examples, defining TME features associated with anti-tumour immunity and ICB response. In each of these signatures or scoring systems, matrisome components are abundant, with commonalities regarding TGFβ signatures, collagen regulation, and immune content.

Table 1.
Stromal signatures associated with prognosis or therapy response.
Table 1.
Stromal signatures associated with prognosis or therapy response.
| Name | Derived | Application | Cancer Focus | Number of Genes |
|---|---|---|---|---|
| Dominguez et al. [39] | Marker genes for a CAF subpopulation | Predicts anti-PD-L1 therapy response | Pancreatic ductal adenocarcinoma | 12 upregulated genes |
| Pearce et al. [8] | Matrix genes associated with disease score and tissue modulus | Predicts poor prognosis | High-grade serous ovarian cancer | 6 upregulated genes, 16 downregulated genes |
| Moffitt et al. [26] | Differential expression of stromal genes between CAFs and tumour cell lines | Predicts poor prognosis | Pancreatic ductal adenocarcinoma | 48 upregulated genes |
| Murray et al. [24] | Transcriptome associated with PKN2 knockout | Predicts poor prognosis | Pancreatic ductal adenocarcinoma | 11 upregulated genes |
| Öhlund et al. [56] | Transcriptome associated with iCAF subtype | CAF subtype discrimination | Pancreatic ductal adenocarcinoma | 200 upregulated genes, 200 downregulated genes |
| Jiang et al. [52] | Transcriptome associated with T-cell exclusion and dysfunction | Predicts immune checkpoint inhibitor response | Multiple | 770 genes |
| Yan et al. [53,54] | TME signatures associated with anti-tumour immunity | Predicts poor prognosis and immune checkpoint inhibitor response | Hepatocellular carcinoma | Four downregulated genes |
| Lin et al. [55] | Gene expression of 27 survival-related immune signatures | Predicts good prognosis and immune checkpoint inhibitor response | Gastric cancer | 463 upregulated genes |
| Brechbuhl et al. [23] | Proteome associated with CD146–CAFs | Associated with increased risk of metastasis | Breast cancer | 12 upregulated genes |
| Wang et al. [57] | Regression in TCGA ESTIMATE scores | Predicts poor prognosis | Gastric cancer | Three upregulated genes, one downregulated gene |
| Jia et al. [58] | Regression in TCGA ESTIMATE scores | Predicts poor prognosis | Colon adenocarcinoma | Three upregulated genes |
| Yue et al. [59] | Transcriptome associated with TCGA lymphovascular space invasion | Predicts poor prognosis | Serous ovarian cancer | Eight upregulated genes |
| Isella et al. [60] | CAF-expressing genes associated with stem/serrated/mesenchymal (SSM) transcriptional subtype | Predicts poor prognosis | Colorectal adenocarcinoma | 130 upregulated genes |
| Farmer et al. [61] | Genes co-expressed with decorin | Predicts poor neoadjuvant chemotherapy response | ER-negative breast cancer | 50 upregulated genes |
| Boersma et al. [62] | Stromal genes differentially expressed between inflammatory and non-inflammatory breast cancer | High inflammation | Inflammatory breast cancer | 2 upregulated genes, 20 downregulated genes |
| Strell et al. [63] | Differential expression on platelet-derived growth factor (PDGF)–activated human fibroblasts | Predicts poor prognosis | Early breast cancer | 55 upregulated genes, 58 downregulated genes |
| Casey et al. [64] | Differential expression between invasive cancer stroma and normal stroma | Associated with invasion | Breast-invasive carcinoma | Nine upregulated genes, five downregulated genes |
| Winslow et al. [65] | Differential expression between LCM stromal TN breast cancer tumours. Correlating gene changes with TCGA | Predicts poor prognosis | Breast-invasive carcinoma | 53 upregulated genes, 26 downregulated genes |
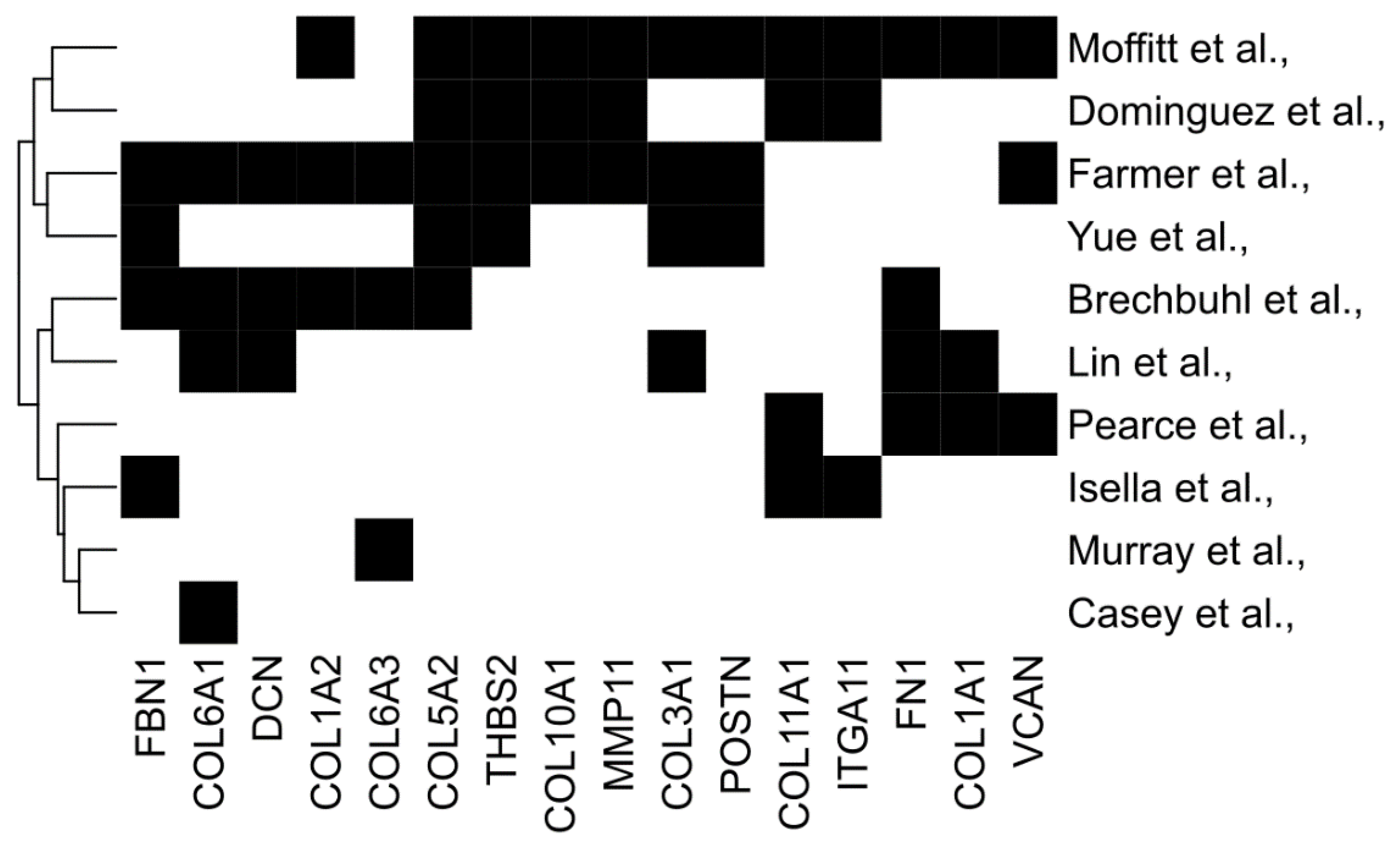
Taking several matrix signatures together, we also examined the overlap between genes to identify commonalities; 16 genes appeared in at least three independent signatures, including COLA1, COL11A1, VCAN, POSTN, and FN1 (Figure 1). In the next sections, we will first consider how distinct CAF subtypes are associated with both matrisome signatures and tumour immunophenotype. Then, we will address the degree to which the matrisome can dictate, rather than correlate with, immunophenotypes by considering both CAF-directed and individual-matrisome-component-directed interventions; we will focus on matrisome genes common to multiple predictive signatures (Figure 1).
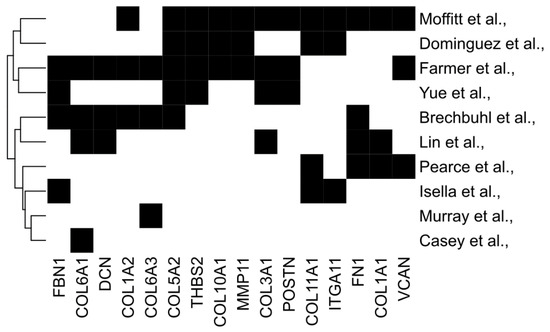
Figure 1.
Commonalities between diverse predictive matrix signatures. Common genes from several stromal signatures associated with prognosis or therapy response. Genes selected participate in three or more stromal signatures.
4. Cancer-Associated Fibroblasts Are Principal Contributors to the Matrisome
CAFs are considered the dominant source of the TGFβ-stimulated pro-tumourigenic matrisome and many recent spatial and single-cell transcriptomic studies have characterized distinct CAF sub-populations, which evolve with advancing malignancy. A seminal study by the Tuveson lab identified a major dichotomisation of CAFs in PDAC into myofibroblast CAFs (myCAFs) and inflammatory CAFs (iCAFs), since confirmed in numerous scRNA-seq studies across many solid cancers [39,56,57,58,59,60,61,62,63,64,65]. Typically, myCAFs are driven through activation of TGFβ signalling, while iCAFs are reported to be driven by inflammatory cytokines [58]. Differential spatial locations within tumours and reciprocal interaction between the TGFβ and IL1 signalling pathways appear to drive distinct lineage trajectories [7,62]. Numerous additional CAF subtypes have since been described, including those defined by high matrix deposition (ecm-CAFs [64] and matrix-CAFs [57]). The cataloguing of fibroblast heterogeneity in solid tumours has supported the idea of tumour-promoting and tumour-supressing CAF populations, but it has also revealed the complexity of targeting these cells due to overlapping function and a lack of discriminatory biomarkers and drug targets [7,62].
4.1. Targeting CAF Subsets to Modify Tumour Biology
Approaches to selectively target CAF subtypes have yielded mixed results. In 2014, a group of important studies, which variously suppressed myofibroblast populations in PDAC, suggested that CAFs may play a tumour-suppressive role [19,20,21]. Recent studies have revealed a more nuanced picture with distinct CAF subsets playing opposing roles in tumours. Exciting recent work from the Turley lab elegantly mapped the evolution of CAF subsets as PDAC progresses in mouse models to define a TGFβ-programmed LRRC15+ CAF population associated with advanced disease [39,40]. Importantly, this CAF signature was identified in many solid tumour types and associated with poor ICB response in trial data. Ablation of LRRC15+ CAFs in mouse models was also able to enhance anti-tumour immunity and ICB response [40]. Interestingly, this study also revealed that both myCAF and iCAF lineages can express abundant collagen I and II, suggesting a more complex relationship between myCAF subtype and fibrosis. Hutton et al. also identified distinct CAF lineages in PDAC with distinct CD105-positive CAF-promoting tumour growth, while CD105-negative CAFs support robust anti-tumour immunity [41]. Interestingly, in the 2014 Ozdemir et al. study in which α-SMA CAF ablation drives more aggressive PDAC growth, tumours were also rendered more sensitive to anti-CTLA4 immunotherapy, perhaps indicating distinct tumour-restraining and immune-suppressive roles for overlapping CAF populations. TGFβ-driven CAF subsets supporting immune evasion have also been defined in mouse models of breast and other cancers [7,38,59,66]. In TNBC, distinct CAF lineages have also been associated with CD8+ T-cell exclusion or functional suppression [60]. The causal, prognostic, predictive association between TGFβ-induced stromal signatures and the suppression of anti-tumour immunity and ICB response has now been robustly established [7,38].
Powerful spatial transcriptomic studies have also demonstrated that individual glands within tumours can harbour highly localised relationships between cancer cells, CAFs, and immune cells [67,68]; advanced tumours can contain many distinct cancer-subtypes and localised TME interactions, which must be taken into account when considering therapy response and resistance [69]. In the next section, rather than focussing on targeting the complex and localised interaction between CAFs and tumour cells, we wish to explore interventions which specifically target matrisome components.
4.2. Targeting TGFβ and CAF Activation
Many TGFβ- and CAF-targeting strategies have been shown to suppress stromal activation and tumour fibrosis, but interpretation is often complex due to multimodal mechanisms of action. This is exemplified by studies targeting the Hedgehog pathway where both positive and negative impacts on tumour progression and therapy response have been reported [9,20,61]. Co-administration of TGFβ-blocking antibodies or inhibitors with anti-PD-L1/PD1 has proven successful in mouse models [38,66]. Indeed, inhibitors of TGFβ signalling, including Galunisertib, NIS793, and LY364947, have all reached clinical testing for a variety of solid tumours, though concerns remain regarding tumour-suppressor roles in cancer cells for this signalling axis [7,70,71,72,73]. The angiotensin antagonist Losartan can suppress TGFβ release and fibrosis in a variety of pathological settings, including pancreatic and other solid tumours. In fibrotic breast and pancreatic cancer models, Losartan has been shown to reduce CAF-derived collagen and HA to enhance perfusion and potentiate chemo- and immunotherapy responses [74,75,76], and this has translated to promising clinical trial results [77,78]. The anti-fibrotic drug Pirfenidone also limits fibrosis by targeting TGFβ signalling and CAFs in both breast and pancreatic cancer models [79,80,81,82]. Numerous activating signalling axes in CAFs have also been the subject of significant studies, including Hedgehog signalling, Vitamin A and D pathways, FAK, and PKN2 [7,9,24,83,84,85]. Clinical trials targeting CAFs and TGFβ, while largely disappointing, have shown some promise in subsets of patients (reviewed [7,86]). The multimodal mechanisms of action of these important interventions lies beyond the scope of this review.
5. Direct Matrix—Interventions
While it has long been established that ECM signatures correlate with tumour progression and therapy responses, therapeutic targeting of specific matrisome components has proved difficult to enact [87]. Is this because ECM signatures are correlative but not causative with advancing malignancy, or can the matrix be used to dictate tumour phenotypes? Here, we will focus primarily on the direct targeting of matrisome components, rather than the targeting of TME cells to remodel the stroma. We will address efforts to both degrade the matrisome to support therapy and efforts to protect the matrix from degradation to prevent invasion and metastasis. The opposing need to prevent cancer cell egress while promoting the ingress of anti-tumour immune populations presents a unique dilemma when targeting the matrisome, which may underlie poor clinical success. The onus is on identifying how altering the matrisome composition can dictate beneficial tumour phenotypes, while maintaining tumour-suppressive functions.
5.1. Is Tumour Fibrosis Good or Bad?
Among the predominant fibrous proteins in the ECM are collagens, elastin, fibronectin, and laminins. Laminins are largely associated with basement membranes, playing key roles in tissue organisation, tumour invasion, and metastasis, as reviewed elsewhere [88,89,90]. The interstitial matrix is largely composed of collagen fibres, crosslinked with elastin fibres, and interspersed with varying levels of HA and proteoglycans (Figure 2). The general impact of high levels of desmoplasia on tumour prognosis is complex, but it is most often associated with poor outcomes in advanced cancer [91,92,93,94] (Figure 2); whether fibrosis is correlative with aggressive disease or causative remains an area of significant debate. Here, we will examine evidence for pro- and anti-tumour roles for central ECM components.
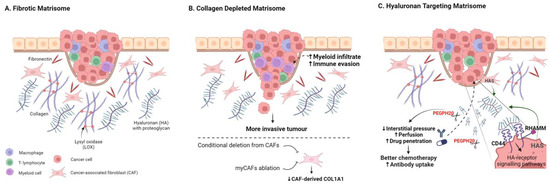
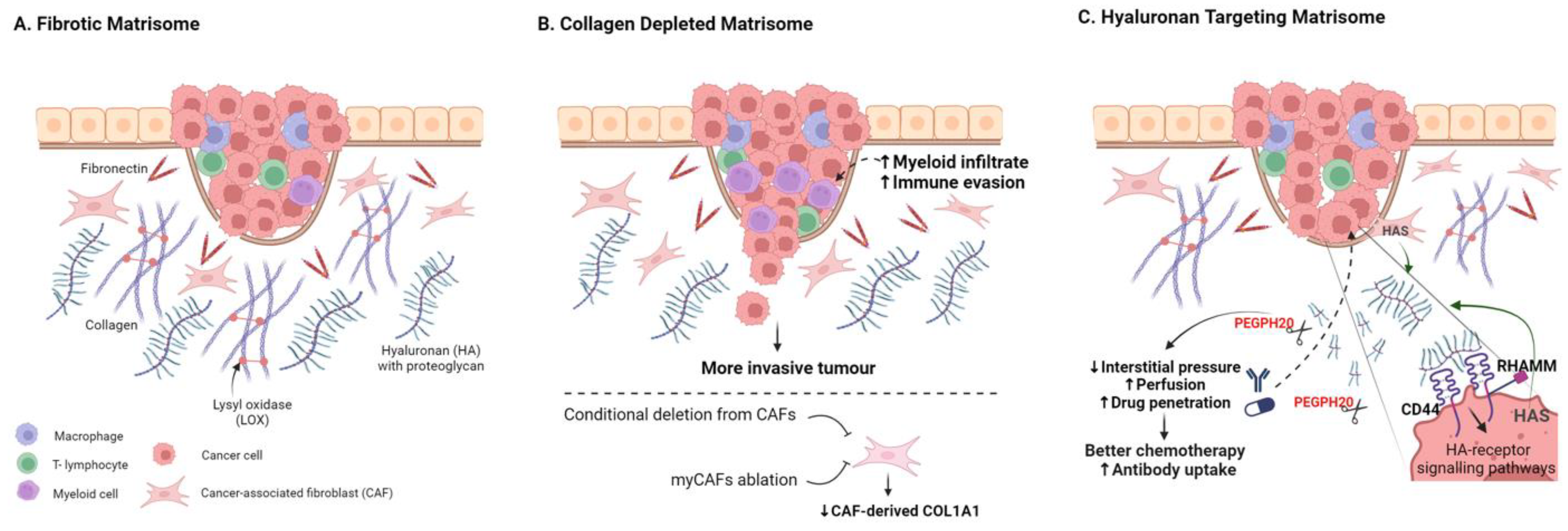
Figure 2.
Targeting the Fibrotic Tumour Matrisome. (A) Schematic highlighting key abundant matrisome components. (B) Targeting stromal-cell-derived collagen 1 fibrosis through conditional deletion of Col1a1 in CAFs or through CAF ablation can result in more invasive tumours and reduced anti-tumour immunity. (C) Targeting hyaluronic acid can enhance drug perfusion and therapy response in many pre-clinical models, with numerous trials underway to assess clinical utility. Created with BioRender.com.
Collagen. The most abundant protein component of the TME, and indeed the human body, is type 1 collagen, and collagen-rich tumour fibrosis has long been considered as a barrier to therapy, particularly in fibrotic tumours such as PDAC [18,75,95,96]. Furthermore, aligned collagen has been identified as a functional promoter of invasion and poor-prognosis biomarker in breast cancer [97,98,99,100]. Perhaps surprisingly, conditional deletion of Col1a1 from α-SMA-positive CAFs (α-SMA-Cre/Col1a1fl/fl) crossed with a KrasG12D/+; Trp53frt/frt; Pdx1-Flp (KPPF) mouse model of pancreatic cancer resulted in reduced fibrosis but more aggressive tumours, lower survival, and myeloid suppression of CD8+ T-cells (Figure 2B) [101]. In various additional mouse models of PDAC, other myofibroblast CAF-targeting approaches have also led to reduced tumour fibrosis, often resulting in more aggressive and immune-suppressive tumours [19,20,21,24,102]. A tumour-restraining role for CAF-derived type 1 collagen has also been reported for growth of PDAC and CRC liver metastases; collagen 1 was shown to physically restrain tumours using a hepatic stellate cell (HSC)-specific Col1a1fl/fl conditional knockout model [102]. The emerging consensus thus suggests a tumour-restraining role for CAF-derived type 1 collagen in multiple mouse models [101,102].
In the same liver metastasis study, Bhattacharjee et al. additionally showed that CAF-derived HA and HGF were pro-tumourigenic and that depletion of α-SMA-positive HS-derived CAFs resulted in slower growth and enhanced survival [102]. This reveals tumour-promoting and -suppressing roles for the same CAF population to highlight the challenges associated with CAF ablation strategies [102]. Adding further nuance, targeted deletion of Col1a1 specifically from the pancreatic cancer cell compartment in KPPC; Col1pdxKO mice (LSL-KrasG12D/+; Trp53loxP/loxP; Pdx1-CreCol1a1loxP/loxP) increased mouse survival by suppressing the formation of oncogenic Col1a1 homotrimers [103]; moreover, Col1 targeting in this context enhances T-cell infiltration and checkpoint response while also impacting the tumour microbiome. Tumour-cell-derived Col1a1 has also been reported as a metastatic driver in breast and liver cancer models [32,97,104]. Cell-type-specific conditional targeting of Col1a1 in mouse models of breast and other solid cancers are yet to be reported. Thus, while collagen 1 genes feature in a broad number of poor-prognosis signatures, they currently do not readily represent a tractable therapeutic target.
While type-1 collagen is the most abundant collagen, at least 28 collagen types have been described with diverse functions in cancer [105]. Some consensus emerges for some collagen types, including type 6 and 11. Col11A1 is a key upregulated component of the Matrix Index, which predicts metastasis across many tumour types, including ovarian, pancreatic, and breast cancers [8]. Col11a1+ CAFs, which co-express LRRC15, are also associated with tumour progression in scRNA-seq and tissue-staining studies [106,107]. Other studies have identified COL11A1 as a functional driver of cancer progression and prognostic biomarker [108,109,110]. A similar body of the literature is emerging supporting pro-tumourigenic functions for COLVI in breast, ovarian [111], pancreatic [112], lung [113], and brain [114] cancers. Clinical intervention here is yet to be explored.
In summary, while fibrosis is a common feature of many aggressive treatment-resistant tumour, and a known activator of oncogenic mechanotransduction signalling axes, relief of collagen-rich fibrosis alone may promote rather than improve outcome.
Fibronectin. PhysiologicallyC, fibronectin is produced by myofibroblasts at sites of injury; as cancers can be considered as “wounds that never heal”, FN is generally upregulated in the tumour ECM, where high expression is often associated with invasive disease and poor prognosis. As FN is upregulated in many cancers, it has been exploited primarily in cancer imaging and for the targeted delivery of therapies; numerous antibodies, peptides, and modified ligands have been used to deliver isotopes, chemotherapeutics, and immunomodulators to tumours in pre-clinical models (reviewed [115,116]). Fibronectin has also emerged as an attractive target for guiding CAR-T therapy, particularly focused on cancer-specific splice variants [117,118,119,120,121]. Mechanistically, fibronectin can promote both growth and invasion through engagement of α5β1 and αv-class integrins, and this provides therapeutic opportunities [122,123,124,125]. Several monoclonal antibodies (mAbs) preventing integrin interaction with FN, including those against αvβ6 and α5β1, have shown excellent pre-clinical promise in breast and glioma models, respectively [126,127], highlighting the value of this axis. Surprisingly, few studies have been published examining the conditional deletion of FN in pre-clinical tumour models, particularly given the well-documented roles in promoting invasive disease. Interestingly, conditional deletion of circulating plasma fibronectin, through targeted deletion in hepatocytes, reduces MDA-MB-231 bone metastatic colonisation in nude mice [128]. Suppression of FN expression in MDA-MB-231 has also been shown to reduce bone metastases [129]. Suppression of FN expression by microRNAs also limits glioma progression in a mouse model, adding further evidence of therapeutic potential [89]. Conditional deletion from CAFs—the predominant source of fibronectin in many tumours—remains to be reported. The importance of matrix proteases in the degradation of FN will be addressed later in this review.
5.2. Degrading Hyaluronic Acid
A direct ECM depletion approach which has gained more traction has been the targeting of hyaluronic acid (HA, Hyaluronan), an abundant glycosaminoglycan ECM component in solid tumours, which can be a dominant contributor to tumour stiffness, hydration, and interstitial pressure [18,130,131]. HA and its two main receptors, CD44 and RHAMM, have been implicated in inflammation in both physiological settings and cancer [132,133]. Dysregulation of HA synthase enzymes (HAS1-3) has also been reported for many cancers including pancreatic, breast, and prostate cancer [134,135,136,137]. Numerous studies have implicated high levels of HA as a prognostic indicator and functional driver of poor outcome in solid tumours (Figure 2C) [138,139,140,141,142]. Importantly, HA can be degraded by hyaluronidases, and the use of exogenous hyaluronidases as drugs to augment cancer therapy has a long history, although early translation was limited by poor drug characteristics [143,144]. PEGylated human hyaluronidase (PEGPH20) was developed to improve pharmacokinetics, showing activity in high-HA preclinical prostate tumour xenografts [145]. PEGPH20 also sensitised pre-clinical PDAC models to chemo-, radio-, and immunotherapy approaches [10,18,145,146,147]. Increased chemotherapy uptake and response in preclinical ovarian cancer mouse models have also been reported [148]. Phase II and phase III trials combining PEGPH20 with chemotherapy to treat advanced PDAC have followed; although failing to meet primary endpoints, some response improvement was observed in HA-rich tumours [149,150,151,152]. Additional combinatorial trials are underway for PDAC and other solid cancers [153]; stratifying for tumours with high-HA content is likely to be critical to improve trial success [145,149,150].
In breast cancer, PEGPH20 has been shown to improve HER2-targeting antibody therapy response [154] and anti-PDL1antibody uptake and therapy responses [155] in mouse xenograft models. In addition to facilitating therapy uptake, depletion of HA may also limit inflammatory HA-CD44 signalling. In breast cancer xenograft models, depletion of HA or loss of CD44 were both shown to reduce CCL2 production in vitro and loss of CD44 impaired macrophage recruitment and tumour induction in vivo [156]. A phase II metastatic breast cancer trial combining Eribulin with PEGPH20 was terminated (NCT02753595) and to date PEGPH20 trial results in breast cancer are yet to be reported. Although translation has been challenging, the positive impact of PEGPH20 on immune infiltration, drug perfusion, and pre-clinical tumour model drug responses provides encouragement that modifying the biophysical properties of the stroma can enhance clinical impact.
6. Matrix Crosslinking
In addition to changes in composition, the stiffness of the ECM is critically regulated by crosslinking, with the lysyl-oxidase (LOX) family of collagen crosslinkers playing a dominant role. LOX and LOX-like (LOXL) enzymes catalyse the crosslinking of collagen and elastin, and this promotes matrix stiffening in pathological and malignant settings. The targeting of LOX enzymes, using function-blocking mAbs, has long shown promise in normalising a pathologically stiff ECM in mouse xenografts and fibrosis models [157,158]; the targeting of LOX can reduce stiffness, desmoplasia, invasion, and metastasis. High levels of LOX family members have also been functionally implicated in disease progression and correlated with poor outcome in many fibrotic tumour types, including pancreatic, breast, and lung cancer [159,160,161,162,163,164,165].
In a pre-clinical MDA-MB-231 xenograft model of TNBC, mAb targeting of LOX was shown to limit chemotherapy resistance by improving drug penetration and through regulation of FN1 and ITGAV expression. Upregulation of LOX, FN1, and ITGAV is also seen in relapsed TNBC patients, providing data to support translation [166]. Rossow et al. further demonstrated that LOX enzymes limit drug access in a range of solid tumour xenograft models, using over-expression studies and the pan-LOX small-molecule inhibitor bAPN [167]. Nanovesicle delivery of anti-LOX mAbs has also been validated in MDA-MB-231 xenografts [168]. Small-molecule inhibitors of LOXL2 have also been developed, with the potential to reduce MDA-MB-231 xenograft metastasis and tissue fibrosis [169,170,171].
As with other interventions to supress stromal fibrosis, conflicting results have emerged in pancreatic cancer models. LOX targeting with mAbs in KPC mice potentiated gemcitabine responses, reduced metastasis, and extended survival; mechanistically, these enhanced responses were associated with enhanced myeloid cell infiltration, reduced fibrillar collagen, and improved vasculature, rather than enhanced levels of gemcitabine delivery [172]. Genetic deletion or cancer cell overexpression of LOXL2 in KPC and KC autochthonous models also support a tumour- and metastasis-promoting role [173]. In contrast, LOX targeting in a syngeneic orthotopic PDAC model resulted in more aggressive disease [174], in line with distinct CAF depletion and collagen-targeting strategies [19,20]. bAPN has, however, been shown to facilitate drug perfusion in PDAC orthotopic xenografts in nude mice [175]. A clinical trial combining the LOXL2-targeting mAb Simtuzumab with gemcitabine did not, however, improve outcome, though this may reflect advanced disease stage and a lack of patient stratification (NCT01472198 [176]). Simtuzumab has also been explored clinically for treatment of pulmonary and liver fibrosis, though results remain disappointing [177,178,179,180].
7. Matrix Proteases
Metalloproteinases, including MMPs, ADAMs, and ADAMTSs, constitute large families of proteases which can degrade various components of the ECM (ECM). They have been broadly implicated as facilitators of invasion and metastasis, by degrading the basement membrane and stromal desmoplastic matrix, and as such have been the subject of extensive drug discovery programs. To date, clinical translation has been largely unsuccessful, which has largely been attributed to a lack of inhibitor specificity, combined with tumour-suppressive functions and the normal physiological roles of related family members. Despite the disappointing results from numerous clinical trials, hope remains that protease-targeting drugs can be of use in cancer (clinical trials reviewed by Coussens et al. [181] and Cathcart et al. [182]). Roles for specific MMPs are nuanced and show both disease- and context-dependent functions. Here, we will cover some recent examples where antagonistic behaviours of close protease family members are likely to confound therapeutic targeting with non-selective small molecules. Alternative strategies, such as mAb or gene-suppression approaches, may present alternative isoform-specific strategies.
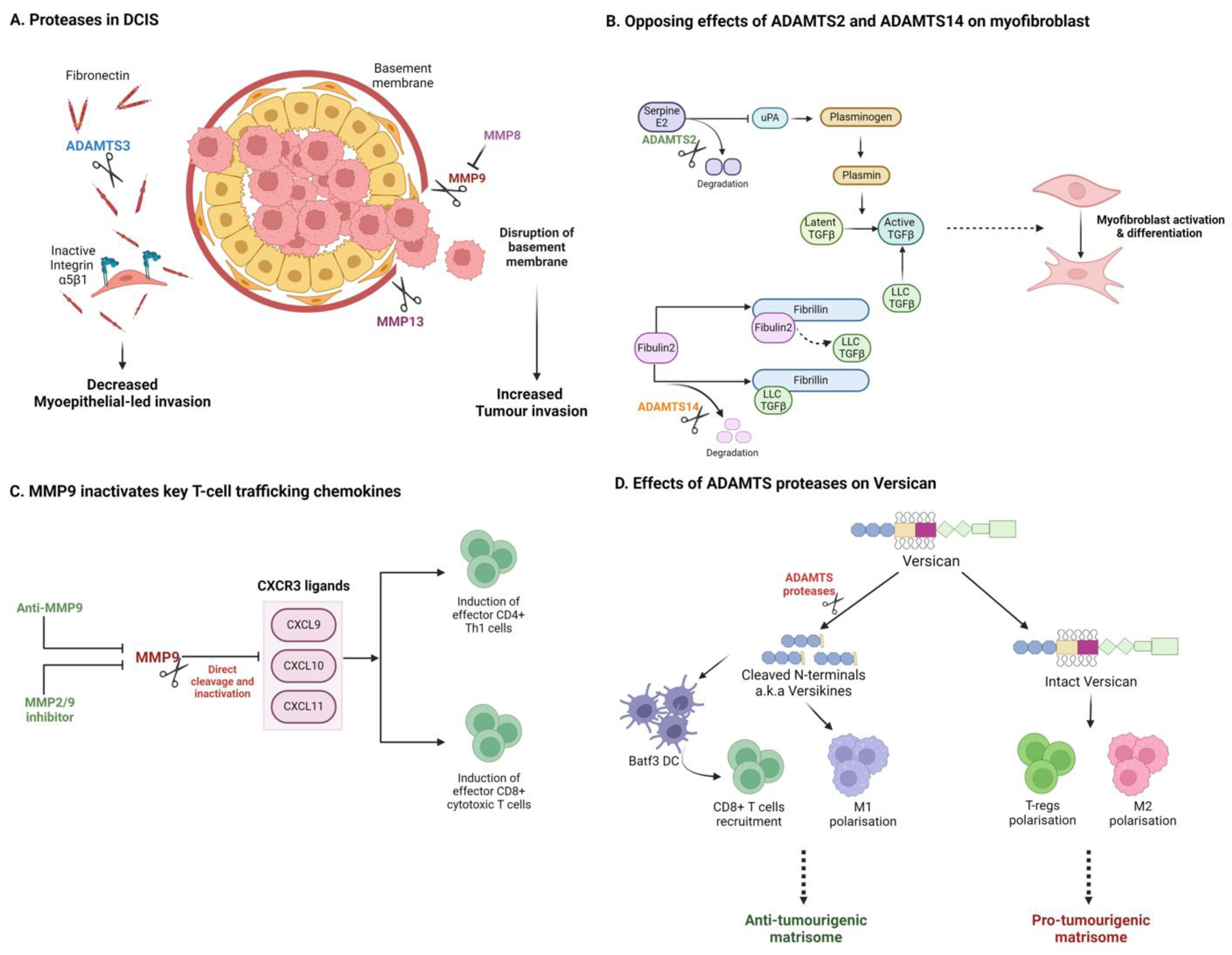
Regarding matrix degradation and remodelling, distinct matrix proteases can either support or impede tumour invasion. While proteases may facilitate invasion, distinct family members which degrade or remodel a pathogenic pro-invasive matrisome can also act as tumour suppressors. DCIS provides some excellent illustrative examples. In DCIS, luminal tumour cells proliferate within breast ducts but are restrained from invading the surrounding stroma by myoepithelial cells and basement membrane surrounding the ducts. In progressive disease, disruption of the basement membrane has been associated with upregulation of pro-invasive MMPs (e.g., MMP9 and MMP13) by stromal cells, including myoepithelial cells and CAFs (Figure 3A) [183,184,185,186,187]. In many DCIS cases, however, progression to invasive carcinoma does not occur. Discriminating between indolent disease and those cases likely to progress to invasive carcinoma remains an unmet clinical demand. Accumulation of fibronectin has been associated with disease progression and poor outcome, potentially by promoting myoepithelial-led invasion [122,188,189,190]. A recent study demonstrated that ADAMTS3, produced by myoepithelial cells, can restrict invasion by degrading fibronectin in a DCIS model [191]. Loss of ADAMTS3 supports the accumulation of fibronectin to promote an integrin-α5β1-directed myoepithelial-led invasion (Figure 3A). Similarly, MMP8, which is lost in invasive disease, has been shown to act as a tumour suppressor through promotion of adhesion and suppression of MMP-9 function [192]. Sparing proteases responsible for fibronectin degradation (e.g., ADAMTS3) may thus be desirable in a therapeutic setting. The challenge of developing inhibitors to discriminate between closely related proteases with antagonistic functions can help explain the poor performance of broad MMP inhibitors in clinical trials.
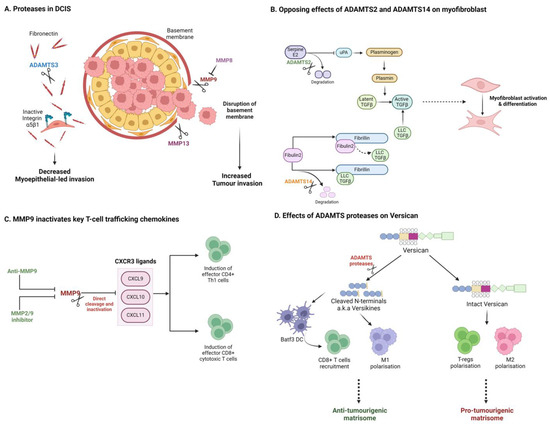
Figure 3.
Matrix proteases can impact tumour progression through diverse and opposing mechanisms. (A) In DCIS, distinct proteases promote or suppress tumour invasion, dependent on their respective substrates. (B) Closely related ADAMTS isoforms can elicit opposing effects on TGFβ and CAF activation in PDAC. (C) Proteases can alter the immune microenvironment by degrading key cytokines and chemokines. (D) Degradation of matrix components can generate tumour and immune modulating matrikines such as versican-derived versikine. Created with BioRender.com.
In pancreatic cancer, the desmoplastic stroma is considered a barrier to therapy, but numerous recent studies also suggest a robust tumour-suppressive function, where suppression of fibrosis has been associated with invasive and metastatic disease. A recent report by Carter et al. highlights antagonistic roles or the closely related collagenases, ADAMTS2 and ADAMTS14 in the promotion of pancreatic cancer invasion (Figure 3B) [193]. Suppression of ADAMTS2 was found to limit pancreatic stellate cell (PSC) activation, while suppression of ADAMTS14 promoted PSC myofibroblast differentiation and cancer cell invasion in heterotypic 3D models. Mechanistically, these two proteases differentially target the distinct matrisome substrates, serpinE2 and fibulin 2, to regulate TGFβ availability. Of note, antagonistic regulation of Fibulin 2 cleavage by ADAMTS5 and ADAMTS12 has been previously reported to promote invasive behaviour in breast cancer cells, potentially through production of bioactive Fibulin 2 proteolytic products [194]. These examples illustrate nuanced, substrate-specific signalling capability for this family of enzymes, akin to behaviours observed in intracellular signalling cascades.
8. Matrix Protease Regulation of the Immune Landscape
While protease modulation of the TME may be pro-invasive in many scenarios, it is also clear that MMPs and other matrix degrading proteases can also play tumour-suppressive roles, not least through their importance in enabling immune cell access [195]. While the matrix can act as a protective barrier preventing tumour cell escape, impeding immune cell access also provides a safe haven for cancer cells. Ager et al. demonstrated that anti-MMP14 mAb DX-2400 alone was able to enhance anti-tumour immunity to impede tumour growth and enhance radiotherapy response in 4T1 nude mouse xenografts and syngeneic E0771 immune-competent models [196]. Increased M1 macrophage infiltration, with downregulation of TGFβ and SMAD pathways, suggests remodelling of the immune-suppressive stroma. Similar results were reported for a second MMP-14-targeting antibody using MDA-MB-231 xenografts and the syngeneic 4T1/BALB-c metastasis model [197]. Targeting MMP-9 has also showed promise for enhancing immunity. Anti-MMP9 was found to slow growth of HER2+ mouse orthotopic breast tumours, while combination with anti-PDL1 enhanced effector T-cell infiltration [198]. Intriguingly, the authors demonstrated that MMP9 was capable of directly cleaving and inactivating T-cell chemokines CXCL9/10/11, and that inhibition with anti-MMP9 could enhance CXCL10 in vivo through this mechanism (Figure 3C) [198]. This again highlights substrate-specific roles for matrix proteases, beyond remodelling the ECM. Owyonng et al. separately confirmed that anti-MMP-9 can enhance CD8+ T-cell content in the MMTV-PyMT breast cancer model [199]. Excitingly, in mouse lung and melanoma models, SB-3CT, a small-molecule MMP2/9 inhibitor, has been shown to enhance PD-1 or CTLA-4 blockade responses by promoting anti-tumour immunity [200].
The ADAMTS proteases have also been associated with immune modulation and in particular with inflammation, although cancer studies remain somewhat limited [201]. ADAMTS1 acts as a tumour promoter in the genetic MMTV-PyMT model, with ADAMTS1 knockout mice having reduced tumour burden, metastasis, and prolonged survival [202], associated with enhanced leukocyte infiltration and cytotoxic immune signatures. Tumour-promoting roles for ADAMTS1 have also been reported in melanoma, and ADAMTS1 deficiency can promote a pro-inflammatory landscape in melanoma [203,204]. Similarly, ADAMTS4 has been associated with high-macrophage and tumour-promotion content in a subcutaneous CRC xenograft model [205]. In contrast, a tumour-suppressor role has also been reported for ADAMTS1 in breast cancer, related primarily in this study to the regulation of tumour vasculature [206].
Among the substrates of ADAMTS proteases, the proteoglycan versican has shown most promise as an immune modulator [207,208]. Interestingly, ADAMTS-derived cleavage fragments of versican (e.g., versikine [209]) have been shown to modulate key aspects of the tumour immune landscape, including modulation of DCs and CD8+ T-cell recruitment in myeloma, lung cancer, and CRC (Figure 3D) [210,211,212,213]. Recent work shows that versikine enhances DC abundance and activity in a Lewis Lung Carcinoma (LLC) model [214]. In CRC, VCAN proteolysis and versikine levels are associated with robust CD8+ T-cell infiltration, also likely driven through modulation of DC numbers and activation status [210]. Furthermore, versikine levels have been correlated with ICB (pembrolizumab) response in a Phase 1 clinical trial for metastatic CRC (NCT02837263). The action of versikine, and other ‘matrikine’ cleavage products, once again reveals complex regulator roles for specific protease–ECM interactions. Matrikine cleavage products with biological function in cancer have been reported for many matrix components including elastin [215,216,217,218]. Matrikine production during remodelling of the ECM is an area of matrix biology which is sure to gather pace with the application of novel degradomics technologies [219,220]. Examples provided in this review suggest complex signalling networks will emerge in the extracellular space, perhaps under-appreciated due to our historic reliance on limited tissue culture models. Therapeutically, the presence of such extra-cellular signalling networks opens numerous opportunities for the development of biological therapies (e.g., mAbs).
Together, these findings suggest isoform-specific protease inhibition may be of value in combination with ICB in a variety of solid tumour settings. Proteases can act through novel immunomodulatory mechanisms beyond depletion of matrix components, including the generation and degradation of immunostimulatory chemokines and matrikines.
9. Matrisome Interventions to Sculpt the Immune Landscape
A recurrent theme in the majority of promising therapeutic interventions targeting the tumour matrisome is promotion of anti-tumour immunity or sensitisation to ICB inhibitors. Attempts to degrade or suppress the collagen-rich matrix have largely failed, due to induction of an immune-suppressive, myeloid-rich TME, although in some cases this does not tell the whole story. Multiple mouse model studies on PDAC suggest that more aggressive inflammatory tumours, promoted by stromal suppression, may be sensitised to ICB inhibitors. As an example, ablation of α-SMA CAFs in a genetic PDAC model (PKT: Ptf1acre/+; LSL-KrasG12D/+; Tgfbr2flox/flox) suppressed fibrosis and accelerated tumour growth, but it also was sensitised to anti-CTLA-4 therapy, associated with increased inflammation. Similarly, targeting the Hedgehog pathway to limit stromal activation has been shown to promote aggressive inflammatory pancreatic tumour growth [20]; Hedgehog inhibitors, nonetheless, also show promise in combination with chemotherapy or immunotherapy regimens in pancreatic [6], ovarian [9,221], and other solid cancers. These approaches are yet to make an impact clinically (reviewed [7,86,222]). Likewise, preclinical successes with hyaluronidase PEGPH20 have been associated with promotion of anti-tumour immunity and ICB responses in pancreatic and breast models [147,155].
While numerous matrisome signatures have been shown to correlate with prognosis and ICB response, increasingly, the evidence supports direct causative functions. The ability of the matrisome to dictate immune cell phenotypes has elegantly been demonstrated using decellularised-tumour-derived matrices. Puttock [223] et al. adopted an ex vivo approach using a decellularised ECM from mouse ovarian cancer metastases to examine immune modulation. Monocytes cultured on a decellularised ECM were found to differentiate into immunoregulatory M0 phenotype macrophages. Furthermore, these matrisome-educated macrophages could promote T-cell proliferation and expression of T-cell activation markers [223].
Versican appears in several poor outcome matrix signatures, including the Matrix Index and C-ECM [8,25]. Versican interacts with many of the ECM components addressed in this review, including HA, FN1, and MMPs, and several studies have delineated roles in the modulation of myeloid cells and inflammation [207,224]. The versikine studies described above indicate a positive functional impact on the ability of DCs to prime adaptive immunity and promote ICB response. Versican can also directly activate macrophages to stimulate TNF-α secretion and lung tumour growth in the LCC model [225]. Versican modulates TAM phenotype in mesothelioma, where high levels correlate with poor outcome [226]. Mechanistically, versican-deficient tumours in mice are less aggressive and have fewer macrophages and neutrophils. Importantly, versican-deficient mesothelioma cells polarised co-cultured macrophages to an M1 phenotype, suggesting VCAN targeting may promote a more favourable immune landscape. Versican may also act as a direct regulator of T-cell trafficking, likely in complex with HA [227]. Indeed, versican predominantly binds to HA in tumours and degradation of the HA matrix with PEGPH20 will act in part by altering versican and versikine distribution, with repercussions on myeloid and lymphocyte recruitment [132,227,228].
10. Summary
The concept that tissue composition, and not just cancer cell properties, is critical in deciding cancer outcome was first proposed in 1889 when Paget described his seed and soil theory of metastasis [229,230,231]. The matrix is now understood to regulate all stages and hallmarks of cancer, from initiation, growth, and metastasis to disease recurrence and therapy resistance. With the emergence of diverse omics technologies, our ability to map the evolution of the matrisome in time and space, as cancers progress, is uncovering a multitude of mechanisms and therapeutic opportunities. Most matrix-directed therapeutic approaches have focussed on attempting to limit cancer spread or improve therapeutic responses; in many cases, these two aims are at odds with one another. The same matrix which limits drug or immune cell access can also suppress tumourigenesis by restraining cancer growth and spread. Most successes in pre-clinical models from diverse cancer types have been associated with improving anti-tumour immunity, although mechanisms are unsurprisingly diverse. Regulating the matrisome to sculpt the immune landscape has enormous promise in combination therapies. Significant challenges remain with clinical translation, not least with spatial heterogeneity within tumours and between patients. Better stratification, examining both tumour and stroma, will help unlock the promise of matrisome interventions, particularly to reawaken anti-tumour immunity.
Author Contributions
All authors contributed to the focus of the review and writing of the manuscript. S.M.A.J. and J.C.H. made the figures with input from A.J.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Majlis Amanah Rakyat (MARA) Graduate Excellence Program (GrEP) for S.M.A.J., the MRC (MR/X018997/1) for J.C.H. and a HEFCE award for A.J.M.C. Research is also supported by Cancer Research UK Centre Grants to Barts Cancer Institute (C355/A25137) and the City of London Centre (C7893/A26233).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
ADAMTS—A disintegrin and metalloproteinase with thrombospondin motifs
bAPN—β-Aminopropionitrile
CAFs—Cancer-associated fibroblasts
COL—Collagen
COMP—Cartilage oligomeric matrix protein
CRC—Colorectal cancer
ECM—Extracellular matrix
FAK—Focal adhesion kinase
FN—Fibronectin
HA—Hyaluronic acid
HAS—Hyaluronic acid synthase
HGF—Hepatocyte growth factor
HSC—Hepatic stellate cell
iCAF—Inflammatory cancer-associated fibroblasts
ICB—Immune checkpoint blockade
ITG—Integrin
KO—Knockout
KPC—KrasG12D/+; Trp53R172H/+; Pdx-Cre mouse model
KC—KrasG12D/+; Pdx-Cre mouse model
LOX—Lysyl oxidase
LOXL2—Lysyl oxidase-like 2
LRRC15—Leucine-rich repeat containing
MI—Matrix Index
MMP—Matrix metalloproteinase
MMTV-PyMT—Mouse mammary tumour virus–polyoma middle tumour antigen
MSI—Microsatellite instability
myCAF—Myofibroblast cancer-associated fibroblasts
PDAC—Pancreatic ductal adenocarcinoma
PD1—Programmed death 1
PD-L1—Programmed death ligand 1
PEGPH20—PEGylated human hyaluronidase
PKN2—Protein kinase N2
PSC—Pancreatic stellate cell
RHAMM—Receptor for Hyaluronan Mediated Motility
TGF-b—Transforming Growth Factor-b
TME—Tumour microenvironment
TNBC—Triple-negative breast cancer
VCAN—Versican
References
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Naba, A.; Clauser, K.R.; Hoersch, S.; Liu, H.; Carr, S.A.; Hynes, R.O. The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteom. MCP 2012, 11, M111.014647. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O.; Naba, A. Overview of the matrisome—An inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 2012, 4, a004903. [Google Scholar] [CrossRef]
- Shao, X.; Taha, I.N.; Clauser, K.R.; Gao, Y.T.; Naba, A. MatrisomeDB: The ECM-protein knowledge database. Nucleic Acids Res. 2020, 48, D1136–D1144. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, Y.; Zhang, S.; Wang, X.; Dou, H.; Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef] [PubMed]
- Menezes, S.; Okail, M.H.; Jalil, S.M.A.; Kocher, H.M.; Cameron, A.J. Cancer-associated fibroblasts in pancreatic cancer: New subtypes, new markers, new targets. J. Pathol. 2022, 257, 526–544. [Google Scholar] [CrossRef] [PubMed]
- Pearce, O.M.T.; Delaine-Smith, R.M.; Maniati, E.; Nichols, S.; Wang, J.; Bohm, S.; Rajeeve, V.; Ullah, D.; Chakravarty, P.; Jones, R.R.; et al. Deconstruction of a Metastatic Tumor Microenvironment Reveals a Common Matrix Response in Human Cancers. Cancer Discov. 2018, 8, 304–319. [Google Scholar] [CrossRef]
- Olive, K.P.; Jacobetz, M.A.; Davidson, C.J.; Gopinathan, A.; McIntyre, D.; Honess, D.; Madhu, B.; Goldgraben, M.A.; Caldwell, M.E.; Allard, D.; et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009, 324, 1457–1461. [Google Scholar] [CrossRef]
- Jacobetz, M.A.; Chan, D.S.; Neesse, A.; Bapiro, T.E.; Cook, N.; Frese, K.K.; Feig, C.; Nakagawa, T.; Caldwell, M.E.; Zecchini, H.I.; et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut 2013, 62, 112–120. [Google Scholar] [CrossRef]
- Colpaert, C.G.; Vermeulen, P.B.; Fox, S.B.; Harris, A.L.; Dirix, L.Y.; Van Marck, E.A. The presence of a fibrotic focus in invasive breast carcinoma correlates with the expression of carbonic anhydrase IX and is a marker of hypoxia and poor prognosis. Breast Cancer Res. Treat. 2003, 81, 137–147. [Google Scholar] [CrossRef]
- Van den Eynden, G.G.; Smid, M.; Van Laere, S.J.; Colpaert, C.G.; Van der Auwera, I.; Bich, T.X.; van Dam, P.; den Bakker, M.A.; Dirix, L.Y.; Van Marck, E.A.; et al. Gene expression profiles associated with the presence of a fibrotic focus and the growth pattern in lymph node-negative breast cancer. Clin. Cancer Res. 2008, 14, 2944–2952. [Google Scholar] [CrossRef]
- Boyd, N.F.; Guo, H.; Martin, L.J.; Sun, L.; Stone, J.; Fishell, E.; Jong, R.A.; Hislop, G.; Chiarelli, A.; Minkin, S.; et al. Mammographic density and the risk and detection of breast cancer. N. Engl. J. Med. 2007, 356, 227–236. [Google Scholar] [CrossRef]
- Martin, L.J.; Boyd, N.F. Mammographic density. Potential mechanisms of breast cancer risk associated with mammographic density: Hypotheses based on epidemiological evidence. Breast Cancer Res. BCR 2008, 10, 201. [Google Scholar] [CrossRef]
- Ekbom, A.; McLaughlin, J.K.; Karlsson, B.M.; Nyren, O.; Gridley, G.; Adami, H.O.; Fraumeni, J.F., Jr. Pancreatitis and pancreatic cancer: A population-based study. J. Natl. Cancer Inst. 1994, 86, 625–627. [Google Scholar] [CrossRef]
- Duell, E.J.; Lucenteforte, E.; Olson, S.H.; Bracci, P.M.; Li, D.; Risch, H.A.; Silverman, D.T.; Ji, B.T.; Gallinger, S.; Holly, E.A.; et al. Pancreatitis and pancreatic cancer risk: A pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4). Ann. Oncol. 2012, 23, 2964–2970. [Google Scholar] [CrossRef]
- Kalaitzakis, E.; Gunnarsdottir, S.A.; Josefsson, A.; Bjornsson, E. Increased risk for malignant neoplasms among patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2011, 9, 168–174. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Cuevas, C.; Chang, A.E.; Goel, V.K.; Von Hoff, D.D.; Hingorani, S.R. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell 2012, 21, 418–429. [Google Scholar] [CrossRef]
- Ozdemir, B.C.; Pentcheva-Hoang, T.; Carstens, J.L.; Zheng, X.; Wu, C.C.; Simpson, T.R.; Laklai, H.; Sugimoto, H.; Kahlert, C.; Novitskiy, S.V.; et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 2014, 25, 719–734. [Google Scholar] [CrossRef]
- Rhim, A.D.; Oberstein, P.E.; Thomas, D.H.; Mirek, E.T.; Palermo, C.F.; Sastra, S.A.; Dekleva, E.N.; Saunders, T.; Becerra, C.P.; Tattersall, I.W.; et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014, 25, 735–747. [Google Scholar] [CrossRef]
- Lee, J.J.; Perera, R.M.; Wang, H.; Wu, D.C.; Liu, X.S.; Han, S.; Fitamant, J.; Jones, P.D.; Ghanta, K.S.; Kawano, S.; et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc. Natl. Acad. Sci. USA 2014, 111, E3091–E3100. [Google Scholar] [CrossRef]
- Kai, F.; Drain, A.P.; Weaver, V.M. The Extracellular Matrix Modulates the Metastatic Journey. Dev. Cell 2019, 49, 332–346. [Google Scholar] [CrossRef]
- Brechbuhl, H.M.; Barrett, A.S.; Kopin, E.; Hagen, J.C.; Han, A.L.; Gillen, A.E.; Finlay-Schultz, J.; Cittelly, D.M.; Owens, P.; Horwitz, K.B.; et al. Fibroblast subtypes define a metastatic matrisome in breast cancer. JCI Insight 2020, 5, e130751. [Google Scholar] [CrossRef]
- Murray, E.R.; Menezes, S.; Henry, J.C.; Williams, J.L.; Alba-Castellon, L.; Baskaran, P.; Quetier, I.; Desai, A.; Marshall, J.J.T.; Rosewell, I.; et al. Disruption of pancreatic stellate cell myofibroblast phenotype promotes pancreatic tumor invasion. Cell Rep. 2022, 38, 110227. [Google Scholar] [CrossRef]
- Chakravarthy, A.; Khan, L.; Bensler, N.P.; Bose, P.; De Carvalho, D.D. TGF-beta-associated extracellular matrix genes link cancer-associated fibroblasts to immune evasion and immunotherapy failure. Nat. Commun. 2018, 9, 4692. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Wishart, A.L.; Conner, S.J.; Guarin, J.R.; Fatherree, J.P.; Peng, Y.; McGinn, R.A.; Crews, R.; Naber, S.P.; Hunter, M.; Greenberg, A.S.; et al. Decellularized extracellular matrix scaffolds identify full-length collagen VI as a driver of breast cancer cell invasion in obesity and metastasis. Sci. Adv. 2020, 6, eabc3175. [Google Scholar] [CrossRef]
- Nolan, J.; Mahdi, A.F.; Dunne, C.P.; Kiely, P.A. Collagen and fibronectin promote an aggressive cancer phenotype in breast cancer cells but drive autonomous gene expression patterns. Gene 2020, 761, 145024. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Zhu, B.; Qiu, Z.; Lin, Z. Systematic identification of the key candidate genes in breast cancer stroma. Cell. Mol. Biol. Lett. 2018, 23, 44. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, C.; Ye, Y.; Wang, Z.; He, Y.; Li, Y.; Mao, H. High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncol. Lett. 2020, 19, 93–102. [Google Scholar] [CrossRef]
- Zhang, X.X.; Luo, J.H.; Wu, L.Q. FN1 overexpression is correlated with unfavorable prognosis and immune infiltrates in breast cancer. Front. Genet. 2022, 13, 913659. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.P.; Chang, H.L.; Bamodu, O.A.; Yadav, V.K.; Huang, T.Y.; Wu, A.T.H.; Yeh, C.T.; Tsai, S.H.; Lee, W.H. Collagen 1A1 (COL1A1) Is a Reliable Biomarker and Putative Therapeutic Target for Hepatocellular Carcinogenesis and Metastasis. Cancers 2019, 11, 786. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Lin, T.; Wang, Y.; Liu, B.; Wang, M. Collagen type 1 alpha 1 chain is a novel predictive biomarker of poor progression-free survival and chemoresistance in metastatic lung cancer. J. Cancer 2021, 12, 5723–5731. [Google Scholar] [CrossRef] [PubMed]
- Geng, Q.; Shen, Z.; Li, L.; Zhao, J. COL1A1 is a prognostic biomarker and correlated with immune infiltrates in lung cancer. PeerJ 2021, 9, e11145. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, S.; Wang, X.; Liu, H.; Zhou, X.; Liu, H. COL1A1 Is a Potential Prognostic Biomarker and Correlated with Immune Infiltration in Mesothelioma. Biomed. Res. Int. 2021, 2021, 5320941. [Google Scholar] [CrossRef]
- Jiang, K.; Liu, H.; Xie, D.; Xiao, Q. Differentially expressed genes ASPN, COL1A1, FN1, VCAN and MUC5AC are potential prognostic biomarkers for gastric cancer. Oncol. Lett. 2019, 17, 3191–3202. [Google Scholar] [CrossRef] [PubMed]
- Papanicolaou, M.; Parker, A.L.; Yam, M.; Filipe, E.C.; Wu, S.Z.; Chitty, J.L.; Wyllie, K.; Tran, E.; Mok, E.; Nadalini, A.; et al. Temporal profiling of the breast tumour microenvironment reveals collagen XII as a driver of metastasis. Nat. Commun. 2022, 13, 4587. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Dominguez, C.X.; Muller, S.; Keerthivasan, S.; Koeppen, H.; Hung, J.; Gierke, S.; Breart, B.; Foreman, O.; Bainbridge, T.W.; Castiglioni, A.; et al. Single-Cell RNA Sequencing Reveals Stromal Evolution into LRRC15(+) Myofibroblasts as a Determinant of Patient Response to Cancer Immunotherapy. Cancer Discov. 2020, 10, 232–253. [Google Scholar] [CrossRef]
- Krishnamurty, A.T.; Shyer, J.A.; Thai, M.; Gandham, V.; Buechler, M.B.; Yang, Y.A.; Pradhan, R.N.; Wang, A.W.; Sanchez, P.L.; Qu, Y.; et al. LRRC15(+) myofibroblasts dictate the stromal setpoint to suppress tumour immunity. Nature 2022, 611, 148–154. [Google Scholar] [CrossRef]
- Hutton, C.; Heider, F.; Blanco-Gomez, A.; Banyard, A.; Kononov, A.; Zhang, X.; Karim, S.; Paulus-Hock, V.; Watt, D.; Steele, N.; et al. Single-cell analysis defines a pancreatic fibroblast lineage that supports anti-tumor immunity. Cancer Cell 2021, 39, 1227–1244. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; Bertos, N.; Pepin, F.; Sadekova, S.; Souleimanova, M.; Zhao, H.; Chen, H.; Omeroglu, G.; Meterissian, S.; Omeroglu, A.; et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 2008, 14, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Mlecnik, B.; Van den Eynde, M.; Bindea, G.; Church, S.E.; Vasaturo, A.; Fredriksen, T.; Lafontaine, L.; Haicheur, N.; Marliot, F.; Debetancourt, D.; et al. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. J. Natl. Cancer Inst. 2018, 110, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Pages, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef] [PubMed]
- Motta, R.; Cabezas-Camarero, S.; Torres-Mattos, C.; Riquelme, A.; Calle, A.; Figueroa, A.; Sotelo, M.J. Immunotherapy in microsatellite instability metastatic colorectal cancer: Current status and future perspectives. J. Clin. Transl. Res. 2021, 7, 511–522. [Google Scholar] [PubMed]
- Yuan, Y.; Zhu, Z.; Lan, Y.; Duan, S.; Zhu, Z.; Zhang, X.; Li, G.; Qu, H.; Feng, Y.; Cai, H.; et al. Development and Validation of a CD8+ T Cell Infiltration-Related Signature for Melanoma Patients. Front. Immunol. 2021, 12, 659444. [Google Scholar] [CrossRef] [PubMed]
- Daley, D.; Zambirinis, C.P.; Seifert, L.; Akkad, N.; Mohan, N.; Werba, G.; Barilla, R.; Torres-Hernandez, A.; Hundeyin, M.; Mani, V.R.K.; et al. Gammadelta T Cells Support Pancreatic Oncogenesis by Restraining alphabeta T Cell Activation. Cell 2016, 166, 1485–1499.e15. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Nishino, M.; Ramaiya, N.H.; Hatabu, H.; Hodi, F.S. Monitoring immune-checkpoint blockade: Response evaluation and biomarker development. Nat. Rev. Clin. Oncol. 2017, 14, 655–668. [Google Scholar] [CrossRef]
- Joyce, J.A.; Fearon, D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, L.; Zhang, H.; Zhao, Y.; Zhang, H.; Wu, S.; Xu, B. A risk model developed based on tumor microenvironment predicts overall survival and associates with tumor immunity of patients with lung adenocarcinoma. Oncogene 2021, 40, 4413–4424. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.J.; Yu, C.T.; Chen, L.; Wang, H.Y. Development of a TMErisk model based on immune infiltration in tumour microenvironment to predict prognosis of immune checkpoint inhibitor treatment in hepatocellular carcinoma. Brief. Bioinform. 2023, 24, bbad067. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, R.; He, D.; Zhang, Y.; Chen, H.; Zhu, Y.; Cheng, Y.; Liu, R.; Zhu, R.; Gong, L.; et al. CD8+ T effector and immune checkpoint signatures predict prognosis and responsiveness to immunotherapy in bladder cancer. Oncogene 2021, 40, 6223–6234. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell 2018, 33, 463–479.e10. [Google Scholar] [CrossRef] [PubMed]
- Bartoschek, M.; Oskolkov, N.; Bocci, M.; Lovrot, J.; Larsson, C.; Sommarin, M.; Madsen, C.D.; Lindgren, D.; Pekar, G.; Karlsson, G.; et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 2018, 9, 5150. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Oni, T.E.; Spielman, B.; Hao, Y.; Elyada, E.; Park, Y.; Preall, J.; Tuveson, D.A. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFbeta to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2019, 9, 282–301. [Google Scholar] [CrossRef] [PubMed]
- Grauel, A.L.; Nguyen, B.; Ruddy, D.; Laszewski, T.; Schwartz, S.; Chang, J.; Chen, J.; Piquet, M.; Pelletier, M.; Yan, Z.; et al. TGFbeta-blockade uncovers stromal plasticity in tumors by revealing the existence of a subset of interferon-licensed fibroblasts. Nat. Commun. 2020, 11, 6315. [Google Scholar] [CrossRef]
- Wu, S.Z.; Roden, D.L.; Wang, C.; Holliday, H.; Harvey, K.; Cazet, A.S.; Murphy, K.J.; Pereira, B.; Al-Eryani, G.; Bartonicek, N.; et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 2020, 39, e104063. [Google Scholar] [CrossRef]
- Steele, N.G.; Biffi, G.; Kemp, S.B.; Zhang, Y.; Drouillard, D.; Syu, L.; Hao, Y.; Oni, T.E.; Brosnan, E.; Elyada, E.; et al. Inhibition of Hedgehog Signaling Alters Fibroblast Composition in Pancreatic Cancer. Clin. Cancer Res. 2021, 27, 2023–2037. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tuveson, D.A. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol. Rev. 2021, 101, 147–176. [Google Scholar] [CrossRef]
- Affo, S.; Nair, A.; Brundu, F.; Ravichandra, A.; Bhattacharjee, S.; Matsuda, M.; Chin, L.; Filliol, A.; Wen, W.; Song, X.; et al. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell 2021, 39, 866–882.e11. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, Z.; Peng, G.; Xiao, Y.; Guo, J.; Wu, B.; Li, X.; Zhou, W.; Li, J.; Li, Z.; et al. Single-cell RNA sequencing reveals a pro-invasive cancer-associated fibroblast subgroup associated with poor clinical outcomes in patients with gastric cancer. Theranostics 2022, 12, 620–638. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, Y.; Hocine, H.R.; Gentric, G.; Pelon, F.; Bernard, C.; Bourachot, B.; Lameiras, S.; Albergante, L.; Bonneau, C.; Guyard, A.; et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020, 10, 1330–1351. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Canellas, A.; Hernando-Momblona, X.; et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Grunwald, B.T.; Devisme, A.; Andrieux, G.; Vyas, F.; Aliar, K.; McCloskey, C.W.; Macklin, A.; Jang, G.H.; Denroche, R.; Romero, J.M.; et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell 2021, 184, 5577–5592.e18. [Google Scholar] [CrossRef]
- Ligorio, M.; Sil, S.; Malagon-Lopez, J.; Nieman, L.T.; Misale, S.; Di Pilato, M.; Ebright, R.Y.; Karabacak, M.N.; Kulkarni, A.S.; Liu, A.; et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell 2019, 178, 160–175.e27. [Google Scholar] [CrossRef]
- Lee, J.J.; Bernard, V.; Semaan, A.; Monberg, M.E.; Huang, J.; Stephens, B.M.; Lin, D.; Rajapakshe, K.I.; Weston, B.R.; Bhutani, M.S.; et al. Elucidation of Tumor-Stromal Heterogeneity and the Ligand-Receptor Interactome by Single-Cell Transcriptomics in Real-world Pancreatic Cancer Biopsies. Clin. Cancer Res. 2021, 27, 5912–5921. [Google Scholar] [CrossRef]
- Herbertz, S.; Sawyer, J.S.; Stauber, A.J.; Gueorguieva, I.; Driscoll, K.E.; Estrem, S.T.; Cleverly, A.L.; Desaiah, D.; Guba, S.C.; Benhadji, K.A.; et al. Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-beta signaling pathway. Drug Des. Dev. Ther. 2015, 9, 4479–4499. [Google Scholar] [CrossRef]
- Sow, H.S.; Ren, J.; Camps, M.; Ossendorp, F.; Ten Dijke, P. Combined Inhibition of TGF-beta Signaling and the PD-L1 Immune Checkpoint Is Differentially Effective in Tumor Models. Cells 2019, 8, 320. [Google Scholar] [CrossRef] [PubMed]
- Tschernia, N.P.; Gulley, J.L. Tumor in the Crossfire: Inhibiting TGF-beta to Enhance Cancer Immunotherapy. BioDrugs 2022, 36, 153–180. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Hoffman, M.T.; Ali, L.R.; Castillo, J.I.; Kageler, L.; Temesgen, A.; Lenehan, P.; Wang, S.J.; Bello, E.; Cardot-Ruffino, V.; et al. Transforming Growth Factor-beta Blockade in Pancreatic Cancer Enhances Sensitivity to Combination Chemotherapy. Gastroenterology 2023, 165, 874–890.e10. [Google Scholar] [CrossRef] [PubMed]
- Diop-Frimpong, B.; Chauhan, V.P.; Krane, S.; Boucher, Y.; Jain, R.K. Losartan inhibits collagen I synthesis and improves the distribution and efficacy of nanotherapeutics in tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 2909–2914. [Google Scholar] [CrossRef]
- Chauhan, V.P.; Martin, J.D.; Liu, H.; Lacorre, D.A.; Jain, S.R.; Kozin, S.V.; Stylianopoulos, T.; Mousa, A.S.; Han, X.; Adstamongkonkul, P.; et al. Angiotensin inhibition enhances drug delivery and potentiates chemotherapy by decompressing tumour blood vessels. Nat. Commun. 2013, 4, 2516. [Google Scholar] [CrossRef]
- Zhao, Q.; He, X.; Qin, X.; Liu, Y.; Jiang, H.; Wang, J.; Wu, S.; Zhou, R.; Yu, C.; Liu, S.; et al. Enhanced Therapeutic Efficacy of Combining Losartan and Chemo-Immunotherapy for Triple Negative Breast Cancer. Front. Immunol. 2022, 13, 938439. [Google Scholar] [CrossRef] [PubMed]
- Boucher, Y.; Posada, J.M.; Subudhi, S.; Kumar, A.S.; Rosario, S.R.; Gu, L.; Kumra, H.; Mino-Kenudson, M.; Talele, N.P.; Duda, D.G.; et al. Addition of Losartan to FOLFIRINOX and Chemoradiation Reduces Immunosuppression-Associated Genes, Tregs, and FOXP3+ Cancer Cells in Locally Advanced Pancreatic Cancer. Clin. Cancer Res. 2023, 29, 1605–1619. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.E.; Wo, J.Y.; Ryan, D.P.; Clark, J.W.; Jiang, W.; Yeap, B.Y.; Drapek, L.C.; Ly, L.; Baglini, C.V.; Blaszkowsky, L.S.; et al. Total Neoadjuvant Therapy with FOLFIRINOX in Combination with Losartan Followed by Chemoradiotherapy for Locally Advanced Pancreatic Cancer: A Phase 2 Clinical Trial. JAMA Oncol. 2019, 5, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Kozono, S.; Ohuchida, K.; Eguchi, D.; Ikenaga, N.; Fujiwara, K.; Cui, L.; Mizumoto, K.; Tanaka, M. Pirfenidone inhibits pancreatic cancer desmoplasia by regulating stellate cells. Cancer Res. 2013, 73, 2345–2356. [Google Scholar] [CrossRef]
- Polydorou, C.; Mpekris, F.; Papageorgis, P.; Voutouri, C.; Stylianopoulos, T. Pirfenidone normalizes the tumor microenvironment to improve chemotherapy. Oncotarget 2017, 8, 24506–24517. [Google Scholar] [CrossRef]
- Es, H.A.; Cox, T.R.; Sarafraz-Yazdi, E.; Thiery, J.P.; Warkiani, M.E. Pirfenidone Reduces Epithelial-Mesenchymal Transition and Spheroid Formation in Breast Carcinoma through Targeting Cancer-Associated Fibroblasts (CAFs). Cancers 2021, 13, 5118. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Zeng, X.; Zhang, S.; Li, D.; Cheng, Z.; Wang, Y.; Long, J.; Hu, Z.; Long, S.; Zhou, J.; et al. Pirfenidone suppressed triple-negative breast cancer metastasis by inhibiting the activity of the TGF-beta/SMAD pathway. J. Cell. Mol. Med. 2023, 27, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Kocher, H.M.; Basu, B.; Froeling, F.E.M.; Sarker, D.; Slater, S.; Carlin, D.; deSouza, N.M.; De Paepe, K.N.; Goulart, M.R.; Hughes, C.; et al. Phase I clinical trial repurposing all-trans retinoic acid as a stromal targeting agent for pancreatic cancer. Nat. Commun. 2020, 11, 4841. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.H.; Yu, R.T.; Engle, D.D.; Ding, N.; Atkins, A.R.; Tiriac, H.; Collisson, E.A.; Connor, F.; Van Dyke, T.; Kozlov, S.; et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 2014, 159, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Hegde, S.; Knolhoff, B.L.; Zhu, Y.; Herndon, J.M.; Meyer, M.A.; Nywening, T.M.; Hawkins, W.G.; Shapiro, I.M.; Weaver, D.T.; et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 2016, 22, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Egeblad, M.; Rasch, M.G.; Weaver, V.M. Dynamic interplay between the collagen scaffold and tumor evolution. Curr. Opin. Cell Biol. 2010, 22, 697–706. [Google Scholar] [CrossRef]
- Hamill, K.J.; Kligys, K.; Hopkinson, S.B.; Jones, J.C. Laminin deposition in the extracellular matrix: A complex picture emerges. J. Cell Sci. 2009, 122, 4409–4417. [Google Scholar] [CrossRef]
- Hohenester, E.; Yurchenco, P.D. Laminins in basement membrane assembly. Cell Adhes. Migr. 2013, 7, 56–63. [Google Scholar] [CrossRef]
- Anastassiades, O.T.; Pryce, D.M. Fibrosis as in indication of time in infiltrating breast cancer and its importance in prognosis. Br. J. Cancer 1974, 29, 232–239. [Google Scholar] [CrossRef]
- Erkan, M.; Michalski, C.W.; Rieder, S.; Reiser-Erkan, C.; Abiatari, I.; Kolb, A.; Giese, N.A.; Esposito, I.; Friess, H.; Kleeff, J. The activated stroma index is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Clin. Gastroenterol. Hepatol. 2008, 6, 1155–1161. [Google Scholar] [CrossRef]
- Ding, J.H.; Xiao, Y.; Zhao, S.; Xu, Y.; Xiao, Y.L.; Shao, Z.M.; Jiang, Y.Z.; Di, G.H. Integrated analysis reveals the molecular features of fibrosis in triple-negative breast cancer. Mol. Ther. Oncolytics 2022, 24, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Chandler, C.; Liu, T.; Buckanovich, R.; Coffman, L.G. The double edge sword of fibrosis in cancer. Transl. Res. 2019, 209, 55–67. [Google Scholar] [CrossRef]
- Nieskoski, M.D.; Marra, K.; Gunn, J.R.; Hoopes, P.J.; Doyley, M.M.; Hasan, T.; Trembly, B.S.; Pogue, B.W. Collagen Complexity Spatially Defines Microregions of Total Tissue Pressure in Pancreatic Cancer. Sci. Rep. 2017, 7, 10093. [Google Scholar] [CrossRef] [PubMed]
- Voutouri, C.; Polydorou, C.; Papageorgis, P.; Gkretsi, V.; Stylianopoulos, T. Hyaluronan-Derived Swelling of Solid Tumors, the Contribution of Collagen and Cancer Cells, and Implications for Cancer Therapy. Neoplasia 2016, 18, 732–741. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Conklin, M.W.; Eickhoff, J.C.; Riching, K.M.; Pehlke, C.A.; Eliceiri, K.W.; Provenzano, P.P.; Friedl, A.; Keely, P.J. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011, 178, 1221–1232. [Google Scholar] [CrossRef]
- Conklin, M.W.; Gangnon, R.E.; Sprague, B.L.; Van Gemert, L.; Hampton, J.M.; Eliceiri, K.W.; Bredfeldt, J.S.; Liu, Y.; Surachaicharn, N.; Newcomb, P.A.; et al. Collagen Alignment as a Predictor of Recurrence after Ductal Carcinoma In Situ. Cancer Epidemiol. Biomark. Prev. 2018, 27, 138–145. [Google Scholar] [CrossRef]
- Koorman, T.; Jansen, K.A.; Khalil, A.; Haughton, P.D.; Visser, D.; Ratze, M.A.K.; Haakma, W.E.; Sakalauskaite, G.; van Diest, P.J.; de Rooij, J.; et al. Spatial collagen stiffening promotes collective breast cancer cell invasion by reinforcing extracellular matrix alignment. Oncogene 2022, 41, 2458–2469. [Google Scholar] [CrossRef]
- Chen, Y.; Kim, J.; Yang, S.; Wang, H.; Wu, C.J.; Sugimoto, H.; LeBleu, V.S.; Kalluri, R. Type I collagen deletion in alphaSMA(+) myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell 2021, 39, 548–565.e6. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Hamberger, F.; Ravichandra, A.; Miller, M.; Nair, A.; Affo, S.; Filliol, A.; Chin, L.; Savage, T.M.; Yin, D.; et al. Tumor restriction by type I collagen opposes tumor-promoting effects of cancer-associated fibroblasts. J. Clin. Investig. 2021, 131, e146987. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, S.; Tavormina, J.; Tampe, D.; Zeisberg, M.; Wang, H.; Mahadevan, K.K.; Wu, C.J.; Sugimoto, H.; Chang, C.C.; et al. Oncogenic collagen I homotrimers from cancer cells bind to alpha3beta1 integrin and impact tumor microbiome and immunity to promote pancreatic cancer. Cancer Cell 2022, 40, 818–834.e9. [Google Scholar] [CrossRef]
- Liu, J.; Shen, J.X.; Wu, H.T.; Li, X.L.; Wen, X.F.; Du, C.W.; Zhang, G.J. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov. Med. 2018, 25, 211–223. [Google Scholar] [PubMed]
- Xu, S.; Xu, H.; Wang, W.; Li, S.; Li, H.; Li, T.; Zhang, W.; Yu, X.; Liu, L. The role of collagen in cancer: From bench to bedside. J. Transl. Med. 2019, 17, 309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, S.; Lu, T.; Han, D.; Zhang, K.; Gan, L.; Wu, X.; Li, Y.; Zhao, X.; Li, Z.; et al. Single-cell analysis reveals the COL11A1(+) fibroblasts are cancer-specific fibroblasts that promote tumor progression. Front. Pharmacol. 2023, 14, 1121586. [Google Scholar] [CrossRef]
- Vazquez-Villa, F.; Garcia-Ocana, M.; Galvan, J.A.; Garcia-Martinez, J.; Garcia-Pravia, C.; Menendez-Rodriguez, P.; Gonzalez-del Rey, C.; Barneo-Serra, L.; de Los Toyos, J.R. COL11A1/(pro)collagen 11A1 expression is a remarkable biomarker of human invasive carcinoma-associated stromal cells and carcinoma progression. Tumour Biol. 2015, 36, 2213–2222. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Meng, K. COL11A1 is Downregulated by miR-339-5p and Promotes Colon Carcinoma Progression. Can. J. Gastroenterol. Hepatol. 2022, 2022, 8116990. [Google Scholar] [CrossRef] [PubMed]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.J. Collagen Type XI Alpha 1 (COL11A1): A Novel Biomarker and a Key Player in Cancer. Cancers 2021, 13, 935. [Google Scholar] [CrossRef]
- Lee, C.S.; Siprashvili, Z.; Mah, A.; Bencomo, T.; Elcavage, L.E.; Che, Y.; Shenoy, R.M.; Aasi, S.Z.; Khavari, P.A. Mutant collagen COL11A1 enhances cancerous invasion. Oncogene 2021, 40, 6299–6307. [Google Scholar] [CrossRef]
- Pietila, E.A.; Gonzalez-Molina, J.; Moyano-Galceran, L.; Jamalzadeh, S.; Zhang, K.; Lehtinen, L.; Turunen, S.P.; Martins, T.A.; Gultekin, O.; Lamminen, T.; et al. Co-evolution of matrisome and adaptive adhesion dynamics drives ovarian cancer chemoresistance. Nat. Commun. 2021, 12, 3904. [Google Scholar] [CrossRef] [PubMed]
- Arafat, H.; Lazar, M.; Salem, K.; Chipitsyna, G.; Gong, Q.; Pan, T.C.; Zhang, R.Z.; Yeo, C.J.; Chu, M.L. Tumor-specific expression and alternative splicing of the COL6A3 gene in pancreatic cancer. Surgery 2011, 150, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Voiles, L.; Lewis, D.E.; Han, L.; Lupov, I.P.; Lin, T.L.; Robertson, M.J.; Petrache, I.; Chang, H.C. Overexpression of type VI collagen in neoplastic lung tissues. Oncol. Rep. 2014, 32, 1897–1904. [Google Scholar] [CrossRef] [PubMed]
- Cescon, M.; Rampazzo, E.; Bresolin, S.; Da Ros, F.; Manfreda, L.; Cani, A.; Della Puppa, A.; Braghetta, P.; Bonaldo, P.; Persano, L. Collagen VI sustains cell stemness and chemotherapy resistance in glioblastoma. Cell. Mol. Life Sci. 2023, 80, 233. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Lu, Z.R. Targeting Fibronectin for Cancer Imaging and Therapy. J. Mater. Chem. B 2017, 5, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Wang, Y.; Sims, M.; Cai, C.; Pfeffer, L.M. MicroRNA-1 suppresses glioblastoma in preclinical models by targeting fibronectin. Cancer Lett. 2019, 465, 59–67. [Google Scholar] [CrossRef]
- Xie, Y.J.; Dougan, M.; Jailkhani, N.; Ingram, J.; Fang, T.; Kummer, L.; Momin, N.; Pishesha, N.; Rickelt, S.; Hynes, R.O.; et al. Nanobody-based CAR T cells that target the tumor microenvironment inhibit the growth of solid tumors in immunocompetent mice. Proc. Natl. Acad. Sci. USA 2019, 116, 7624–7631. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Wickman, E.; Shaw, T.I.; Anido, A.A.; Langfitt, D.; Zhang, J.; Porter, S.N.; Pruett-Miller, S.M.; Tillman, H.; Krenciute, G.; et al. Antitumor Effects of CAR T Cells Redirected to the EDB Splice Variant of Fibronectin. Cancer Immunol. Res. 2021, 9, 279–290. [Google Scholar] [CrossRef]
- Martin-Otal, C.; Lasarte-Cia, A.; Serrano, D.; Casares, N.; Conde, E.; Navarro, F.; Sanchez-Moreno, I.; Gorraiz, M.; Sarrion, P.; Calvo, A.; et al. Targeting the extra domain A of fibronectin for cancer therapy with CAR-T cells. J. Immunother. Cancer 2022, 10, e004479. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, C.; Yang, Z.; Yin, H. CAR-T-Cell Therapy for Solid Tumors Positive for Fibronectin Extra Domain B. Cells 2022, 11, 2863. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, C.; Wang, M.; Sun, R.; Yang, Z.; Hua, Z.; Wu, Y.; Wu, M.; Wang, H.; Qiu, W.; et al. Treating solid tumors with TCR-based chimeric antigen receptor targeting extra domain B-containing fibronectin. J. Immunother. Cancer 2023, 11, e007199. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.K.; Kim, A.; Kim, M.K.; Choi, J.E.; Kang, S.H.; Lee, S.J. Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum. Pathol. 2013, 44, 2028–2037. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liu, X.; Lu, W.; Chen, Y.; Wu, X.; Li, M.; Wang, X.A.; Zhang, F.; Jiang, L.; Zhang, Y.; et al. Fibronectin promotes cell proliferation and invasion through mTOR signaling pathway activation in gallbladder cancer. Cancer Lett. 2015, 360, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Kanaji, N.; Kita, N.; Kadowaki, N.; Bandoh, S. Fibronectin and Hepatocyte Growth Factor Produced by Lung Fibroblasts Augment Migration and Invasion of Malignant Pleural Mesothelioma Cells. Anticancer Res. 2017, 37, 2393–2400. [Google Scholar] [CrossRef] [PubMed]
- Gaggioli, C.; Robert, G.; Bertolotto, C.; Bailet, O.; Abbe, P.; Spadafora, A.; Bahadoran, P.; Ortonne, J.P.; Baron, V.; Ballotti, R.; et al. Tumor-derived fibronectin is involved in melanoma cell invasion and regulated by V600E B-Raf signaling pathway. J. Investig. Dermatol. 2007, 127, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.M.; Thomas, G.J.; Duffy, S.W.; Warwick, J.; Gabe, R.; Chou, P.; Ellis, I.O.; Green, A.R.; Haider, S.; Brouilette, K.; et al. Therapeutic targeting of integrin alphavbeta6 in breast cancer. J. Natl. Cancer Inst. 2014, 106, dju169. [Google Scholar] [CrossRef] [PubMed]
- Martinkova, E.; Maglott, A.; Leger, D.Y.; Bonnet, D.; Stiborova, M.; Takeda, K.; Martin, S.; Dontenwill, M. alpha5beta1 integrin antagonists reduce chemotherapy-induced premature senescence and facilitate apoptosis in human glioblastoma cells. Int. J. Cancer 2010, 127, 1240–1248. [Google Scholar] [CrossRef]
- von Au, A.; Vasel, M.; Kraft, S.; Sens, C.; Hackl, N.; Marx, A.; Stroebel, P.; Hennenlotter, J.; Todenhofer, T.; Stenzl, A.; et al. Circulating fibronectin controls tumor growth. Neoplasia 2013, 15, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Ghura, H.; Keimer, M.; von Au, A.; Hackl, N.; Klemis, V.; Nakchbandi, I.A. Inhibition of fibronectin accumulation suppresses tumor growth. Neoplasia 2021, 23, 837–850. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan promotes the malignant phenotype. Glycobiology 2002, 12, 37R–42R. [Google Scholar] [CrossRef]
- Reeves, E.L.; Li, J.; Zormpas-Petridis, K.; Boult, J.K.R.; Sullivan, J.; Cummings, C.; Blouw, B.; Kang, D.; Sinkus, R.; Bamber, J.C.; et al. Investigating the contribution of hyaluronan to the breast tumour microenvironment using multiparametric MRI and MR elastography. Mol. Oncol. 2023, 17, 1076–1092. [Google Scholar] [CrossRef] [PubMed]
- Petrey, A.C.; de la Motte, C.A. Hyaluronan, a crucial regulator of inflammation. Front. Immunol. 2014, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Kultti, A.; Zhao, C.; Singha, N.C.; Zimmerman, S.; Osgood, R.J.; Symons, R.; Jiang, P.; Li, X.; Thompson, C.B.; Infante, J.R.; et al. Accumulation of extracellular hyaluronan by hyaluronan synthase 3 promotes tumor growth and modulates the pancreatic cancer microenvironment. Biomed. Res. Int. 2014, 2014, 817613. [Google Scholar] [CrossRef] [PubMed]
- Kimata, K.; Honma, Y.; Okayama, M.; Oguri, K.; Hozumi, M.; Suzuki, S. Increased synthesis of hyaluronic acid by mouse mammary carcinoma cell variants with high metastatic potential. Cancer Res. 1983, 43, 1347–1354. [Google Scholar] [PubMed]
- Itano, N.; Sawai, T.; Miyaishi, O.; Kimata, K. Relationship between hyaluronan production and metastatic potential of mouse mammary carcinoma cells. Cancer Res. 1999, 59, 2499–2504. [Google Scholar] [PubMed]
- Simpson, M.A.; Reiland, J.; Burger, S.R.; Furcht, L.T.; Spicer, A.P.; Oegema, T.R., Jr.; McCarthy, J.B. Hyaluronan synthase elevation in metastatic prostate carcinoma cells correlates with hyaluronan surface retention, a prerequisite for rapid adhesion to bone marrow endothelial cells. J. Biol. Chem. 2001, 276, 17949–17957. [Google Scholar] [CrossRef]
- Ropponen, K.; Tammi, M.; Parkkinen, J.; Eskelinen, M.; Tammi, R.; Lipponen, P.; Agren, U.; Alhava, E.; Kosma, V.M. Tumor cell-associated hyaluronan as an unfavorable prognostic factor in colorectal cancer. Cancer Res. 1998, 58, 342–347. [Google Scholar]
- Setala, L.P.; Tammi, M.I.; Tammi, R.H.; Eskelinen, M.J.; Lipponen, P.K.; Agren, U.M.; Parkkinen, J.; Alhava, E.M.; Kosma, V.M. Hyaluronan expression in gastric cancer cells is associated with local and nodal spread and reduced survival rate. Br. J. Cancer 1999, 79, 1133–1138. [Google Scholar] [CrossRef]
- Auvinen, P.; Tammi, R.; Kosma, V.M.; Sironen, R.; Soini, Y.; Mannermaa, A.; Tumelius, R.; Uljas, E.; Tammi, M. Increased hyaluronan content and stromal cell CD44 associate with HER2 positivity and poor prognosis in human breast cancer. Int. J. Cancer 2013, 132, 531–539. [Google Scholar] [CrossRef]
- Auvinen, P.; Tammi, R.; Parkkinen, J.; Tammi, M.; Agren, U.; Johansson, R.; Hirvikoski, P.; Eskelinen, M.; Kosma, V.M. Hyaluronan in peritumoral stroma and malignant cells associates with breast cancer spreading and predicts survival. Am. J. Pathol. 2000, 156, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Whatcott, C.J.; Diep, C.H.; Jiang, P.; Watanabe, A.; LoBello, J.; Sima, C.; Hostetter, G.; Shepard, H.M.; Von Hoff, D.D.; Han, H. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin. Cancer Res. 2015, 21, 3561–3568. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, G.; Gomar-Hoss, C.; Sakr, L.; Ulsperger, E.; Wogritsch, C. The impact of extracellular matrix on the chemoresistance of solid tumors—Experimental and clinical results of hyaluronidase as additive to cytostatic chemotherapy. Cancer Lett. 1998, 131, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Pillwein, K.; Fuiko, R.; Slavc, I.; Czech, T.; Hawliczek, G.; Bernhardt, G.; Nirnberger, G.; Koller, U. Hyaluronidase additional to standard chemotherapy improves outcome for children with malignant brain tumors. Cancer Lett. 1998, 131, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.B.; Shepard, H.M.; O’Connor, P.M.; Kadhim, S.; Jiang, P.; Osgood, R.J.; Bookbinder, L.H.; Li, X.; Sugarman, B.J.; Connor, R.J.; et al. Enzymatic depletion of tumor hyaluronan induces antitumor responses in preclinical animal models. Mol. Cancer Ther. 2010, 9, 3052–3064. [Google Scholar] [CrossRef] [PubMed]
- Seki, T.; Saida, Y.; Kishimoto, S.; Lee, J.; Otowa, Y.; Yamamoto, K.; Chandramouli, G.V.; Devasahayam, N.; Mitchell, J.B.; Krishna, M.C.; et al. PEGPH20, a PEGylated human hyaluronidase, induces radiosensitization by reoxygenation in pancreatic cancer xenografts. A molecular imaging study. Neoplasia 2022, 30, 100793. [Google Scholar] [CrossRef] [PubMed]
- Blair, A.B.; Wang, J.; Davelaar, J.; Baker, A.; Li, K.; Niu, N.; Wang, J.; Shao, Y.; Funes, V.; Li, P.; et al. Dual Stromal Targeting Sensitizes Pancreatic Adenocarcinoma for Anti-Programmed Cell Death Protein 1 Therapy. Gastroenterology 2022, 163, 1267–1280.e7. [Google Scholar] [CrossRef] [PubMed]
- Morosi, L.; Meroni, M.; Ubezio, P.; Fuso Nerini, I.; Minoli, L.; Porcu, L.; Panini, N.; Colombo, M.; Blouw, B.; Kang, D.W.; et al. PEGylated recombinant human hyaluronidase (PEGPH20) pre-treatment improves intra-tumour distribution and efficacy of paclitaxel in preclinical models. J. Exp. Clin. Cancer Res. 2021, 40, 286. [Google Scholar] [CrossRef] [PubMed]
- Hingorani, S.R.; Zheng, L.; Bullock, A.J.; Seery, T.E.; Harris, W.P.; Sigal, D.S.; Braiteh, F.; Ritch, P.S.; Zalupski, M.M.; Bahary, N.; et al. HALO 202: Randomized Phase II Study of PEGPH20 Plus Nab-Paclitaxel/Gemcitabine Versus Nab-Paclitaxel/Gemcitabine in Patients with Untreated, Metastatic Pancreatic Ductal Adenocarcinoma. J. Clin. Oncol. 2018, 36, 359–366. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Tempero, M.A.; Sigal, D.; Oh, D.Y.; Fazio, N.; Macarulla, T.; Hitre, E.; Hammel, P.; Hendifar, A.E.; Bates, S.E.; et al. Randomized Phase III Trial of Pegvorhyaluronidase Alfa with Nab-Paclitaxel Plus Gemcitabine for Patients with Hyaluronan-High Metastatic Pancreatic Adenocarcinoma. J. Clin. Oncol. 2020, 38, 3185–3194. [Google Scholar] [CrossRef]
- Ramanathan, R.K.; McDonough, S.L.; Philip, P.A.; Hingorani, S.R.; Lacy, J.; Kortmansky, J.S.; Thumar, J.; Chiorean, E.G.; Shields, A.F.; Behl, D.; et al. Phase IB/II Randomized Study of FOLFIRINOX Plus Pegylated Recombinant Human Hyaluronidase Versus FOLFIRINOX Alone in Patients with Metastatic Pancreatic Adenocarcinoma: SWOG S1313. J. Clin. Oncol. 2019, 37, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Doherty, G.J.; Tempero, M.; Corrie, P.G. HALO-109-301: A Phase III trial of PEGPH20 (with gemcitabine and nab-paclitaxel) in hyaluronic acid-high stage IV pancreatic cancer. Future Oncol. 2018, 14, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Infante, J.R.; Korn, R.L.; Rosen, L.S.; LoRusso, P.; Dychter, S.S.; Zhu, J.; Maneval, D.C.; Jiang, P.; Shepard, H.M.; Frost, G.; et al. Phase 1 trials of PEGylated recombinant human hyaluronidase PH20 in patients with advanced solid tumours. Br. J. Cancer 2018, 118, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Singha, N.C.; Nekoroski, T.; Zhao, C.; Symons, R.; Jiang, P.; Frost, G.I.; Huang, Z.; Shepard, H.M. Tumor-associated hyaluronan limits efficacy of monoclonal antibody therapy. Mol. Cancer Ther. 2015, 14, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Clift, R.; Souratha, J.; Garrovillo, S.A.; Zimmerman, S.; Blouw, B. Remodeling the Tumor Microenvironment Sensitizes Breast Tumors to Anti-Programmed Death-Ligand 1 Immunotherapy. Cancer Res. 2019, 79, 4149–4159. [Google Scholar] [CrossRef] [PubMed]
- Witschen, P.M.; Chaffee, T.S.; Brady, N.J.; Huggins, D.N.; Knutson, T.P.; LaRue, R.S.; Munro, S.A.; Tiegs, L.; McCarthy, J.B.; Nelson, A.C.; et al. Tumor Cell Associated Hyaluronan-CD44 Signaling Promotes Pro-Tumor Inflammation in Breast Cancer. Cancers 2020, 12, 1325. [Google Scholar] [CrossRef] [PubMed]
- Barry-Hamilton, V.; Spangler, R.; Marshall, D.; McCauley, S.; Rodriguez, H.M.; Oyasu, M.; Mikels, A.; Vaysberg, M.; Ghermazien, H.; Wai, C.; et al. Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nat. Med. 2010, 16, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R.; Bird, D.; Baker, A.M.; Barker, H.E.; Ho, M.W.; Lang, G.; Erler, J.T. LOX-mediated collagen crosslinking is responsible for fibrosis-enhanced metastasis. Cancer Res. 2013, 73, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.M.; Cox, T.R.; Bird, D.; Lang, G.; Murray, G.I.; Sun, X.F.; Southall, S.M.; Wilson, J.R.; Erler, J.T. The role of lysyl oxidase in SRC-dependent proliferation and metastasis of colorectal cancer. J. Natl. Cancer Inst. 2011, 103, 407–424. [Google Scholar] [CrossRef]
- Wiel, C.; Augert, A.; Vincent, D.F.; Gitenay, D.; Vindrieux, D.; Le Calve, B.; Arfi, V.; Lallet-Daher, H.; Reynaud, C.; Treilleux, I.; et al. Lysyl oxidase activity regulates oncogenic stress response and tumorigenesis. Cell Death Dis. 2013, 4, e855. [Google Scholar] [CrossRef]
- Tanaka, N.; Yamada, S.; Sonohara, F.; Suenaga, M.; Hayashi, M.; Takami, H.; Niwa, Y.; Hattori, N.; Iwata, N.; Kanda, M.; et al. Clinical Implications of Lysyl Oxidase-Like Protein 2 Expression in Pancreatic Cancer. Sci. Rep. 2018, 8, 9846. [Google Scholar] [CrossRef]
- Zhan, P.; Lv, X.J.; Ji, Y.N.; Xie, H.; Yu, L.K. Increased lysyl oxidase-like 2 associates with a poor prognosis in non-small cell lung cancer. Clin. Respir. J. 2018, 12, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, J.; You, F.; Li, W.; Zou, Z. Prognostic and clinicopathological significance of LOXL2 in cancers: A systematic review and meta-analysis. J. Cell. Physiol. 2019, 234, 21369–21379. [Google Scholar] [CrossRef]
- Wang, C.; Xu, S.; Tian, Y.; Ju, A.; Hou, Q.; Liu, J.; Fu, Y.; Luo, Y. Lysyl Oxidase-Like Protein 2 Promotes Tumor Lymphangiogenesis and Lymph Node Metastasis in Breast Cancer. Neoplasia 2019, 21, 413–427. [Google Scholar] [CrossRef]
- Ramos, S.; Ferreira, S.; Fernandes, A.S.; Saraiva, N. Lysyl Oxidases Expression and Breast Cancer Progression: A Bioinformatic Analysis. Front. Pharmacol. 2022, 13, 883998. [Google Scholar] [CrossRef] [PubMed]
- Saatci, O.; Kaymak, A.; Raza, U.; Ersan, P.G.; Akbulut, O.; Banister, C.E.; Sikirzhytski, V.; Tokat, U.M.; Aykut, G.; Ansari, S.A.; et al. Targeting lysyl oxidase (LOX) overcomes chemotherapy resistance in triple negative breast cancer. Nat. Commun. 2020, 11, 2416. [Google Scholar] [CrossRef] [PubMed]
- Rossow, L.; Veitl, S.; Vorlova, S.; Wax, J.K.; Kuhn, A.E.; Maltzahn, V.; Upcin, B.; Karl, F.; Hoffmann, H.; Gatzner, S.; et al. LOX-catalyzed collagen stabilization is a proximal cause for intrinsic resistance to chemotherapy. Oncogene 2018, 37, 4921–4940. [Google Scholar] [CrossRef] [PubMed]
- De Vita, A.; Liverani, C.; Molinaro, R.; Martinez, J.O.; Hartman, K.A.; Spadazzi, C.; Miserocchi, G.; Taraballi, F.; Evangelopoulos, M.; Pieri, F.; et al. Lysyl oxidase engineered lipid nanovesicles for the treatment of triple negative breast cancer. Sci. Rep. 2021, 11, 5107. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Lucas, M.C.; Leonte, L.E.; Garcia-Montolio, M.; Singh, L.B.; Findlay, A.D.; Deodhar, M.; Foot, J.S.; Jarolimek, W.; Timpson, P.; et al. Pre-clinical evaluation of small molecule LOXL2 inhibitors in breast cancer. Oncotarget 2017, 8, 26066–26078. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, J.H.; Rowbottom, M.W.; Lonergan, D.; Darlington, J.; Prodanovich, P.; King, C.D.; Evans, J.F.; Bain, G. Small Molecule Lysyl Oxidase-like 2 (LOXL2) Inhibitors: The Identification of an Inhibitor Selective for LOXL2 over LOX. ACS Med. Chem. Lett. 2017, 8, 423–427. [Google Scholar] [CrossRef]
- Schilter, H.; Findlay, A.D.; Perryman, L.; Yow, T.T.; Moses, J.; Zahoor, A.; Turner, C.I.; Deodhar, M.; Foot, J.S.; Zhou, W.; et al. The lysyl oxidase like 2/3 enzymatic inhibitor, PXS-5153A, reduces crosslinks and ameliorates fibrosis. J. Cell. Mol. Med. 2019, 23, 1759–1770. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.W.; Morton, J.P.; Pinese, M.; Saturno, G.; Jamieson, N.B.; McGhee, E.; Timpson, P.; Leach, J.; McGarry, L.; Shanks, E.; et al. Targeting the LOX/hypoxia axis reverses many of the features that make pancreatic cancer deadly: Inhibition of LOX abrogates metastasis and enhances drug efficacy. EMBO Mol. Med. 2015, 7, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Nocelo, M.; Ruiz-Canas, L.; Sancho, P.; Gorgulu, K.; Alcala, S.; Pedrero, C.; Vallespinos, M.; Lopez-Gil, J.C.; Ochando, M.; Garcia-Garcia, E.; et al. Macrophages direct cancer cells through a LOXL2-mediated metastatic cascade in pancreatic ductal adenocarcinoma. Gut 2023, 72, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Torphy, R.J.; Steiger, K.; Hongo, H.; Ritchie, A.J.; Kriegsmann, M.; Horst, D.; Umetsu, S.E.; Joseph, N.M.; McGregor, K.; et al. Pancreatic ductal adenocarcinoma progression is restrained by stromal matrix. J. Clin. Investig. 2020, 130, 4704–4709. [Google Scholar] [CrossRef] [PubMed]
- Le Calve, B.; Griveau, A.; Vindrieux, D.; Marechal, R.; Wiel, C.; Svrcek, M.; Gout, J.; Azzi, L.; Payen, L.; Cros, J.; et al. Lysyl oxidase family activity promotes resistance of pancreatic ductal adenocarcinoma to chemotherapy by limiting the intratumoral anticancer drug distribution. Oncotarget 2016, 7, 32100–32112. [Google Scholar] [CrossRef]
- Benson, A.B., 3rd; Wainberg, Z.A.; Hecht, J.R.; Vyushkov, D.; Dong, H.; Bendell, J.; Kudrik, F. A Phase II Randomized, Double-Blind, Placebo-Controlled Study of Simtuzumab or Placebo in Combination with Gemcitabine for the First-Line Treatment of Pancreatic Adenocarcinoma. Oncologist 2017, 22, 241.e15. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Brown, K.K.; Collard, H.R.; Cottin, V.; Gibson, K.F.; Kaner, R.J.; Lederer, D.J.; Martinez, F.J.; Noble, P.W.; Song, J.W.; et al. Efficacy of simtuzumab versus placebo in patients with idiopathic pulmonary fibrosis: A randomised, double-blind, controlled, phase 2 trial. Lancet Respir. Med. 2017, 5, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Abdelmalek, M.F.; Caldwell, S.; Shiffman, M.L.; Diehl, A.M.; Ghalib, R.; Lawitz, E.J.; Rockey, D.C.; Schall, R.A.; Jia, C.; et al. Simtuzumab Is Ineffective for Patients with Bridging Fibrosis or Compensated Cirrhosis Caused by Nonalcoholic Steatohepatitis. Gastroenterology 2018, 155, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Muir, A.J.; Levy, C.; Janssen, H.L.A.; Montano-Loza, A.J.; Shiffman, M.L.; Caldwell, S.; Luketic, V.; Ding, D.; Jia, C.; McColgan, B.J.; et al. Simtuzumab for Primary Sclerosing Cholangitis: Phase 2 Study Results with Insights on the Natural History of the Disease. Hepatology 2019, 69, 684–698. [Google Scholar] [CrossRef]
- Chen, W.; Yang, A.; Jia, J.; Popov, Y.V.; Schuppan, D.; You, H. Lysyl Oxidase (LOX) Family Members: Rationale and Their Potential as Therapeutic Targets for Liver Fibrosis. Hepatology 2020, 72, 729–741. [Google Scholar] [CrossRef]
- Coussens, L.M.; Fingleton, B.; Matrisian, L.M. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science 2002, 295, 2387–2392. [Google Scholar] [CrossRef] [PubMed]
- Cathcart, J.; Pulkoski-Gross, A.; Cao, J. Targeting Matrix Metalloproteinases in Cancer: Bringing New Life to Old Ideas. Genes Dis. 2015, 2, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, B.S.; Rank, F.; Lopez, J.M.; Balbin, M.; Vizoso, F.; Lund, L.R.; Dano, K.; Lopez-Otin, C. Collagenase-3 expression in breast myofibroblasts as a molecular marker of transition of ductal carcinoma in situ lesions to invasive ductal carcinomas. Cancer Res. 2001, 61, 7091–7100. [Google Scholar] [PubMed]
- Gonzalez, L.O.; Gonzalez-Reyes, S.; Junquera, S.; Marin, L.; Gonzalez, L.; Del Casar, J.M.; Gonzalez, J.M.; Vizoso, F. Expression of metalloproteases and their inhibitors by tumor and stromal cells in ductal carcinoma in situ of the breast and their relationship with microinvasive events. J. Cancer Res. Clin. Oncol. 2010, 136, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Marshall, J.F.; Jones, J.L. alphavbeta6 Expression in myoepithelial cells: A novel marker for predicting DCIS progression with therapeutic potential. Cancer Res. 2014, 74, 5942–5947. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Allen, M.D.; Thomas, G.J.; Clark, S.; Dawoud, M.M.; Vallath, S.; Payne, S.J.; Gomm, J.J.; Dreger, S.A.; Dickinson, S.; Edwards, D.R.; et al. Altered microenvironment promotes progression of preinvasive breast cancer: Myoepithelial expression of alphavbeta6 integrin in DCIS identifies high-risk patients and predicts recurrence. Clin. Cancer Res. 2014, 20, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Gibson, S.V.; Tomas Bort, E.; Rodriguez-Fernandez, L.; Allen, M.D.; Gomm, J.J.; Goulding, I.; Auf dem Keller, U.; Agnoletto, A.; Brisken, C.; Peck, B.; et al. TGFbeta-mediated MMP13 secretion drives myoepithelial cell dependent breast cancer progression. NPJ Breast Cancer 2023, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Hayward, M.K.; Allen, M.D.; Gomm, J.J.; Goulding, I.; Thompson, C.L.; Knight, M.M.; Marshall, J.F.; Jones, J.L. Mechanostimulation of breast myoepithelial cells induces functional changes associated with DCIS progression to invasion. NPJ Breast Cancer 2022, 8, 109. [Google Scholar] [CrossRef]
- D’Arcy, C.; Zimmermann, C.C.; Espinoza-Sanchez, N.A.; Greve, B.; Schmidt, A.; Kiesel, L.; von Wahlde, M.K.; Gotte, M. The heparan sulphate proteoglycan Syndecan-1 (CD138) regulates tumour progression in a 3D model of ductal carcinoma in situ of the breast. IUBMB Life 2022, 74, 955–968. [Google Scholar] [CrossRef]
- Dawoud, M.M.; Jones, D.T.; Chelala, C.; Abdou, A.G.; Dreger, S.A.; Asaad, N.; Abd El-Wahed, M.; Jones, L. Expression Profile of Myoepithelial Cells in DCIS: Do They Change From Protective Angels to Wicked Witches? Appl. Immunohistochem. Mol. Morphol. 2022, 30, 397–409. [Google Scholar] [CrossRef]
- Gibson, S.V.; Madzharova, E.; Tan, A.C.; Allen, M.D.; Keller, U.A.D.; Louise Jones, J.; Carter, E.P.; Grose, R.P. ADAMTS3 restricts cancer invasion in models of early breast cancer progression through enhanced fibronectin degradation. Matrix Biol. 2023, 121, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Sarper, M.; Allen, M.D.; Gomm, J.; Haywood, L.; Decock, J.; Thirkettle, S.; Ustaoglu, A.; Sarker, S.J.; Marshall, J.; Edwards, D.R.; et al. Loss of MMP-8 in ductal carcinoma in situ (DCIS)-associated myoepithelial cells contributes to tumour promotion through altered adhesive and proteolytic function. Breast Cancer Res. BCR 2017, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.P.; Yoneten, K.K.; Gavara, N.; Tyler, E.J.; Gauthier, V.; Murray, E.R.; Ten Dijke, P.; Cameron, A.J.; Pearce, O.; Grose, R.P. Opposing roles for ADAMTS2 and ADAMTS14 in myofibroblast differentiation and function. J. Pathol. 2023, 262, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Fontanil, T.; Alvarez-Teijeiro, S.; Villaronga, M.A.; Mohamedi, Y.; Solares, L.; Moncada-Pazos, A.; Vega, J.A.; Garcia-Suarez, O.; Perez-Basterrechea, M.; Garcia-Pedrero, J.M.; et al. Cleavage of Fibulin-2 by the aggrecanases ADAMTS-4 and ADAMTS-5 contributes to the tumorigenic potential of breast cancer cells. Oncotarget 2017, 8, 13716–13729. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J. Matrix metalloproteinases as therapeutic targets in breast cancer. Front. Oncol. 2022, 12, 1108695. [Google Scholar] [CrossRef] [PubMed]
- Ager, E.I.; Kozin, S.V.; Kirkpatrick, N.D.; Seano, G.; Kodack, D.P.; Askoxylakis, V.; Huang, Y.; Goel, S.; Snuderl, M.; Muzikansky, A.; et al. Blockade of MMP14 activity in murine breast carcinomas: Implications for macrophages, vessels, and radiotherapy. J. Natl. Cancer Inst. 2015, 107, djv017. [Google Scholar] [CrossRef] [PubMed]
- Ling, B.; Watt, K.; Banerjee, S.; Newsted, D.; Truesdell, P.; Adams, J.; Sidhu, S.S.; Craig, A.W.B. A novel immunotherapy targeting MMP-14 limits hypoxia, immune suppression and metastasis in triple-negative breast cancer models. Oncotarget 2017, 8, 58372–58385. [Google Scholar] [CrossRef] [PubMed]
- Juric, V.; O’Sullivan, C.; Stefanutti, E.; Kovalenko, M.; Greenstein, A.; Barry-Hamilton, V.; Mikaelian, I.; Degenhardt, J.; Yue, P.; Smith, V.; et al. MMP-9 inhibition promotes anti-tumor immunity through disruption of biochemical and physical barriers to T-cell trafficking to tumors. PLoS ONE 2018, 13, e0207255. [Google Scholar] [CrossRef] [PubMed]
- Owyong, M.; Chou, J.; van den Bijgaart, R.J.; Kong, N.; Efe, G.; Maynard, C.; Talmi-Frank, D.; Solomonov, I.; Koopman, C.; Hadler-Olsen, E.; et al. MMP9 modulates the metastatic cascade and immune landscape for breast cancer anti-metastatic therapy. Life Sci. Alliance 2019, 2, e201800226. [Google Scholar] [CrossRef]
- Ye, Y.; Kuang, X.; Xie, Z.; Liang, L.; Zhang, Z.; Zhang, Y.; Ma, F.; Gao, Q.; Chang, R.; Lee, H.H.; et al. Small-molecule MMP2/MMP9 inhibitor SB-3CT modulates tumor immune surveillance by regulating PD-L1. Genome Med. 2020, 12, 83. [Google Scholar] [CrossRef]
- Redondo-Garcia, S.; Peris-Torres, C.; Caracuel-Peramos, R.; Rodriguez-Manzaneque, J.C. ADAMTS proteases and the tumor immune microenvironment: Lessons from substrates and pathologies. Matrix Biol. Plus 2021, 9, 100054. [Google Scholar] [CrossRef] [PubMed]
- Ricciardelli, C.; Frewin, K.M.; Tan Ide, A.; Williams, E.D.; Opeskin, K.; Pritchard, M.A.; Ingman, W.V.; Russell, D.L. The ADAMTS1 protease gene is required for mammary tumor growth and metastasis. Am. J. Pathol. 2011, 179, 3075–3085. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Rodriguez, R.; Rodriguez-Baena, F.J.; Martino-Echarri, E.; Peris-Torres, C.; Del Carmen Plaza-Calonge, M.; Rodriguez-Manzaneque, J.C. Stroma-derived but not tumor ADAMTS1 is a main driver of tumor growth and metastasis. Oncotarget 2016, 7, 34507–34519. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Baena, F.J.; Redondo-Garcia, S.; Peris-Torres, C.; Martino-Echarri, E.; Fernandez-Rodriguez, R.; Plaza-Calonge, M.D.C.; Anderson, P.; Rodriguez-Manzaneque, J.C. ADAMTS1 protease is required for a balanced immune cell repertoire and tumour inflammatory response. Sci. Rep. 2018, 8, 13103. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Luo, Y.; Zhou, Y.; Qin, S.; Qiu, Y.; Cui, R.; Yu, M.; Qin, J.; Zhong, M. Promotion of Tumor Growth by ADAMTS4 in Colorectal Cancer: Focused on Macrophages. Cell. Physiol. Biochem. 2018, 46, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Martino-Echarri, E.; Fernandez-Rodriguez, R.; Rodriguez-Baena, F.J.; Barrientos-Duran, A.; Torres-Collado, A.X.; Plaza-Calonge Mdel, C.; Amador-Cubero, S.; Cortes, J.; Reynolds, L.E.; Hodivala-Dilke, K.M.; et al. Contribution of ADAMTS1 as a tumor suppressor gene in human breast carcinoma. Linking its tumor inhibitory properties to its proteolytic activity on nidogen-1 and nidogen-2. Int. J. Cancer 2013, 133, 2315–2324. [Google Scholar] [CrossRef]
- Papadas, A.; Arauz, G.; Cicala, A.; Wiesner, J.; Asimakopoulos, F. Versican and Versican-matrikines in Cancer Progression, Inflammation, and Immunity. J. Histochem. Cytochem. 2020, 68, 871–885. [Google Scholar] [CrossRef] [PubMed]
- Papadas, A.; Asimakopoulos, F. Versican in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1272, 55–72. [Google Scholar] [CrossRef]
- McCulloch, D.R.; Nelson, C.M.; Dixon, L.J.; Silver, D.L.; Wylie, J.D.; Lindner, V.; Sasaki, T.; Cooley, M.A.; Argraves, W.S.; Apte, S.S. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev. Cell 2009, 17, 687–698. [Google Scholar] [CrossRef]
- Hope, C.; Emmerich, P.B.; Papadas, A.; Pagenkopf, A.; Matkowskyj, K.A.; Van De Hey, D.R.; Payne, S.N.; Clipson, L.; Callander, N.S.; Hematti, P.; et al. Versican-Derived Matrikines Regulate Batf3-Dendritic Cell Differentiation and Promote T Cell Infiltration in Colorectal Cancer. J. Immunol. 2017, 199, 1933–1941. [Google Scholar] [CrossRef]
- Dhakal, B.; Pagenkopf, A.; Mushtaq, M.U.; Cunningham, A.M.; Flietner, E.; Morrow, Z.; Papadas, A.; Hope, C.; Leith, C.; Hematti, P.; et al. Versican proteolysis predicts immune effector infiltration and post-transplant survival in myeloma. Leuk. Lymphoma 2019, 60, 2558–2562. [Google Scholar] [CrossRef] [PubMed]
- Hope, C.; Foulcer, S.; Jagodinsky, J.; Chen, S.X.; Jensen, J.L.; Patel, S.; Leith, C.; Maroulakou, I.; Callander, N.; Miyamoto, S.; et al. Immunoregulatory roles of versican proteolysis in the myeloma microenvironment. Blood 2016, 128, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hirani, P.; Gauthier, V.; Allen, C.E.; Wight, T.N.; Pearce, O.M.T. Targeting Versican as a Potential Immunotherapeutic Strategy in the Treatment of Cancer. Front. Oncol. 2021, 11, 712807. [Google Scholar] [CrossRef] [PubMed]
- Papadas, A.; Deb, G.; Cicala, A.; Officer, A.; Hope, C.; Pagenkopf, A.; Flietner, E.; Morrow, Z.T.; Emmerich, P.; Wiesner, J.; et al. Stromal remodeling regulates dendritic cell abundance and activity in the tumor microenvironment. Cell Rep. 2022, 40, 111201. [Google Scholar] [CrossRef] [PubMed]
- Le Page, A.; Khalil, A.; Vermette, P.; Frost, E.H.; Larbi, A.; Witkowski, J.M.; Fulop, T. The role of elastin-derived peptides in human physiology and diseases. Matrix Biol. 2019, 84, 81–96. [Google Scholar] [CrossRef]
- Nannan, L.; Gsell, W.; Belderbos, S.; Gallet, C.; Wouters, J.; Brassart-Pasco, S.; Himmelreich, U.; Brassart, B. A multimodal imaging study to highlight elastin-derived peptide pro-tumoral effect in a pancreatic xenograft model. Br. J. Cancer 2023, 128, 2000–2012. [Google Scholar] [CrossRef]
- Szychowski, K.A.; Skora, B. The elastin-derived peptide (VGVAPG) activates autophagy in neuroblastoma (SH-SY5Y) cells via peroxisome proliferator-activated receptor gamma (PPARgamma). Mol. Cell. Neurosci. 2023, 127, 103902. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, K.; Shanthi, C. Matrikines for therapeutic and biomedical applications. Life Sci. 2018, 214, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Gaggar, A.; Weathington, N. Bioactive extracellular matrix fragments in lung health and disease. J. Clin. Investig. 2016, 126, 3176–3184. [Google Scholar] [CrossRef]
- Martin, D.R.; Santamaria, S.; Koch, C.D.; Ahnstrom, J.; Apte, S.S. Identification of novel ADAMTS1, ADAMTS4 and ADAMTS5 cleavage sites in versican using a label-free quantitative proteomics approach. J. Proteom. 2021, 249, 104358. [Google Scholar] [CrossRef]
- Cascio, S.; Chandler, C.; Zhang, L.; Sinno, S.; Gao, B.; Onkar, S.; Bruno, T.C.; Vignali, D.A.A.; Mahdi, H.; Osmanbeyoglu, H.U.; et al. Cancer-associated MSC drive tumor immune exclusion and resistance to immunotherapy, which can be overcome by Hedgehog inhibition. Sci. Adv. 2021, 7, eabi5790. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.M.; Cho, J. Hedgehog Pathway Inhibitors as Targeted Cancer Therapy and Strategies to Overcome Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1733. [Google Scholar] [CrossRef] [PubMed]
- Puttock, E.H.; Tyler, E.J.; Manni, M.; Maniati, E.; Butterworth, C.; Burger Ramos, M.; Peerani, E.; Hirani, P.; Gauthier, V.; Liu, Y.; et al. Extracellular matrix educates an immunoregulatory tumor macrophage phenotype found in ovarian cancer metastasis. Nat. Commun. 2023, 14, 2514. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.; Koul, A.M.; Wani, U.M.; Farooq, F.; Amin, B.; Wani, Z.; Lone, A.; Qadri, A.; Qadri, R.A. Dissection of paracrine/autocrine interplay in lung tumor microenvironment mimicking cancer cell-monocyte co-culture models reveals proteins that promote inflammation and metastasis. BMC Cancer 2023, 23, 926. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Takahashi, H.; Lin, W.W.; Descargues, P.; Grivennikov, S.; Kim, Y.; Luo, J.L.; Karin, M. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature 2009, 457, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Pappas, A.G.; Magkouta, S.; Pateras, I.S.; Skianis, I.; Moschos, C.; Vazakidou, M.E.; Psarra, K.; Gorgoulis, V.G.; Kalomenidis, I. Versican modulates tumor-associated macrophage properties to stimulate mesothelioma growth. Oncoimmunology 2019, 8, e1537427. [Google Scholar] [CrossRef] [PubMed]
- Evanko, S.P.; Potter-Perigo, S.; Bollyky, P.L.; Nepom, G.T.; Wight, T.N. Hyaluronan and versican in the control of human T-lymphocyte adhesion and migration. Matrix Biol. 2012, 31, 90–100. [Google Scholar] [CrossRef]
- Wight, T.N. Provisional matrix: A role for versican and hyaluronan. Matrix Biol. 2017, 60–61, 38–56. [Google Scholar] [CrossRef] [PubMed]
- Paget, S. The distribution of secondary growths in cancer of the breast. 1889 Cancer Metastasis Rev. 1989, 8, 98–101. [Google Scholar] [CrossRef]
- Akhtar, M.; Haider, A.; Rashid, S.; Al-Nabet, A. Paget’s “Seed and Soil” Theory of Cancer Metastasis: An Idea Whose Time has Come. Adv. Anat. Pathol. 2019, 26, 69–74. [Google Scholar] [CrossRef]
- Langley, R.R.; Fidler, I.J. The seed and soil hypothesis revisited—The role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer 2011, 128, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).