Genetic Prognostic Factors in Adult Diffuse Gliomas: A 10-Year Experience at a Single Institution
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
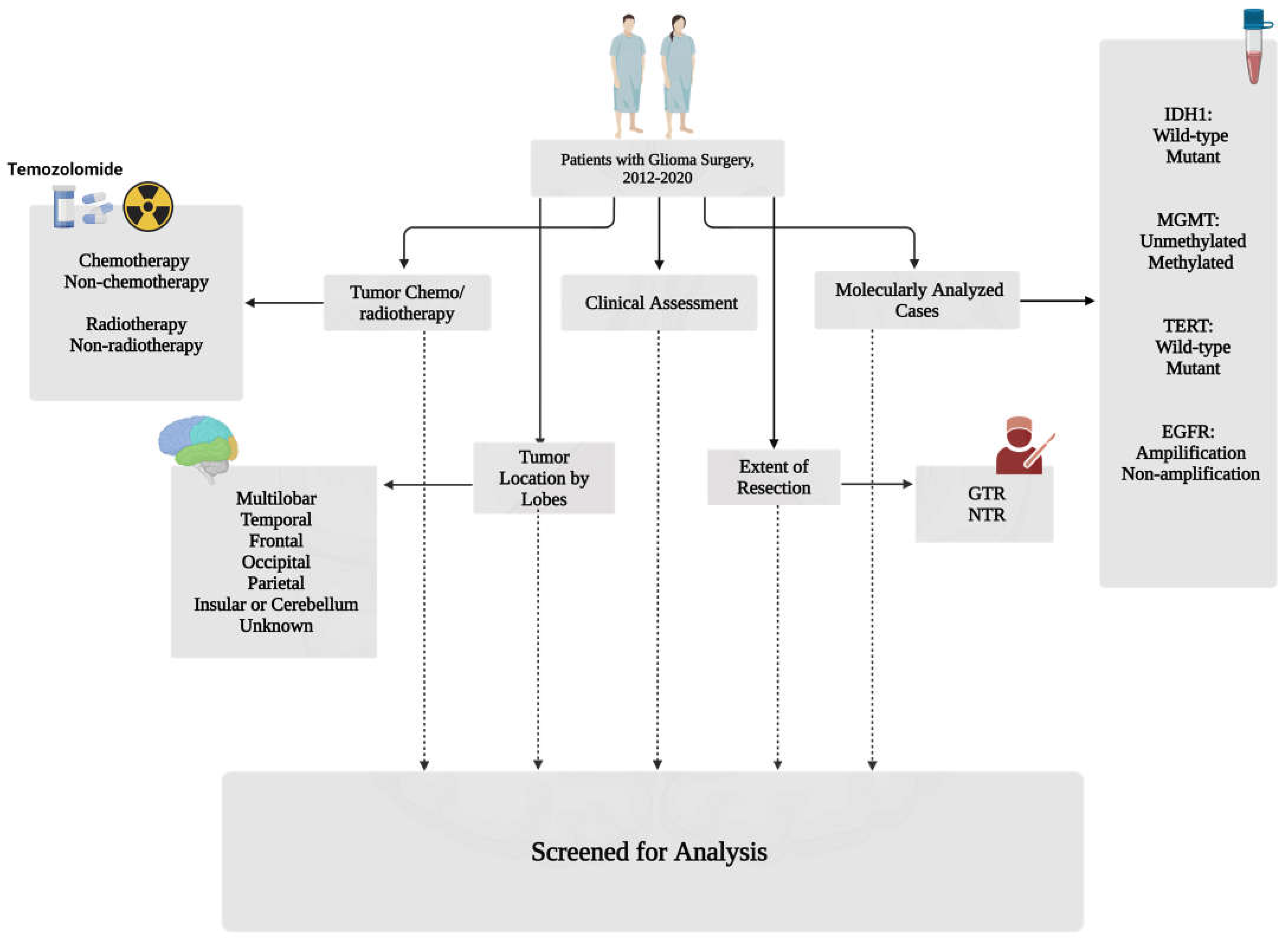
2.1. Population Characteristics and Study Design
2.2. Genetic Profile
2.3. Statistical Analysis
3. Results
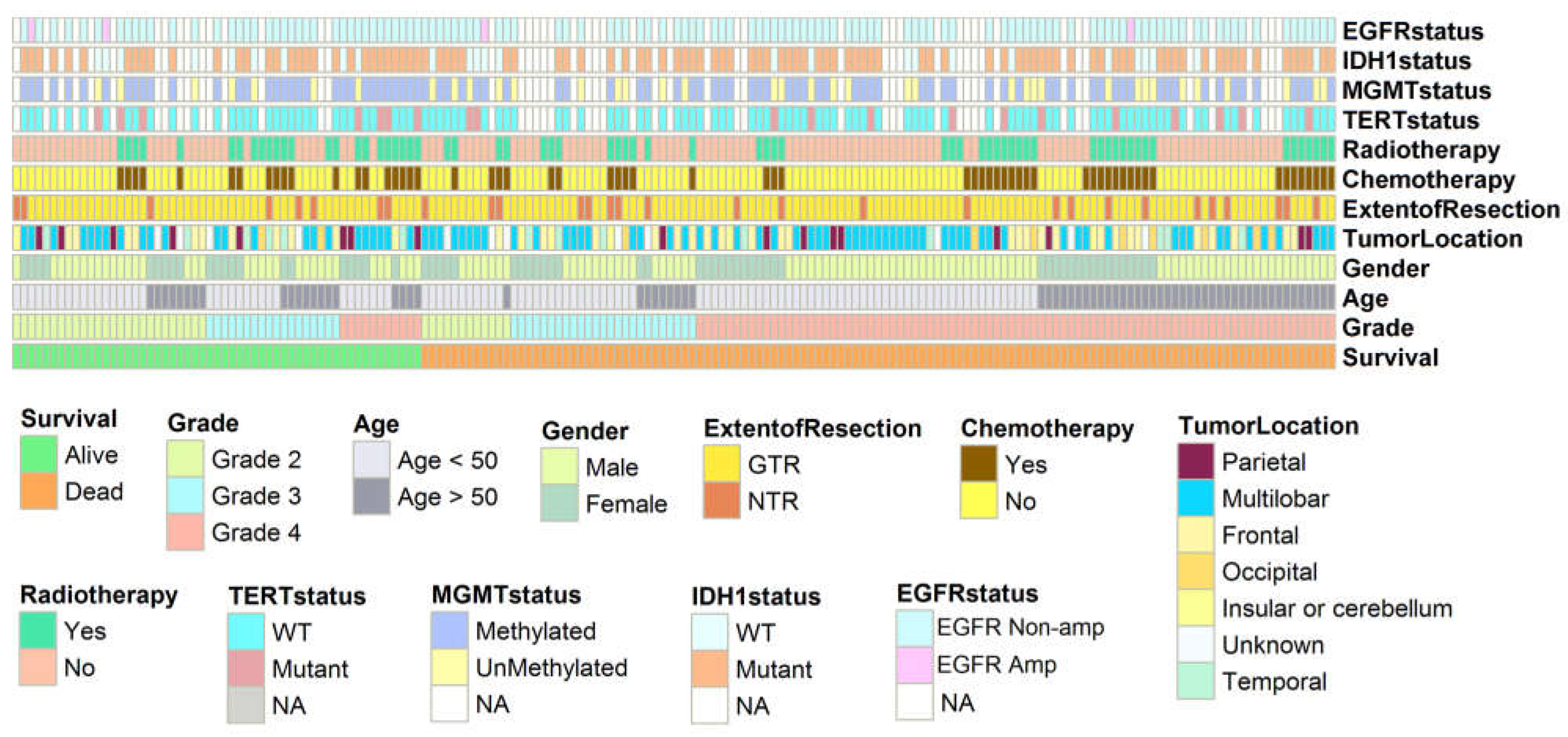
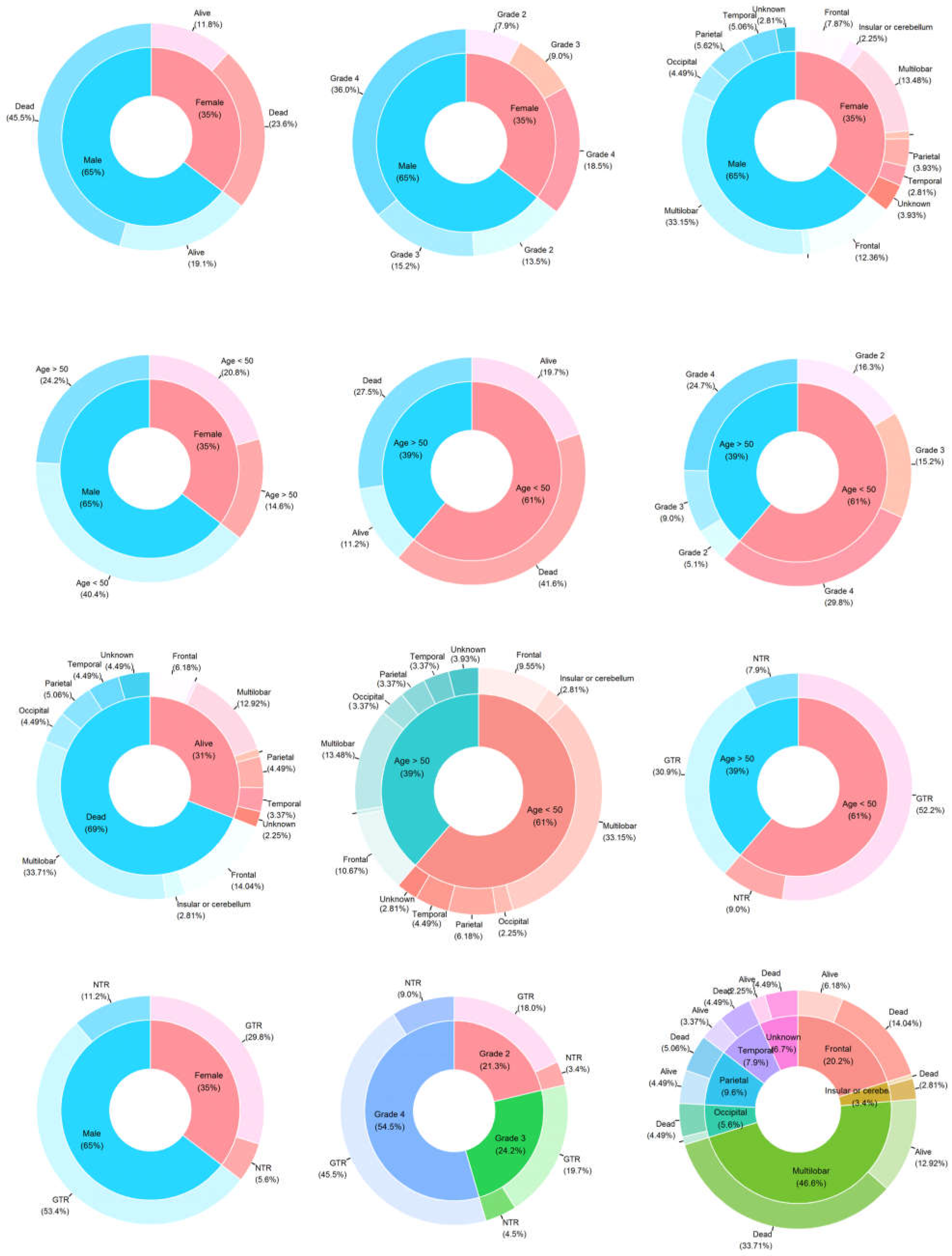
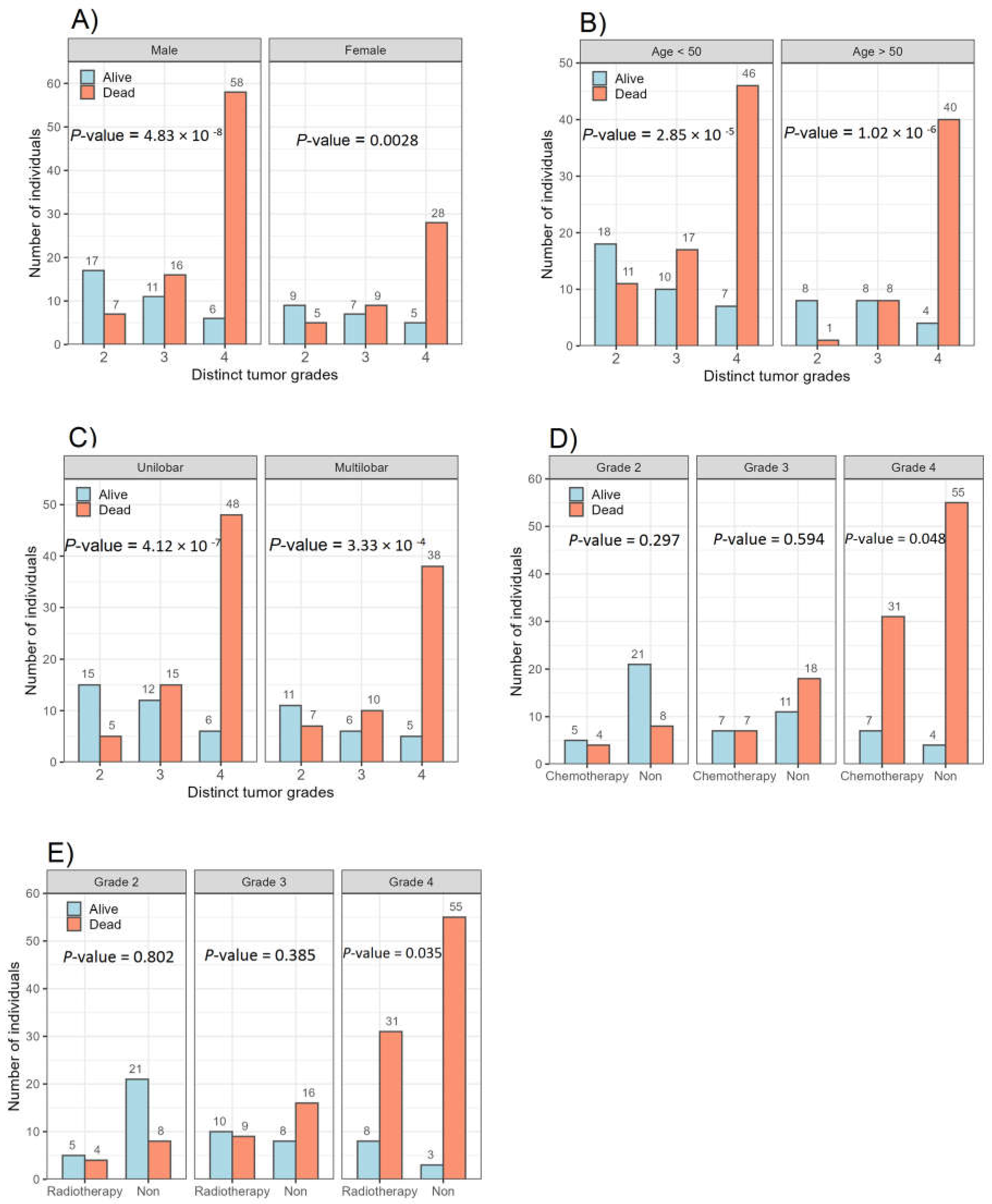
3.1. Association between Glioma Grades and Parameters Studied
3.2. IDH1 and EGFR Status
3.3. TERT and MGMT Genes and Clinical Response in Patients
3.4. Survival Analysis
3.5. Factors Associated with Overall Survival
4. Discussion
5. Conclusions and Future Direction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dunn, G.P.; Rinne, M.L.; Wykosky, J.; Genovese, G.; Quayle, S.N.; Dunn, I.F.; Agarwalla, P.K.; Chheda, M.G.; Campos, B.; Wang, A.; et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012, 26, 756–784. [Google Scholar] [CrossRef] [PubMed]
- Amini Harandi, A.; Zarifi, E.; Shahmohammadi, M.R. Prevalence of primary central nervous system tumors in Iran: A retrospective study. Universa Med. 2023, 42, 206–213. [Google Scholar] [CrossRef]
- Hajiahmadi, S.; Lorzadeh, S.; Iranpour, R.; Karima, S.; Rajabibazl, M.; Shahsavari, Z.; Ghavami, S. Temozolomide, Simvastatin and Acetylshikonin Combination Induces Mitochondrial-Dependent Apoptosis in GBM Cells, Which Is Regulated by Autophagy. Biology 2023, 12, 302. [Google Scholar] [CrossRef] [PubMed]
- Minniti, G.; Lombardi, G.; Paolini, S. Glioblastoma in Elderly Patients: Current Management and Future Perspectives. Cancers 2019, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- da Silva Rosa, S.C.; Barzegar Behrooz, A.; Guedes, S.; Vitorino, R.; Ghavami, S. Prioritization of genes for translation: A computational approach. Expert. Rev. Proteom. 2024, 21, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Dastghaib, S.; Shojaei, S.; Mostafavi-Pour, Z.; Sharma, P.; Patterson, J.B.; Samali, A.; Mokarram, P.; Ghavami, S. Simvastatin Induces Unfolded Protein Response and Enhances Temozolomide-Induced Cell Death in Glioblastoma Cells. Cells 2020, 9, 2339. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Li, Z.; Zeng, T.; Pan, X.; Chen, L.; Liu, D.; Li, H.; Huang, T.; Cai, Y.D. Distinguishing Glioblastoma Subtypes by Methylation Signatures. Front. Genet. 2020, 11, 604336. [Google Scholar] [CrossRef] [PubMed]
- Pour-Rashidi, A.; Yazdanpanah, N.; Rezaei, N. Introduction on Cancer Modifiable Risk Factors and Prevention. In Handbook of Cancer and Immunology; Rezaei, N., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–15. [Google Scholar] [CrossRef]
- Jacobs, J.; Iranpour, R.; Behrooz, A.B.; da Silva Rosa, S.C.; Ghavami, S. The role of BCL2L13 in glioblastoma: Turning a need into a target. Biochem. Cell Biol. 2024, 102, 127–134. [Google Scholar] [CrossRef]
- Dubrow, R.; Darefsky, A.S. Demographic variation in incidence of adult glioma by subtype, United States, 1992–2007. BMC Cancer 2011, 11, 325. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Dinesan, M.; Ajayakumar, T. Survival and quality of life analysis in glioblastoma multiforme with adjuvant chemoradiotherapy: A retrospective study. Rep. Pract. Oncol. Radiother. 2022, 27, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Pour-Rashidi, A.; Aarabi, J. The Principles of Successful Awake Craniotomy: Perioperative Tips and Tricks, 1st ed.; Springer: Singapore, 2023. [Google Scholar] [CrossRef]
- Behrooz, A.B.; Latifi-Navid, H.; Nezhadi, A.; Swiat, M.; Los, M.; Jamalpoor, Z.; Ghavami, S. Molecular mechanisms of microRNAs in glioblastoma pathogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2023, 1870, 119482. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ward, P.S.; Kapoor, G.S.; Rohle, D.; Turcan, S.; Abdel-Wahab, O.; Edwards, C.R.; Khanin, R.; Figueroa, M.E.; Melnick, A.; et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Buckner, J.; Giannini, C.; Eckel-Passow, J.; Lachance, D.; Parney, I.; Laack, N.; Jenkins, R. Management of diffuse low-grade gliomas in adults—Use of molecular diagnostics. Nat. Rev. Neurol. 2017, 13, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Masui, K.; Komori, T.; Kato, Y.; Masutomi, K.; Ichimura, K.; Ogasawara, S.; Kaneko, M.K.; Oki, H.; Suzuki, H.; Nitta, M.; et al. Elevated TERT Expression in TERT-Wildtype Adult Diffuse Gliomas: Histological Evaluation with a Novel TERT-Specific Antibody. Biomed. Res. Int. 2018, 2018, 7945845. [Google Scholar] [CrossRef] [PubMed]
- Szylberg, M.; Sokal, P.; Śledzińska, P.; Bebyn, M.; Krajewski, S.; Szylberg, Ł.; Szylberg, A.; Szylberg, T.; Krystkiewicz, K.; Birski, M.; et al. MGMT Promoter Methylation as a Prognostic Factor in Primary Glioblastoma: A Single-Institution Observational Study. Biomedicines 2022, 10, 2030. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Guo, D. EGFR mutation: Novel prognostic factor associated with immune infiltration in lower-grade glioma; an exploratory study. BMC Cancer 2019, 19, 1184. [Google Scholar] [CrossRef]
- Baid, U.; Rane, S.U.; Talbar, S.; Gupta, S.; Thakur, M.H.; Moiyadi, A.; Mahajan, A. Overall Survival Prediction in Glioblastoma With Radiomic Features Using Machine Learning. Front. Comput. Neurosci. 2020, 14, 61. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, S.; Watanabe, T.; Kleihues, P.; Ohgaki, H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin. Cancer Res. 2009, 15, 6002–6007. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Narita, Y.; Fukushima, S.; Tateishi, K.; Matsushita, Y.; Yoshida, A.; Miyakita, Y.; Ohno, M.; Collins, V.P.; Kawahara, N.; et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013, 126, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, R.; Huang, Z.; Li, J.; Wu, H.; Zhou, Y.; Zhu, J.; Wang, X. A Qualitative Signature to Identify TERT Promoter Mutant High-Risk Tumors in Low-Grade Gliomas. Front. Mol. Biosci. 2022, 9, 806727. [Google Scholar] [CrossRef] [PubMed]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A., Jr.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Rachakonda, P.S.; Hosen, I.; Volz, F.; Hemminki, K.; Weyerbrock, A.; Kumar, R. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 2015, 6, 10617–10633. [Google Scholar] [CrossRef] [PubMed]
- Bollam, S.R.; Berens, M.E.; Dhruv, H.D. When the Ends Are Really the Beginnings: Targeting Telomerase for Treatment of GBM. Curr. Neurol. Neurosci. Rep. 2018, 18, 15. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.; Blanco-Prieto, M.; Sousa, J.; Pais, A.; Vitorino, C. Breaching barriers in glioblastoma. Part I: Molecular pathways and novel treatment approaches. Int. J. Pharm. 2017, 531, 372–388. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Cheng, G.; Zhang, J.; Li, X. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide-based chemoradiotherapy. Clin. Neurol. Neurosurg. 2016, 151, 31–36. [Google Scholar] [CrossRef]
- Shojaei, S.; Koleini, N.; Samiei, E.; Aghaei, M.; Cole, L.K.; Alizadeh, J.; Islam, M.I.; Vosoughi, A.R.; Albokashy, M.; Butterfield, Y.; et al. Simvastatin increases temozolomide-induced cell death by targeting the fusion of autophagosomes and lysosomes. FEBS J. 2020, 287, 1005–1034. [Google Scholar] [CrossRef]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Kim, M.; Ladomersky, E.; Mozny, A.; Kocherginsky, M.; O’Shea, K.; Reinstein, Z.Z.; Zhai, L.; Bell, A.; Lauing, K.L.; Bollu, L.; et al. Glioblastoma as an age-related neurological disorder in adults. Neurooncol. Adv. 2021, 3, vdab125. [Google Scholar] [CrossRef]
- Yuan, G.; Niu, L.; Zhang, Y.; Wang, X.; Ma, K.; Yin, H.; Dai, J.; Zhou, W.; Pan, Y. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J. Neurooncol. 2017, 133, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Joudi Mashhad, M.; Hassanian, S.M.; Anvari, K.; Avan, A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J. Cell Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Francis, J.M.; Zhang, C.Z.; Maire, C.L.; Jung, J.; Manzo, V.E.; Adalsteinsson, V.A.; Homer, H.; Haidar, S.; Blumenstiel, B.; Pedamallu, C.S.; et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014, 4, 956–971. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Aldape, K.; Colman, H.; Holland, E.C.; Louis, D.N.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.K.; Perry, A.; Reifenberger, G.; Stupp, R.; et al. cIMPACT-NOW update 3: Recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018, 136, 805–810. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Higa, N.; Akahane, T.; Hamada, T.; Yonezawa, H.; Uchida, H.; Makino, R.; Watanabe, S.; Takajo, T.; Yokoyama, S.; Kirishima, M.; et al. Distribution and favorable prognostic implication of genomic EGFR alterations in IDH-wildtype glioblastoma. Cancer Med. 2023, 12, 49–60. [Google Scholar] [CrossRef]
- Reifenberger, G.; Collins, V.P. Pathology and molecular genetics of astrocytic gliomas. J. Mol. Med. 2004, 82, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Möllemann, M.; Wolter, M.; Felsberg, J.; Collins, V.P.; Reifenberger, G. Frequent promoter hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int. J. Cancer 2005, 113, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.; Pusch, S.; Balss, J.; Capper, D.; Mueller, W.; Christians, A.; Hartmann, C.; von Deimling, A. PCR- and restriction endonuclease-based detection of IDH1 mutations. Brain Pathol. 2010, 20, 298–300. [Google Scholar] [CrossRef]
- Capper, D.; Weissert, S.; Balss, J.; Habel, A.; Meyer, J.; Jäger, D.; Ackermann, U.; Tessmer, C.; Korshunov, A.; Zentgraf, H.; et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010, 20, 245–254. [Google Scholar] [CrossRef]
- Lyon, E.; Wittwer, C.T. LightCycler technology in molecular diagnostics. J. Mol. Diagn. 2009, 11, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J.M.; Skog, J.; Akers, J.; Li, H.; Komotar, R.; Jensen, R.; Ringel, F.; Yang, I.; Kalkanis, S.; Thompson, R.; et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 2017, 19, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Ostrom, Q.T.; Kruchko, C.; Patil, N.; Tihan, T.; Cioffi, G.; Fuchs, H.E.; Waite, K.A.; Jemal, A.; Siegel, R.L.; et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J. Clin. 2021, 71, 381–406. [Google Scholar] [CrossRef]
- Rabab’h, O.; Al-Ramadan, A.; Shah, J.; Lopez-Negrete, H.; Gharaibeh, A. Twenty Years after Glioblastoma Multiforme Diagnosis: A Case of Long-Term Survival. Cureus 2021, 13, e16061. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro Oncol. 2018, 20, iv1–iv86. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Verbaan, D.; Lamba, S.; Zanon, C.; Jeuken, J.W.; Boots-Sprenger, S.H.; Wesseling, P.; Hulsebos, T.J.; Troost, D.; van Tilborg, A.A.; et al. The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol. 2014, 16, 1263–1273. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Claus, E.B.; Cannataro, V.L.; Gaffney, S.G.; Townsend, J.P. Environmental and sex-specific molecular signatures of glioma causation. Neuro Oncol. 2022, 24, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Vienne-Jumeau, A.; Tafani, C.; Ricard, D. Environmental risk factors of primary brain tumors: A review. Rev. Neurol. 2019, 175, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Ho, V.K.; Reijneveld, J.C.; Enting, R.H.; Bienfait, H.P.; Robe, P.; Baumert, B.G.; Visser, O. Changing incidence and improved survival of gliomas. Eur. J. Cancer 2014, 50, 2309–2318. [Google Scholar] [CrossRef] [PubMed]
- Aldape, K.; Zadeh, G.; Mansouri, S.; Reifenberger, G.; von Deimling, A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 829–848. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; DeAngelis, L.M. Glioblastoma and other malignant gliomas: A clinical review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Jovčevska, I. Genetic secrets of long-term glioblastoma survivors. Bosn. J. Basic Med. Sci. 2019, 19, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Qi, C.; Maling, G.; Xiang, W.; Yanhui, L.; Ruofei, L.; Yunhe, M.; Jiewen, L.; Qing, M. TERT mutation in glioma: Frequency, prognosis and risk. J. Clin. Neurosci. 2016, 26, 57–62. [Google Scholar] [CrossRef]
- Powter, B.; Jeffreys, S.A.; Sareen, H.; Cooper, A.; Brungs, D.; Po, J.; Roberts, T.; Koh, E.S.; Scott, K.F.; Sajinovic, M.; et al. Human TERT promoter mutations as a prognostic biomarker in glioma. J. Cancer Res. Clin. Oncol. 2021, 147, 1007–1017. [Google Scholar] [CrossRef]
- Yang, W.; Warrington, N.M.; Taylor, S.J.; Whitmire, P.; Carrasco, E.; Singleton, K.W.; Wu, N.; Lathia, J.D.; Berens, M.E.; Kim, A.H.; et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci. Transl. Med. 2019, 11, eaao5253. [Google Scholar] [CrossRef]
- Tian, M.; Ma, W.; Chen, Y.; Yu, Y.; Zhu, D.; Shi, J.; Zhang, Y. Impact of gender on the survival of patients with glioblastoma. Biosci. Rep. 2018, 38, BSR20180752. [Google Scholar] [CrossRef]
- Bello-Alvarez, C.; Camacho-Arroyo, I. Impact of sex in the prevalence and progression of glioblastomas: The role of gonadal steroid hormones. Biol. Sex Differ. 2021, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Tseng, M.Y.; Tseng, J.H. Survival analysis for adult glioma in England and Wales. J. Formos. Med. Assoc. 2005, 104, 341–348. [Google Scholar]
- Claus, E.B.; Black, P.M. Survival rates and patterns of care for patients diagnosed with supratentorial low-grade gliomas: Data from the SEER program, 1973–2001. Cancer 2006, 106, 1358–1363. [Google Scholar] [CrossRef]
- Barone, T.A.; Gorski, J.W.; Greenberg, S.J.; Plunkett, R.J. Estrogen increases survival in an orthotopic model of glioblastoma. J. Neurooncol. 2009, 95, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jedlicka, A.; Ahuja, N.; Gibbons, M.C.; Baylin, S.B.; Burger, P.C.; Issa, J.P. Concordant methylation of the ER and N33 genes in glioblastoma multiforme. Oncogene 1998, 16, 3197–3202. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, Y.; Wei, W.; Cong, P.; Ding, Y.; Xiang, L.; Wu, K. Androgen receptor signaling regulates growth of glioblastoma multiforme in men. Tumour Biol. 2015, 36, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.G.; Nguyen, T.Q.; Ngo, T.N.M.; Nguyen, H.C.; Fung, K.M.; Dunn, I.F. The interaction between TERT promoter mutation and MGMT promoter methylation on overall survival of glioma patients: A meta-analysis. BMC Cancer 2020, 20, 897. [Google Scholar] [CrossRef]
- Kikuchi, Z.; Shibahara, I.; Yamaki, T.; Yoshioka, E.; Shofuda, T.; Ohe, R.; Matsuda, K.I.; Saito, R.; Kanamori, M.; Kanemura, Y.; et al. TERT promoter mutation associated with multifocal phenotype and poor prognosis in patients with IDH wild-type glioblastoma. Neurooncol. Adv. 2020, 2, vdaa114. [Google Scholar] [CrossRef]
- Hölzl, D.; Hutarew, G.; Zellinger, B.; Alinger-Scharinger, B.; Schlicker, H.U.; Schwartz, C.; Sotlar, K.; Kraus, T.F.J. EGFR Amplification Is a Phenomenon of IDH Wildtype and TERT Mutated High-Grade Glioma: An Integrated Analysis Using Fluorescence In Situ Hybridization and DNA Methylome Profiling. Biomedicines 2022, 10, 794. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Li, X.; Khan, S.; Zhang, C.; Gao, F.; Sen, S.; Wasylishen, A.R.; Zhao, Y.; Lozano, G.; Koul, D.; et al. EGFR suppresses p53 function by promoting p53 binding to DNA-PKcs: A noncanonical regulatory axis between EGFR and wild-type p53 in glioblastoma. Neuro Oncol. 2022, 24, 1712–1725. [Google Scholar] [CrossRef] [PubMed]
- AbdelFatah, M.A.R.; Kotb, A.; Said, M.A.; Abouelmaaty, E.M.H. Impact of extent of resection of newly diagnosed glioblastomas on survival: A meta-analysis. Egypt. J. Neurosurg. 2022, 37, 3. [Google Scholar] [CrossRef]
- Han, Q.; Liang, H.; Cheng, P.; Yang, H.; Zhao, P. Gross Total vs. Subtotal Resection on Survival Outcomes in Elderly Patients With High-Grade Glioma: A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Barak, T.; Vetsa, S.; Nadar, A.; Jin, L.; Gupte, T.P.; Fomchenko, E.I.; Miyagishima, D.F.; Yalcin, K.; Vasandani, S.; Gorelick, E.; et al. Surgical strategies for older patients with glioblastoma. J. Neurooncol. 2021, 155, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Hervey-Jumper, S.; Morshed, R.A.; Young, J.; Han, S.J.; Chunduru, P.; Zhang, Y.; Phillips, J.J.; Shai, A.; Lafontaine, M.; et al. Association of Maximal Extent of Resection of Contrast-Enhanced and Non-Contrast-Enhanced Tumor With Survival Within Molecular Subgroups of Patients With Newly Diagnosed Glioblastoma. JAMA Oncol. 2020, 6, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Ellingson, B.M.; Lai, A.; Harris, R.J.; Selfridge, J.M.; Yong, W.H.; Das, K.; Pope, W.B.; Nghiemphu, P.L.; Vinters, H.V.; Liau, L.M.; et al. Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR Am. J. Neuroradiol. 2013, 34, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Fyllingen, E.H.; Bø, L.E.; Reinertsen, I.; Jakola, A.S.; Sagberg, L.M.; Berntsen, E.M.; Salvesen, Ø.; Solheim, O. Survival of glioblastoma in relation to tumor location: A statistical tumor atlas of a population-based cohort. Acta Neurochir. 2021, 163, 1895–1905. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.T.; Achrol, A.S.; Mitchell, L.A.; Du, W.A.; Loya, J.J.; Rodriguez, S.A.; Feroze, A.; Westbroek, E.M.; Yeom, K.W.; Stuart, J.M.; et al. Computational Identification of Tumor Anatomic Location Associated with Survival in 2 Large Cohorts of Human Primary Glioblastomas. AJNR Am. J. Neuroradiol. 2016, 37, 621–628. [Google Scholar] [CrossRef]
- Sun, Z.L.; Chan, A.K.; Chen, L.C.; Tang, C.; Zhang, Z.Y.; Ding, X.J.; Wang, Y.; Sun, C.R.; Ng, H.K.; Yao, Y.; et al. TERT promoter mutated WHO grades II and III gliomas are located preferentially in the frontal lobe and avoid the midline. Int. J. Clin. Exp. Pathol. 2015, 8, 11485–11494. [Google Scholar]
- Fan, X.; Wang, Y.; Liu, Y.; Liu, X.; Zhang, C.; Wang, L.; Li, S.; Ma, J.; Jiang, T. Brain regions associated with telomerase reverse transcriptase promoter mutations in primary glioblastomas. J. Neurooncol. 2016, 128, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Yu, L.; Li, H.; Ou, Y.; Qiu, X.; Ding, Y.; Han, H.; Zhang, X. Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol. Lett. 2014, 7, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
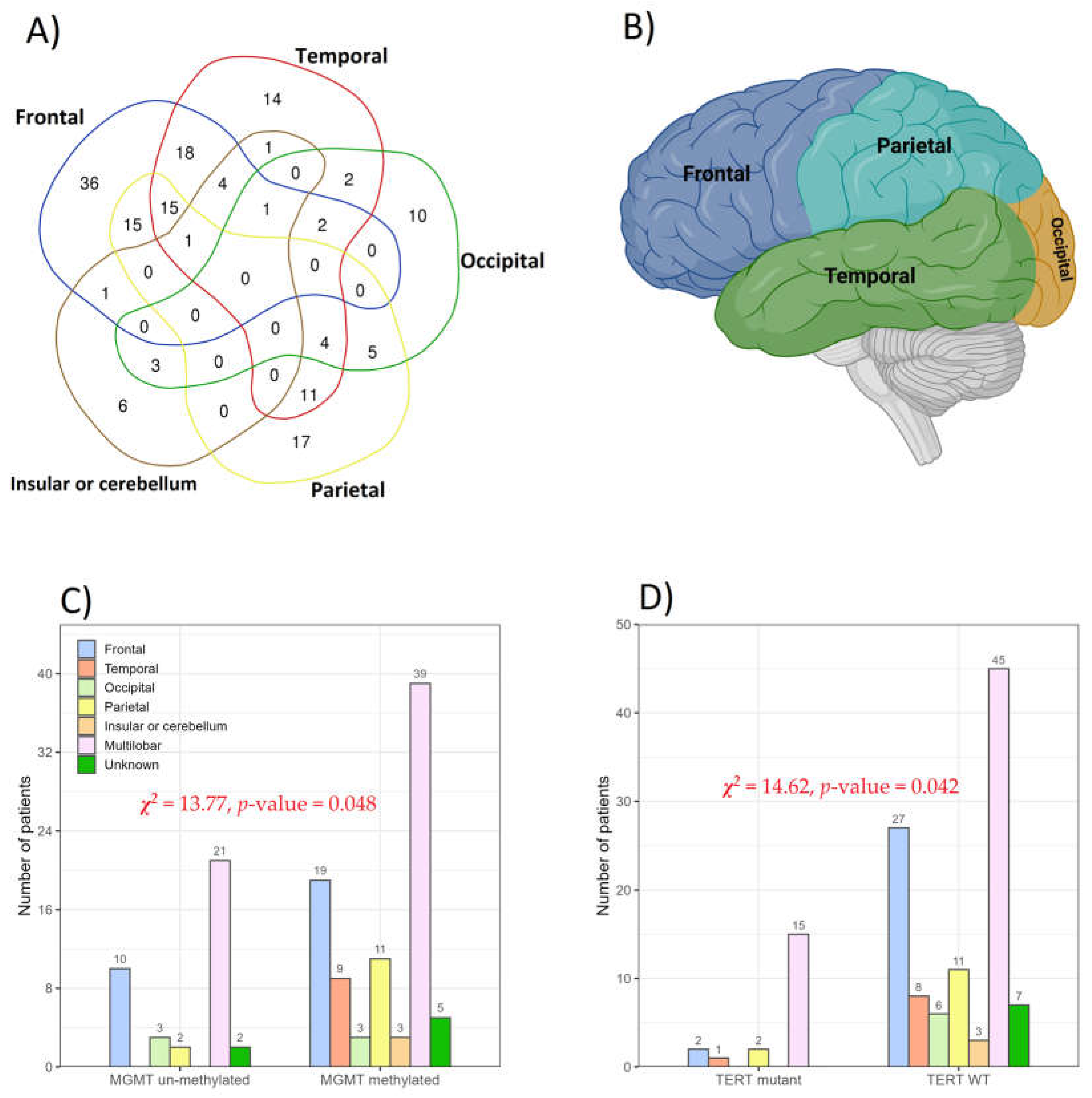
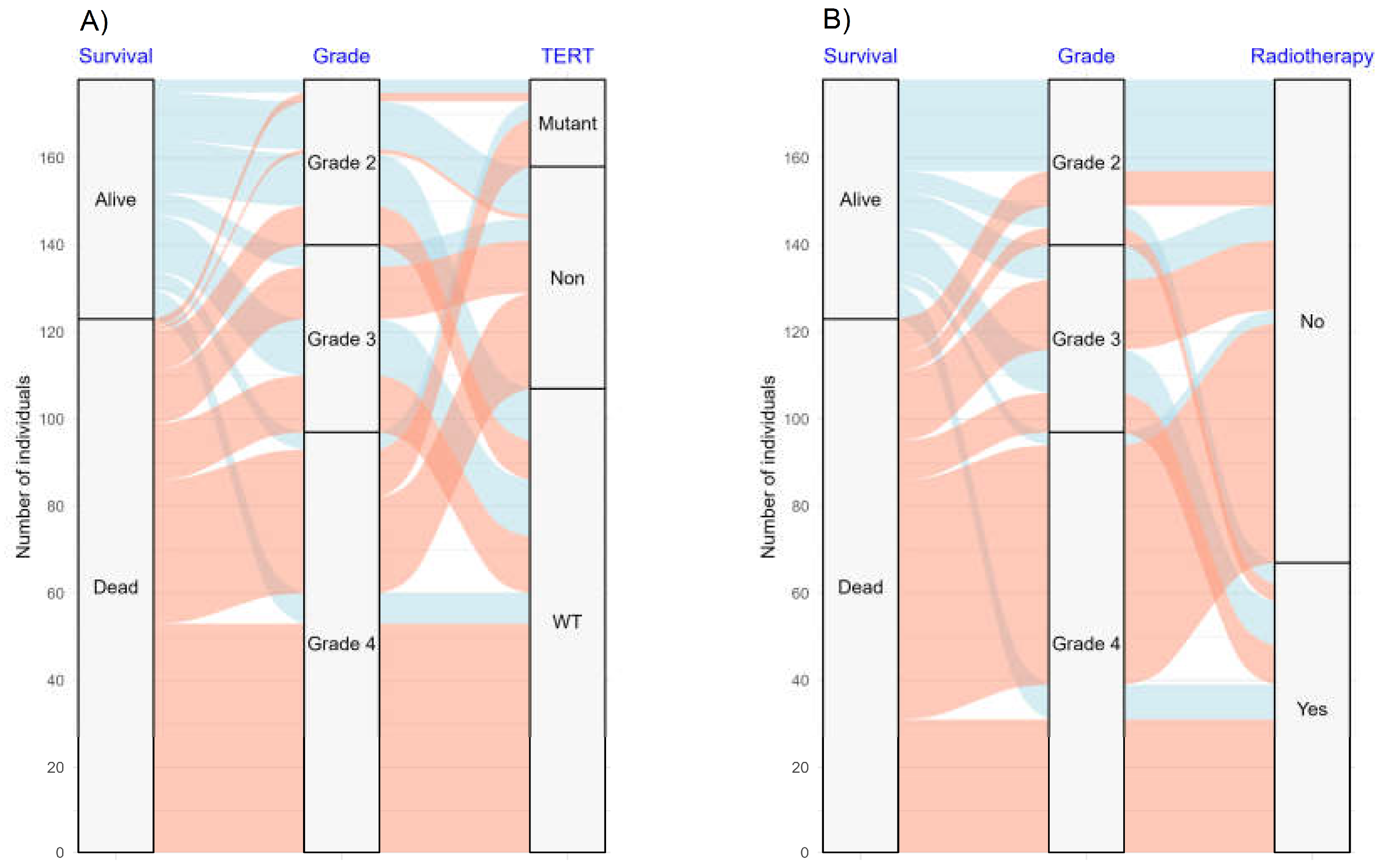
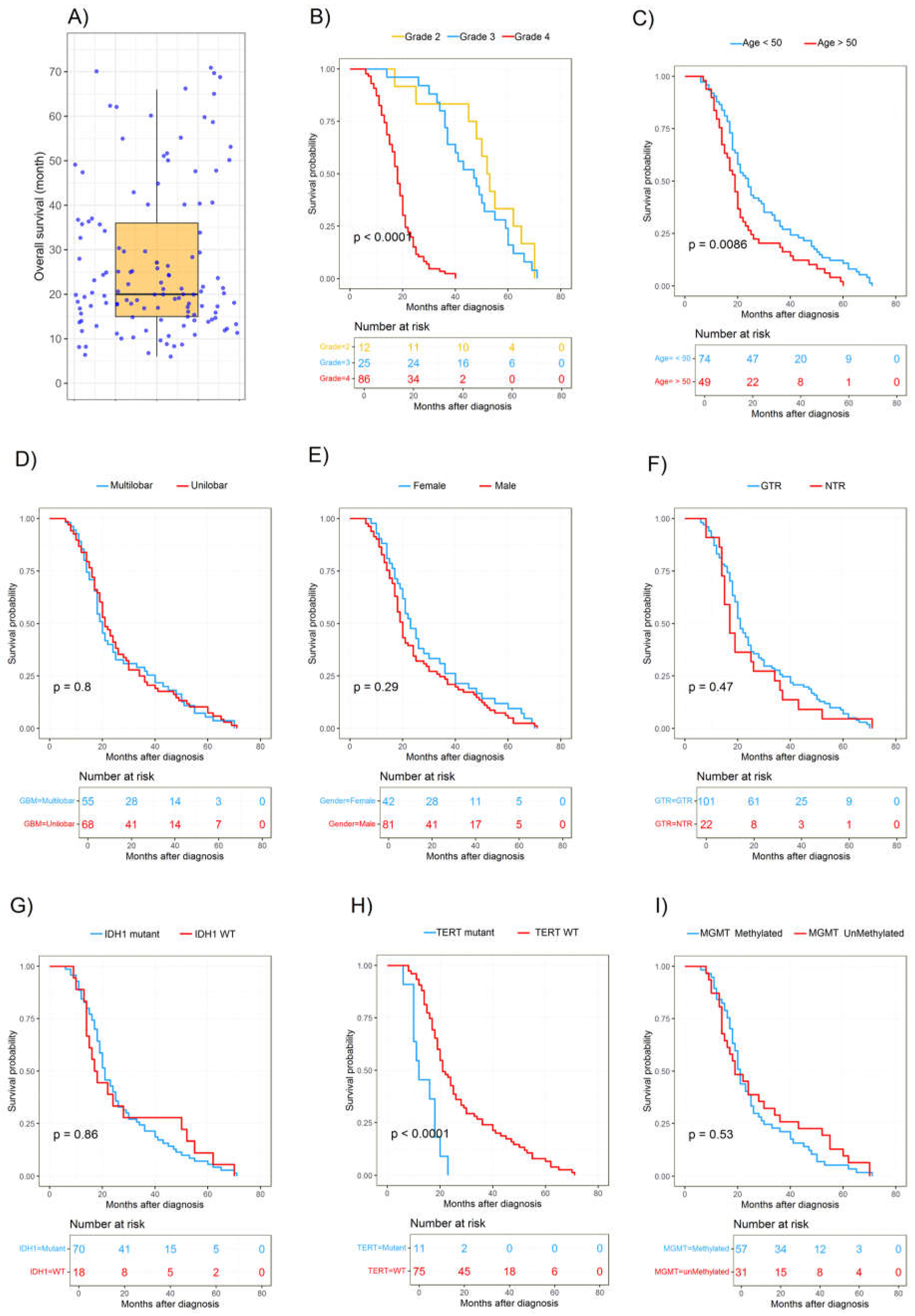
| Characteristics | Cases | % |
|---|---|---|
| Age group | ||
| <50 years | 109 | 61.2 |
| ≥50 years | 69 | 38.8 |
| Median age (range); years | 45 (22–81) | |
| Gender | ||
| Male | 115 | 64.6 |
| Female | 63 | 35.4 |
| Distinct tumor grades | ||
| Grade 2 | 38 | 21.3 |
| Grade 3 | 43 | 24.2 |
| Grade 4 | 97 | 54.5 |
| Extent of resection (EOR) | ||
| Gross-total resection (GTR) | 148 | 83.1 |
| Near-total resection (NTR) | 30 | 16.9 |
| Tumor location by lobe | ||
| Multilobar | 83 | 46.6 |
| Temporal | 14 | 7.9 |
| Frontal | 36 | 20.2 |
| Occipital | 10 | 5.6 |
| Parietal | 17 | 9.1 |
| Insular or cerebellum | 6 | 3.4 |
| Unknown | 12 | 6.7 |
| Survival | ||
| Alive | 55 | 30.9 |
| Dead | 123 | 69.1 |
| Survival ≤ 12 | 19 | 15.4 |
| 12 < Survival ≤ 24 | 56 | 45.5 |
| 24 < Survival ≤ 36 | 18 | 14.6 |
| Survival ≥ 36 | 30 | 24.4 |
| Multiple surgeries | ||
| Yes | 60 | 33.7 |
| No | 118 | 66.3 |
| Anesthesia | ||
| Awake | 20 | 12.1 |
| General | 146 | 87.9 |
| Chemotherapy | ||
| Yes | 61 | 34.3 |
| No | 117 | 65.7 |
| Adjuvant radiotherapy | ||
| Yes | 67 | 37.6 |
| No | 111 | 62.4 |
| Radio/chemotherapy | ||
| Yes | 56 | 31.5 |
| No | 122 | 68.5 |
| Type 2 diabetes mellitus | ||
| Yes | 14 | 7.9 |
| No | 164 | 92.1 |
| Molecular Assay | Different Grades of Glioma Tumor | Total | ||
|---|---|---|---|---|
| Grade 2 (26) | Grade 3 (26) | Grade 4 (75) | ||
| IDH1 status | ||||
| Wild type | 9 | 4 | 14 | 27 |
| Mutant | 17 | 22 | 61 | 100 |
| MGMT methylation status | ||||
| Methylated | 19 | 17 | 53 | 89 |
| Unmethylated | 7 | 9 | 22 | 38 |
| TERT promoter status | ||||
| Wild type | 21 | 26 | 60 | 107 |
| Mutant | 5 | 0 | 15 | 20 |
| EGFR amplification status | ||||
| Non-amplified | 23 | 26 | 74 | 123 |
| Amplified | 3 | 0 | 1 | 4 |
| Characteristic | Patients Number | Univariate | Multivariate | |||
|---|---|---|---|---|---|---|
| p-Value | p-Value | HR | 95% CI for HR | |||
| Lower | Upper | |||||
| Age, per year (<50 vs. >50) | 123 | 0.010 * | 0.224 | 1.370 | 0.866 | 1.845 |
| Gender (Male vs. Female) | 123 | 0.288 | 0.530 | 1.148 | 0.773 | 1.707 |
| Histological grade (Grades 2, 3, and 4) | 123 | 2.2 × 10−16 * | 8.57 × 10−11 * | 5.823 | 3.539 | 9.580 |
| Extent of resection (GTR vs. NTR) | 123 | 0.513 | 0.331 | 1.291 | 0.787 | 2.118 |
| Glioma (unilobar vs. multilobar) | 123 | 0.764 | 0.311 | 0.806 | 0.531 | 1.224 |
| Chemotherapy (Yes vs. No) | 123 | 0.791 | 0.539 | 1.297 | 0.569 | 2.957 |
| Adjuvant radiotherapy (Yes vs. No) | 123 | 0.0057 * | 0.0181 * | 1.807 | 0.375 | 2.705 |
| IDH1 status (WT vs. Mutant) | 88 | 0.430 | 0.043 * | 2.255 | 0.990 | 3.863 |
| TERT mutation (WT vs. Mutant) | 88 | 0.00041 * | 0.0143 * | 0.889 | 0.290 | 1.196 |
| MGMT methylation (WT vs. Mutant) | 88 | 0.769 | 0.553 | 0.848 | 0.493 | 1.461 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barzegar Behrooz, A.; Darzi Ramandi, H.; Latifi-Navid, H.; Peymani, P.; Tarharoudi, R.; Momeni, N.; Sabaghpour Azarian, M.M.; Eltonsy, S.; Pour-Rashidi, A.; Ghavami, S. Genetic Prognostic Factors in Adult Diffuse Gliomas: A 10-Year Experience at a Single Institution. Cancers 2024, 16, 2121. https://doi.org/10.3390/cancers16112121
Barzegar Behrooz A, Darzi Ramandi H, Latifi-Navid H, Peymani P, Tarharoudi R, Momeni N, Sabaghpour Azarian MM, Eltonsy S, Pour-Rashidi A, Ghavami S. Genetic Prognostic Factors in Adult Diffuse Gliomas: A 10-Year Experience at a Single Institution. Cancers. 2024; 16(11):2121. https://doi.org/10.3390/cancers16112121
Chicago/Turabian StyleBarzegar Behrooz, Amir, Hadi Darzi Ramandi, Hamid Latifi-Navid, Payam Peymani, Rahil Tarharoudi, Nasrin Momeni, Mohammad Mehdi Sabaghpour Azarian, Sherif Eltonsy, Ahmad Pour-Rashidi, and Saeid Ghavami. 2024. "Genetic Prognostic Factors in Adult Diffuse Gliomas: A 10-Year Experience at a Single Institution" Cancers 16, no. 11: 2121. https://doi.org/10.3390/cancers16112121
APA StyleBarzegar Behrooz, A., Darzi Ramandi, H., Latifi-Navid, H., Peymani, P., Tarharoudi, R., Momeni, N., Sabaghpour Azarian, M. M., Eltonsy, S., Pour-Rashidi, A., & Ghavami, S. (2024). Genetic Prognostic Factors in Adult Diffuse Gliomas: A 10-Year Experience at a Single Institution. Cancers, 16(11), 2121. https://doi.org/10.3390/cancers16112121








