Pro-Oxidant Auranofin and Glutathione-Depleting Combination Unveils Synergistic Lethality in Glioblastoma Cells with Aberrant Epidermal Growth Factor Receptor Expression
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Cell Vitality
2.3. Total Count and Viability Assay
2.4. Clonogenic Assay
2.5. Western Blot Analysis
2.6. Microscopy Fluorescence Imaging
2.7. Annexin-V
2.8. Measurement of Intracellular General ROS and Superoxide Anions
2.9. Combination Index Analysis
2.10. Determination of TrxR Activity
2.11. Glutathione (GSH) Assay
2.12. EGFR and GCLC Co-Expression Correlation in Brain Cancer and GBM Datasets
2.13. Statistical Analysis
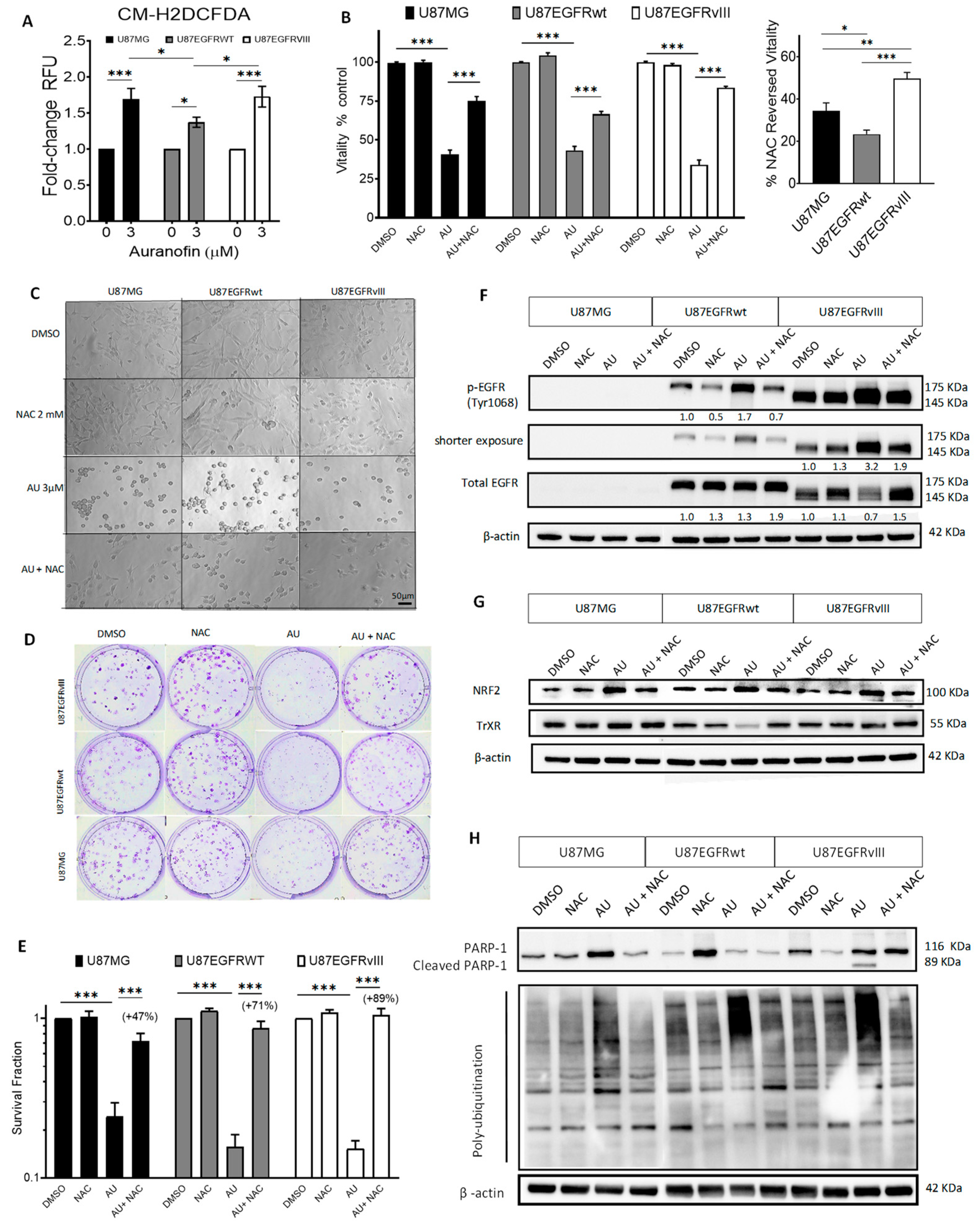
3. Results
3.1. Auranofin-Induced Cytotoxicity in Both Parental and Isogenic EGFRwt or EGFRvIII GBM Cell Lines
3.2. Auranofin Increased ROS along with ROS-Dependent Cytotoxicity and Molecular Effects in GBM U87/EGFR Isogenic Cell Lines
3.3. EGFR Alterations Are Associated with Reduced GSH in GBM Cell Lines and Decreased Correlation between EGFR and GCLC Expression in Patient Datasets
3.4. Co-Targeting TrxR and GCLC in U87/EGFR Isogenic Cell Lines Induced Synergistic Cytotoxicity Associated with Intracellular GSH Decrease
3.5. Auranofin and L-BSO Combination Synergistically Increased ROS-Dependent Cytotoxicity and Induced Differential Molecular Effects in U87/EGFR Isogenic Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 7-AAD | 7-aminoactinomycin D |
| AKT | Protein kinase B (PKB) |
| ATP | Adenosine triphosphate |
| ATR | Ataxia telangiectasia and Rad3-related protein |
| AU | Auranofin |
| BCA | Bicinchoninic acid |
| Cbl | Casitas B-lineage Lymphoma |
| CI | Combination index |
| CM-H2DCFDA | 2′,7′-dichlorodihydrofluorescein diacetate |
| CO2 | Carbon dioxide |
| CUSP9v3 | Coordinated Undermining of Survival Paths combining 9 repurposed non-oncological drugs with metronomic temozolomide-version 3 |
| Cys | Cysteine |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DHE | Dihydroethidium |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DMSO | Dimethyl sulfoxide |
| DNA | Deoxyribonucleic acid |
| DTNB | 5,5′-dithiobis (2-nitrobenzoic) acid |
| DUBs | Deubiquitinases |
| ECL | Enhanced chemiluminescence |
| EGFR | Epidermal growth factor receptor |
| ERK | Extracellular signal-regulated kinase |
| FBS | Fetal bovine serum |
| FDA | US Food and Drug Administration |
| GBM | Glioblastoma |
| GCLC | Glutamate-cysteine ligase catalytic |
| Grb2 | Growth Factor Receptor Bound Protein 2 |
| GSH | Glutathione |
| H2AX | Histone variant H2AX |
| H2O2 | Hydrogen Peroxide |
| HRP | Horseradish peroxidase |
| IC50 | Half-maximal inhibitory concentration |
| IDH | Isocitrate dehydrogenase |
| kDa | Kilodaltons |
| L-BSO | L-buthionine-sulfoximine |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NAC | N-acetylcysteine |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NOX3 | NADPH Oxidase 3 |
| NRF2 | Nuclear factor erythroid 2–related factor 2 |
| OXPHOS | Oxidative phosphorylation |
| PAGE | Polyacrylamide gel electrophoresis |
| PARP | Poly(ADP-ribose) polymerase |
| PBS | Phosphate Buffered Saline |
| PI3K | Phosphoinositide 3-kinase |
| PTEN | Phosphatase and tensin homolog |
| PTPs | Protein tyrosine phosphatases |
| PVDF | Polyvinylidene fluoride |
| Ras | Rat sarcoma virus (protein) |
| ROS | Reactive oxygen species |
| SDS | Sodium dodecyl sulfate |
| SEM | Standard error of the mean |
| STAT | Signal transducer and activator of transcription |
| TBS | Tris-Buffered Saline |
| TCGA | The Cancer Genome Atlas |
| TGX | Tris-Glycine eXtended |
| TMZ | Temozolomide |
| TNB | 2-nitro-5-thiobenzoate |
| Trx | Thioredoxin |
| TrxR | Thioredoxin reductase |
| Tyr | Tyrosine |
| UBA1 | Ubiquitin-like modifier activating enzyme 1 |
| USPs | Ubiquitin-specific proteases |
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Camacho, A.; Flores-Vazquez, J.G.; Moscardini-Martelli, J.; Torres-Rios, J.A.; Olmos-Guzman, A.; Ortiz-Arce, C.S.; Cid-Sanchez, D.R.; Perez, S.R.; Macias-Gonzalez, M.D.S.; Hernandez-Sanchez, L.C.; et al. Glioblastoma Treatment: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2022, 23, 7207. [Google Scholar] [CrossRef] [PubMed]
- Rong, L.; Li, N.; Zhang, Z. Emerging therapies for glioblastoma: Current state and future directions. J. Exp. Clin. Cancer Res. 2022, 41, 142. [Google Scholar] [CrossRef] [PubMed]
- Poon, M.T.C.; Sudlow, C.L.M.; Figueroa, J.D.; Brennan, P.M. Longer-term (≥2 years) survival in patients with glioblastoma in population-based studies pre- and post-2005: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 11622. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zong, H.; Ma, C.; Ming, X.; Shang, M.; Li, K.; He, X.; Du, H.; Cao, L. Epidermal growth factor receptor in glioblastoma. Oncol. Lett. 2017, 14, 512–516. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Hatanpaa, K.J.; Burma, S.; Zhao, D.; Habib, A.A. Epidermal growth factor receptor in glioma: Signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 2010, 12, 675–684. [Google Scholar] [CrossRef]
- Yamoutpour, F.; Bodempudi, V.; Park, S.E.; Pan, W.; Mauzy, M.J.; Kratzke, R.A.; Dudek, A.; Potter, D.A.; Woo, R.A.; O’Rourke, D.M.; et al. Gene silencing for epidermal growth factor receptor variant III induces cell-specific cytotoxicity. Mol. Cancer Ther. 2008, 7, 3586–3597. [Google Scholar] [CrossRef][Green Version]
- Gan, H.K.; Cvrljevic, A.N.; Johns, T.G. The epidermal growth factor receptor variant III (EGFRvIII): Where wild things are altered. FEBS J. 2013, 280, 5350–5370. [Google Scholar] [CrossRef]
- Nagane, M.; Coufal, F.; Lin, H.; Bögler, O.; Cavenee, W.K.; Huang, H.J. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996, 56, 5079–5086. [Google Scholar]
- Aldaz, P.; Arozarena, I. Tyrosine Kinase Inhibitors in Adult Glioblastoma: An (Un)Closed Chapter? Cancers 2021, 13, 5799. [Google Scholar] [CrossRef]
- Kim, G.; Ko, Y.T. Small molecule tyrosine kinase inhibitors in glioblastoma. Arch. Pharm. Res. 2020, 43, 385–394. [Google Scholar] [CrossRef]
- Van Loenhout, J.; Peeters, M.; Bogaerts, A.; Smits, E.; Deben, C. Oxidative Stress-Inducing Anticancer Therapies: Taking a Closer Look at Their Immunomodulating Effects. Antioxidants 2020, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Nizami, Z.N.; Aburawi, H.E.; Semlali, A.; Muhammad, K.; Iratni, R. Oxidative Stress Inducers in Cancer Therapy: Preclinical and Clinical Evidence. Antioxidants 2023, 12, 1159. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef]
- Kong, Q.; Beel, J.A.; Lillehei, K.O. A threshold concept for cancer therapy. Med. Hypotheses 2000, 55, 29–35. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Zhou, L.; Xie, N.; Nice, E.C.; Zhang, H.; Huang, C.; Lei, Y. Cancer drug resistance: Redox resetting renders a way. Oncotarget 2016, 7, 42740–42761. [Google Scholar] [CrossRef]
- Ren, X.; Zou, L.; Zhang, X.; Branco, V.; Wang, J.; Carvalho, C.; Holmgren, A.; Lu, J. Redox Signaling Mediated by Thioredoxin and Glutathione Systems in the Central Nervous System. Antioxid. Redox Signal. 2017, 27, 989–1010. [Google Scholar] [CrossRef]
- Mohammadi, F.; Soltani, A.; Ghahremanloo, A.; Javid, H.; Hashemy, S.I. The thioredoxin system and cancer therapy: A review. Cancer Chemother. Pharmacol. 2019, 84, 925–935. [Google Scholar] [CrossRef]
- Gencheva, R.; Arnér, E.S.J. Thioredoxin Reductase Inhibition for Cancer Therapy. Annu. Rev. Pharmacol. Toxicol. 2022, 62, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Gandin, V.; Fernandes, A.P. Metal- and Semimetal-Containing Inhibitors of Thioredoxin Reductase as Anticancer Agents. Molecules 2015, 20, 12732–12756. [Google Scholar] [CrossRef]
- Weng, M.S.; Chang, J.H.; Hung, W.Y.; Yang, Y.C.; Chien, M.H. The interplay of reactive oxygen species and the epidermal growth factor receptor in tumor progression and drug resistance. J. Exp. Clin. Cancer Res. 2018, 37, 61. [Google Scholar] [CrossRef] [PubMed]
- Gamou, S.; Shimizu, N. Hydrogen peroxide preferentially enhances the tyrosine phosphorylation of epidermal growth factor receptor. FEBS Lett. 1995, 357, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.S.; Kang, S.W.; Seo, M.S.; Baines, I.C.; Tekle, E.; Chock, P.B.; Rhee, S.G. Epidermal Growth Factor (EGF)-induced Generation of Hydrogen Peroxide: Role In EGF Receptor-Mediated Tyrosine Phosphorylation. J. Biol. Chem. 1997, 272, 217–221. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Truong, T.H.; Garcia, F.J.; Homann, A.; Gupta, V.; Leonard, S.E.; Carroll, K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2011, 8, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Truong, T.H.; Carroll, K.S. Redox regulation of epidermal growth factor receptor signaling through cysteine oxidation. Biochemistry 2012, 51, 9954–9965. [Google Scholar] [CrossRef]
- Ravid, T.; Sweeney, C.; Gee, P.; Carraway, K.L., III; Goldkorn, T. Epidermal Growth Factor Receptor Activation under Oxidative Stress Fails to Promote c-Cbl Mediated Down-regulation. J. Biol. Chem. 2002, 277, 31214–31219. [Google Scholar] [CrossRef] [PubMed]
- Nitta, M.; Kozono, D.; Kennedy, R.; Stommel, J.; Ng, K.; Zinn, P.O.; Kushwaha, D.; Kesari, S.; Inda, M.M.; Wykosky, J.; et al. Targeting EGFR induced oxidative stress by PARP1 inhibition in glioblastoma therapy. PLoS ONE 2010, 5, e10767. [Google Scholar] [CrossRef]
- Wu, S.; Gao, F.; Zheng, S.; Zhang, C.; Martinez-Ledesma, E.; Ezhilarasan, R.; Ding, J.; Li, X.; Feng, N.; Multani, A.; et al. EGFR Amplification Induces Increased DNA Damage Response and Renders Selective Sensitivity to Talazoparib (PARP Inhibitor) in Glioblastoma. Clin. Cancer Res. 2020, 26, 1395–1407. [Google Scholar] [CrossRef]
- Okamura, T.; Antoun, G.; Keir, S.T.; Friedman, H.; Bigner, D.D.; Ali-Osman, F. Phosphorylation of Glutathione S-Transferase P1 (GSTP1) by Epidermal Growth Factor Receptor (EGFR) Promotes Formation of the GSTP1-c-Jun N-terminal kinase (JNK) Complex and Suppresses JNK Downstream Signaling and Apoptosis in Brain Tumor Cells. J. Biol. Chem. 2015, 290, 30866–30878. [Google Scholar] [CrossRef] [PubMed]
- Okamura, T.; Singh, S.; Buolamwini, J.; Haystead, T.; Friedman, H.; Bigner, D.; Ali-Osman, F. Tyrosine phosphorylation of the human glutathione S-transferase P1 by epidermal growth factor receptor. J. Biol. Chem. 2009, 284, 16979–16989. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Xia, Y.; Wang, X.C.; Ahn, E.H.; Jin, L.; Ye, K. C/EBPbeta mediates NQO1 and GSTP1 anti-oxidative reductases expression in glioblastoma, promoting brain tumor proliferation. Redox Biol. 2020, 34, 101578. [Google Scholar] [CrossRef] [PubMed]
- Inda, M.M.; Bonavia, R.; Mukasa, A.; Narita, Y.; Sah, D.W.; Vandenberg, S.; Brennan, C.; Johns, T.G.; Bachoo, R.; Hadwiger, P.; et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010, 24, 1731–1745. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, R.; Ji, X.D.; Harmon, R.C.; Lazar, C.S.; Gill, G.N.; Cavenee, W.K.; Huang, H.J. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc. Natl. Acad. Sci. USA 1994, 91, 7727–7731. [Google Scholar] [CrossRef] [PubMed]
- Watt, H.L.; Rachid, Z.; Jean-Claude, B.J. The Concept of Divergent Targeting through the Activation and Inhibition of Receptors as a Novel Chemotherapeutic Strategy: Signaling Responses to Strong DNA-Reactive Combinatorial Mimicries. J. Signal Transduct. 2012, 2012, 282050. [Google Scholar] [CrossRef] [PubMed]
- Watt, H.L.; Rachid, Z.; Jean-Claude, B.J. Receptor activation and inhibition in cellular response to chemotherapeutic combinational mimicries: The concept of divergent targeting. J. Neuro-Oncol. 2010, 100, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Forgie, B.N.; Prakash, R.; Goyeneche, A.A.; Telleria, C.M. Vitality, viability, long-term clonogenic survival, cytotoxicity, cytostasis and lethality: What do they mean when testing new investigational oncology drugs? Discov. Oncol. 2024, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Tada, H.; Shiho, O.; Kuroshima, K.; Koyama, M.; Tsukamoto, K. An improved colorimetric assay for interleukin 2. J. Immunol. Methods 1986, 93, 157–165. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Abdalbari, F.H.; Martinez-Jaramillo, E.; Forgie, B.N.; Tran, E.; Zorychta, E.; Goyeneche, A.A.; Sabri, S.; Telleria, C.M. Auranofin induces lethality driven by reactive oxygen species in high-grade serous ovarian cancer cells. Cancers 2023, 15, 5136. [Google Scholar] [CrossRef]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Miller, H.E.; Bishop, A.J.R. Correlation AnalyzeR: Functional predictions from gene co-expression correlations. BMC Bioinform. 2021, 22, 206. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Gamberi, T.; Chiappetta, G.; Fiaschi, T.; Modesti, A.; Sorbi, F.; Magherini, F. Upgrade of an old drug: Auranofin in innovative cancer therapies to overcome drug resistance and to increase drug effectiveness. Med. Res. Rev. 2022, 42, 1111–1146. [Google Scholar] [CrossRef]
- Krabbendam, I.E.; Honrath, B.; Bothof, L.; Silva-Pavez, E.; Huerta, H.; Penaranda Fajardo, N.M.; Dekker, F.; Schmidt, M.; Culmsee, C.; Cesar Cardenas, J.; et al. SK channel activation potentiates auranofin-induced cell death in glio- and neuroblastoma cells. Biochem. Pharmacol. 2020, 171, 113714. [Google Scholar] [CrossRef]
- Szeliga, M.; Rola, R. Menadione Potentiates Auranofin-Induced Glioblastoma Cell Death. Int. J. Mol. Sci. 2022, 23, 15712. [Google Scholar] [CrossRef]
- Van Loenhout, J.; Freire Boullosa, L.; Quatannens, D.; De Waele, J.; Merlin, C.; Lambrechts, H.; Lau, H.W.; Hermans, C.; Lin, A.; Lardon, F.; et al. Auranofin and Cold Atmospheric Plasma Synergize to Trigger Distinct Cell Death Mechanisms and Immunogenic Responses in Glioblastoma. Cells 2021, 10, 2936. [Google Scholar] [CrossRef]
- Seo, M.J.; Kim, I.Y.; Lee, D.M.; Park, Y.J.; Cho, M.Y.; Jin, H.J.; Choi, K.S. Dual inhibition of thioredoxin reductase and proteasome is required for auranofin-induced paraptosis in breast cancer cells. Cell Death Dis. 2023, 14, 42. [Google Scholar] [CrossRef]
- Zhang, X.; Selvaraju, K.; Saei, A.A.; D’Arcy, P.; Zubarev, R.A.; Arner, E.S.; Linder, S. Repurposing of auranofin: Thioredoxin reductase remains a primary target of the drug. Biochimie 2019, 162, 46–54. [Google Scholar] [CrossRef]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12827. [Google Scholar] [CrossRef]
- Liu, W.M.; Dalgleish, A.G. MTT assays can underestimate cell numbers. Cancer Chemother. Pharmacol. 2009, 64, 861–862. [Google Scholar] [CrossRef]
- Sze, J.H.; Raninga, P.V.; Nakamura, K.; Casey, M.; Khanna, K.K.; Berners-Price, S.J.; Di Trapani, G.; Tonissen, K.F. Anticancer activity of a Gold(I) phosphine thioredoxin reductase inhibitor in multiple myeloma. Redox Biol. 2020, 28, 101310. [Google Scholar] [CrossRef]
- Cui, X.Y.; Park, S.H.; Park, W.H. Anti-Cancer Effects of Auranofin in Human Lung Cancer Cells by Increasing Intracellular ROS Levels and Depleting GSH Levels. Molecules 2022, 27, 5207. [Google Scholar] [CrossRef]
- You, B.R.; Shin, H.R.; Han, B.R.; Kim, S.H.; Park, W.H. Auranofin induces apoptosis and necrosis in HeLa cells via oxidative stress and glutathione depletion. Mol. Med. Rep. 2015, 11, 1428–1434. [Google Scholar] [CrossRef]
- Fiskus, W.; Saba, N.; Shen, M.; Ghias, M.; Liu, J.; Gupta, S.D.; Chauhan, L.; Rao, R.; Gunewardena, S.; Schorno, K.; et al. Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res. 2014, 74, 2520–2532. [Google Scholar] [CrossRef]
- You, B.R.; Park, W.H. Auranofin induces mesothelioma cell death through oxidative stress and GSH depletion. Oncol. Rep. 2016, 35, 546–551. [Google Scholar] [CrossRef]
- Kalyanaraman, B. NAC, NAC, Knockin’ on Heaven’s door: Interpreting the mechanism of action of N-acetylcysteine in tumor and immune cells. Redox Biol. 2022, 57, 102497. [Google Scholar] [CrossRef] [PubMed]
- Prigent, S.A.; Nagane, M.; Lin, H.; Huvar, I.; Boss, G.R.; Feramisco, J.R.; Cavenee, W.K.; Huang, H.S. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J. Biol. Chem. 1996, 271, 25639–25645. [Google Scholar] [CrossRef]
- Fan, Z.; Wirth, A.K.; Chen, D.; Wruck, C.J.; Rauh, M.; Buchfelder, M.; Savaskan, N. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 2017, 6, e371. [Google Scholar] [CrossRef]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Guo, Z.; Xia, X.; Liao, Y.; Zhang, F.; Huang, C.; Liu, Y.; Deng, X.; Jiang, L.; Wang, X.; et al. Auranofin lethality to prostate cancer includes inhibition of proteasomal deubiquitinases and disrupted androgen receptor signaling. Eur. J. Pharmacol. 2019, 846, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Huang, H.; Dou, Q.P.; Liu, J. Inhibition of 19S proteasome-associated deubiquitinases by metal-containing compounds. Oncoscience 2015, 2, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Li, X.; Huang, H.; Zhao, C.; Liao, S.; Yang, C.; Liu, S.; Song, W.; Lu, X.; Lan, X.; et al. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget 2014, 5, 5453–5471. [Google Scholar] [CrossRef] [PubMed]
- Habermann, K.J.; Grunewald, L.; van Wijk, S.; Fulda, S. Targeting redox homeostasis in rhabdomyosarcoma cells: GSH-depleting agents enhance auranofin-induced cell death. Cell Death Dis. 2017, 8, e3067. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, M.; Cui, X.; Lv, D.; Jin, L.; Khan, M.; Ma, T. Brevilin A promotes oxidative stress and induces mitochondrial apoptosis in U87 glioblastoma cells. Onco Targets Ther. 2018, 11, 7031–7040. [Google Scholar] [CrossRef] [PubMed]
- Schonberg, S.A.; Rudra, P.K.; Noding, R.; Skorpen, F.; Bjerve, K.S.; Krokan, H.E. Evidence that changes in Se-glutathione peroxidase levels affect the sensitivity of human tumour cell lines to n-3 fatty acids. Carcinogenesis 1997, 18, 1897–1904. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, P.; Zhang, Y.; Lu, W.; Ding, W.; Luo, Y.; Wen, S.; Xu, R.; Liu, P.; Huang, P. Synergy between Auranofin and Celecoxib against Colon Cancer In Vitro and In Vivo through a Novel Redox-Mediated Mechanism. Cancers 2019, 11, 931. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.S.; Steinauer, K.K.; Hornung, B.; Irish, J.M.; Lecane, P.; Birrell, G.W.; Peehl, D.M.; Knox, S.J. Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ. 2002, 9, 252–263. [Google Scholar] [CrossRef]
- Zhu, Z.; Du, S.; Du, Y.; Ren, J.; Ying, G.; Yan, Z. Glutathione reductase mediates drug resistance in glioblastoma cells by regulating redox homeostasis. J. Neurochem. 2018, 144, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Marengo, B.; De Ciucis, C.; Verzola, D.; Pistoia, V.; Raffaghello, L.; Patriarca, S.; Balbis, E.; Traverso, N.; Cottalasso, D.; Pronzato, M.A.; et al. Mechanisms of BSO (L-buthionine-S,R-sulfoximine)-induced cytotoxic effects in neuroblastoma. Free Radic. Biol. Med. 2008, 44, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Harris, I.S.; Treloar, A.E.; Inoue, S.; Sasaki, M.; Gorrini, C.; Lee, K.C.; Yung, K.Y.; Brenner, D.; Knobbe-Thomsen, C.B.; Cox, M.A.; et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 2015, 27, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhang, H.; Zhang, X.; Lu, J.; Holmgren, A. Thioredoxin 1 is inactivated due to oxidation induced by peroxiredoxin under oxidative stress and reactivated by the glutaredoxin system. J. Biol. Chem. 2013, 288, 32241–32247. [Google Scholar] [CrossRef]
- Muri, J.; Thut, H.; Heer, S.; Krueger, C.C.; Bornkamm, G.W.; Bachmann, M.F.; Kopf, M. The thioredoxin-1 and glutathione/glutaredoxin-1 systems redundantly fuel murine B-cell development and responses. Eur. J. Immunol. 2019, 49, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Schneider, M.; Kolle, P.; Kuhlencordt, P.; Forster, H.; Beck, H.; Bornkamm, G.W.; Conrad, M. Loss of thioredoxin reductase 1 renders tumors highly susceptible to pharmacologic glutathione deprivation. Cancer Res. 2010, 70, 9505–9514. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.R.D.; Regad, T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct. Target. Ther. 2017, 2, 17040. [Google Scholar] [CrossRef]
- Kast, R.E. Glioblastoma invasion, cathepsin B, and the potential for both to be inhibited by auranofin, an old anti-rheumatoid arthritis drug. Cent. Eur. Neurosurg. 2010, 71, 139–142. [Google Scholar] [CrossRef]
- Yan, W.; Zhong, Y.; Hu, X.; Xu, T.; Zhang, Y.; Kales, S.; Qu, Y.; Talley, D.C.; Baljinnyam, B.; LeClair, C.A.; et al. Auranofin targets UBA1 and enhances UBA1 activity by facilitating ubiquitin trans-thioesterification to E2 ubiquitin-conjugating enzymes. Nat. Commun. 2023, 14, 4798. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, R.S.; Sharma, D.; Sandur, S.K. Thioredoxin reductase: An emerging pharmacologic target for radiosensitization of cancer. Transl. Oncol. 2022, 17, 101341. [Google Scholar] [CrossRef]
- Zhang, S.P.; Zhou, J.; Fan, Q.Z.; Lv, X.M.; Wang, T.; Wang, F.; Chen, Y.; Hong, S.Y.; Liu, X.P.; Xu, B.S.; et al. Discovery of hydroxytyrosol as thioredoxin reductase 1 inhibitor to induce apoptosis and G(1)/S cell cycle arrest in human colorectal cancer cells via ROS generation. Exp. Ther. Med. 2021, 22, 829. [Google Scholar] [CrossRef]
- Saccoccia, F.; Angelucci, F.; Boumis, G.; Carotti, D.; Desiato, G.; Miele, A.E.; Bellelli, A. Thioredoxin reductase and its inhibitors. Curr. Protein Pept. Sci. 2014, 15, 621–646. [Google Scholar] [CrossRef]
- Rigobello, M.P.; Folda, A.; Baldoin, M.C.; Scutari, G.; Bindoli, A. Effect of auranofin on the mitochondrial generation of hydrogen peroxide. Role of thioredoxin reductase. Free Radic. Res. 2005, 39, 687–695. [Google Scholar] [CrossRef]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef]
- Cui, X.; Zhao, J.; Li, G.; Yang, C.; Yang, S.; Zhan, Q.; Zhou, J.; Wang, Y.; Xiao, M.; Hong, B.; et al. Blockage of EGFR/AKT and mevalonate pathways synergize the antitumor effect of temozolomide by reprogramming energy metabolism in glioblastoma. Cancer Commun. 2023, 43, 1326–1353. [Google Scholar] [CrossRef]
- Oprita, A.; Baloi, S.C.; Staicu, G.A.; Alexandru, O.; Tache, D.E.; Danoiu, S.; Micu, E.S.; Sevastre, A.S. Updated Insights on EGFR Signaling Pathways in Glioma. Int. J. Mol. Sci. 2021, 22, 587. [Google Scholar] [CrossRef]
- Leung, E.L.; Fan, X.X.; Wong, M.P.; Jiang, Z.H.; Liu, Z.Q.; Yao, X.J.; Lu, L.L.; Zhou, Y.L.; Yau, L.F.; Tin, V.P.; et al. Targeting Tyrosine Kinase Inhibitor-Resistant Non-Small Cell Lung Cancer by Inducing Epidermal Growth Factor Receptor Degradation via Methionine 790 Oxidation. Antioxid. Redox Signal 2016, 24, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Lee, Y.M.; Chang, G.C.; Yu, S.L.; Hsieh, W.Y.; Chen, J.J.; Chen, H.W.; Yang, P.C. Curcumin induces EGFR degradation in lung adenocarcinoma and modulates p38 activation in intestine: The versatile adjuvant for gefitinib therapy. PLoS ONE 2011, 6, e23756. [Google Scholar] [CrossRef]
- Mao, J.; Ma, L.; Shen, Y.; Zhu, K.; Zhang, R.; Xi, W.; Ruan, Z.; Luo, C.; Chen, Z.; Xi, X.; et al. Arsenic circumvents the gefitinib resistance by binding to P62 and mediating autophagic degradation of EGFR in non-small cell lung cancer. Cell Death Dis. 2018, 9, 963. [Google Scholar] [CrossRef]
- Huang, H.S.; Nagane, M.; Klingbeil, C.K.; Lin, H.; Nishikawa, R.; Ji, X.D.; Huang, C.M.; Gill, G.N.; Wiley, H.S.; Cavenee, W.K. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 1997, 272, 2927–2935. [Google Scholar] [CrossRef]
- Tong, J.; Taylor, P.; Moran, M.F. Proteomic analysis of the epidermal growth factor receptor (EGFR) interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Mol. Cell Proteom. 2014, 13, 1644–1658. [Google Scholar] [CrossRef]
- Lee, S.R.; Kwon, K.S.; Kim, S.R.; Rhee, S.G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998, 273, 15366–15372. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.R.; Tischer, C.; Verveer, P.J.; Rocks, O.; Bastiaens, P.I. EGFR activation coupled to inhibition of tyrosine phosphatases causes lateral signal propagation. Nat. Cell Biol. 2003, 5, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.C.; Fukada, T.; Tonks, N.K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 2002, 9, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tan, L.J.; Grachtchouk, V.; Voorhees, J.J.; Fisher, G.J. Receptor-type protein-tyrosine phosphatase-kappa regulates epidermal growth factor receptor function. J. Biol. Chem. 2005, 280, 42694–42700. [Google Scholar] [CrossRef] [PubMed]
- Dagnell, M.; Pace, P.E.; Cheng, Q.; Frijhoff, J.; Ostman, A.; Arner, E.S.J.; Hampton, M.B.; Winterbourn, C.C. Thioredoxin reductase 1 and NADPH directly protect protein tyrosine phosphatase 1B from inactivation during H2O2 exposure. J. Biol. Chem. 2017, 292, 14371–14380. [Google Scholar] [CrossRef] [PubMed]
- Khan, E.M.; Heidinger, J.M.; Levy, M.; Lisanti, M.P.; Ravid, T.; Goldkorn, T. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J. Biol. Chem. 2006, 281, 14486–14493. [Google Scholar] [CrossRef]
- Ravid, T.; Heidinger, J.M.; Gee, P.; Khan, E.M.; Goldkorn, T. c-Cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J. Biol. Chem. 2004, 279, 37153–37162. [Google Scholar] [CrossRef]
- Grandal, M.V.; Zandi, R.; Pedersen, M.W.; Willumsen, B.M.; van Deurs, B.; Poulsen, H.S. EGFRvIII escapes down-regulation due to impaired internalization and sorting to lysosomes. Carcinogenesis 2007, 28, 1408–1417. [Google Scholar] [CrossRef]
- Davies, G.C.; Ryan, P.E.; Rahman, L.; Zajac-Kaye, M.; Lipkowitz, S. EGFRvIII undergoes activation-dependent downregulation mediated by the Cbl proteins. Oncogene 2006, 25, 6497–6509. [Google Scholar] [CrossRef][Green Version]
- Weihua, Z.; Tsan, R.; Huang, W.C.; Wu, Q.; Chiu, C.H.; Fidler, I.J.; Hung, M.C. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell 2008, 13, 385–393. [Google Scholar] [CrossRef]
- Cao, X.; Zhu, H.; Ali-Osman, F.; Lo, H.W. EGFR and EGFRvIII undergo stress- and EGFR kinase inhibitor-induced mitochondrial translocalization: A potential mechanism of EGFR-driven antagonism of apoptosis. Mol. Cancer 2011, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, P.; Beusch, C.M.; Gencheva, R.; Cheng, Q.; Zubarev, R.; Arner, E.S.J. Comprehensive chemical proteomics analyses reveal that the new TRi-1 and TRi-2 compounds are more specific thioredoxin reductase 1 inhibitors than auranofin. Redox Biol. 2021, 48, 102184. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Stokes, W.; Chater, E.; Roy, R.; de Bruin, E.; Hu, Y.; Liu, Z.; Smit, E.F.; Heynen, G.J.; Downward, J.; et al. Decreased glutathione biosynthesis contributes to EGFR T790M-driven erlotinib resistance in non-small cell lung cancer. Cell Discov. 2016, 2, 16031. [Google Scholar] [CrossRef]
- Fang, Q.; Zou, C.; Zhong, P.; Lin, F.; Li, W.; Wang, L.; Zhang, Y.; Zheng, C.; Wang, Y.; Li, X.; et al. EGFR mediates hyperlipidemia-induced renal injury via regulating inflammation and oxidative stress: The detrimental role and mechanism of EGFR activation. Oncotarget 2016, 7, 24361–24373. [Google Scholar] [CrossRef] [PubMed]
- Furnari, F.B.; Lin, H.; Huang, H.S.; Cavenee, W.K. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc. Natl. Acad. Sci. USA 1997, 94, 12479–12484. [Google Scholar] [CrossRef] [PubMed]
- Mellinghoff, I.K.; Wang, M.Y.; Vivanco, I.; Haas-Kogan, D.A.; Zhu, S.; Dia, E.Q.; Lu, K.V.; Yoshimoto, K.; Huang, J.H.; Chute, D.J.; et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005, 353, 2012–2024. [Google Scholar] [CrossRef] [PubMed]
- Prast-Nielsen, S.; Cebula, M.; Pader, I.; Arner, E.S. Noble metal targeting of thioredoxin reductase--covalent complexes with thioredoxin and thioredoxin-related protein of 14 kDa triggered by cisplatin. Free Radic. Biol. Med. 2010, 49, 1765–1778. [Google Scholar] [CrossRef]
- Jean-Claude, B.J. Chemosensitization to Platinum-Based Anticancer Drugs. In DNA Repair in Cancer Therapy; Panasci, L.C., Alaoui-Jamali, M.A., Eds.; Humana Press: Totowa, NJ, USA, 2004; pp. 51–71. [Google Scholar]
- Benhar, M.; Engelberg, D.; Levitzki, A. Cisplatin-induced activation of the EGF receptor. Oncogene 2002, 21, 8723–8731. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.R.; Garcia, C.C.; Vieira, D.B.; Quinet, A.; de Andrade-Lima, L.C.; Munford, V.; Belizario, J.E.; Menck, C.F. Glutathione depletion sensitizes cisplatin- and temozolomide-resistant glioma cells in vitro and in vivo. Cell Death Dis. 2014, 5, e1505. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, Y.; Wang, X.; Qi, C.; Tian, J.; Zou, Z. Synergistic lethality between auranofin-induced oxidative DNA damage and ATR inhibition in cancer cells. Life Sci. 2023, 332, 122131. [Google Scholar] [CrossRef]
- Erasimus, H.; Gobin, M.; Niclou, S.; Van Dyck, E. DNA repair mechanisms and their clinical impact in glioblastoma. Mutat. Res. Rev. Mutat. Res. 2016, 769, 19–35. [Google Scholar] [CrossRef]
- Huang, R.; Chen, H.; Liang, J.; Li, Y.; Yang, J.; Luo, C.; Tang, Y.; Ding, Y.; Liu, X.; Yuan, Q.; et al. Dual Role of Reactive Oxygen Species and their Application in Cancer Therapy. J. Cancer 2021, 12, 5543–5561. [Google Scholar] [CrossRef]
- Jiang, H.; Zuo, J.; Li, B.; Chen, R.; Luo, K.; Xiang, X.; Lu, S.; Huang, C.; Liu, L.; Tang, J.; et al. Drug-induced oxidative stress in cancer treatments: Angel or devil? Redox Biol. 2023, 63, 102754. [Google Scholar] [CrossRef] [PubMed]
- Walz, D.T.; DiMartino, M.J.; Griswold, D.E.; Intoccia, A.P.; Flanagan, T.L. Biologic actions and pharmacokinetic studies of auranofin. Am. J. Med. 1983, 75, 90–108. [Google Scholar] [CrossRef]
- Li, H.; Hu, J.; Wu, S.; Wang, L.; Cao, X.; Zhang, X.; Dai, B.; Cao, M.; Shao, R.; Zhang, R.; et al. Auranofin-mediated inhibition of PI3K/AKT/mTOR axis and anticancer activity in non-small cell lung cancer cells. Oncotarget 2016, 7, 3548–3558. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, X.; Wang, L.; Zhang, R.; Pu, X.; Wu, S.; Li, L.; Tong, P.; Wang, J.; Meng, Q.H.; et al. Inhibition of Thioredoxin/Thioredoxin Reductase Induces Synthetic Lethality in Lung Cancers with Compromised Glutathione Homeostasis. Cancer Res. 2019, 79, 125–132. [Google Scholar] [CrossRef]
- Marzano, C.; Gandin, V.; Folda, A.; Scutari, G.; Bindoli, A.; Rigobello, M.P. Inhibition of thioredoxin reductase by auranofin induces apoptosis in cisplatin-resistant human ovarian cancer cells. Free Radic. Biol. Med. 2007, 42, 872–881. [Google Scholar] [CrossRef]
- Park, N.; Chun, Y.J. Auranofin promotes mitochondrial apoptosis by inducing annexin A5 expression and translocation in human prostate cancer cells. J. Toxicol. Environ. Health A 2014, 77, 1467–1476. [Google Scholar] [CrossRef]
- Goenka, S.; Simon, S.R. Organogold drug Auranofin exhibits anti-melanogenic activity in B16F10 and MNT-1 melanoma cells. Arch. Dermatol. Res. 2020, 312, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Abdalbari, F.H.; Telleria, C.M. The gold complex auranofin: New perspectives for cancer therapy. Discov. Oncol. 2021, 12, 42. [Google Scholar] [CrossRef]
- Halatsch, M.E.; Kast, R.E.; Karpel-Massler, G.; Mayer, B.; Zolk, O.; Schmitz, B.; Scheuerle, A.; Maier, L.; Bullinger, L.; Mayer-Steinacker, R.; et al. A phase Ib/IIa trial of 9 repurposed drugs combined with temozolomide for the treatment of recurrent glioblastoma: CUSP9v3. Neurooncol. Adv. 2021, 3, vdab075. [Google Scholar] [CrossRef]
- Fazzari, F.; Chow, S.; Cheung, M.; Barghout, S.H.; Schimmer, A.D.; Chang, Q.; Hedley, D. Combined Targeting of the Glutathione and Thioredoxin Antioxidant Systems in Pancreatic Cancer. ACS Pharmacol. Transl. Sci. 2022, 5, 1070–1078. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Jaramillo, E.; Jamali, F.; Abdalbari, F.H.; Abdulkarim, B.; Jean-Claude, B.J.; Telleria, C.M.; Sabri, S. Pro-Oxidant Auranofin and Glutathione-Depleting Combination Unveils Synergistic Lethality in Glioblastoma Cells with Aberrant Epidermal Growth Factor Receptor Expression. Cancers 2024, 16, 2319. https://doi.org/10.3390/cancers16132319
Martinez-Jaramillo E, Jamali F, Abdalbari FH, Abdulkarim B, Jean-Claude BJ, Telleria CM, Sabri S. Pro-Oxidant Auranofin and Glutathione-Depleting Combination Unveils Synergistic Lethality in Glioblastoma Cells with Aberrant Epidermal Growth Factor Receptor Expression. Cancers. 2024; 16(13):2319. https://doi.org/10.3390/cancers16132319
Chicago/Turabian StyleMartinez-Jaramillo, Elvis, Fatemeh Jamali, Farah H. Abdalbari, Bassam Abdulkarim, Bertrand J. Jean-Claude, Carlos M. Telleria, and Siham Sabri. 2024. "Pro-Oxidant Auranofin and Glutathione-Depleting Combination Unveils Synergistic Lethality in Glioblastoma Cells with Aberrant Epidermal Growth Factor Receptor Expression" Cancers 16, no. 13: 2319. https://doi.org/10.3390/cancers16132319
APA StyleMartinez-Jaramillo, E., Jamali, F., Abdalbari, F. H., Abdulkarim, B., Jean-Claude, B. J., Telleria, C. M., & Sabri, S. (2024). Pro-Oxidant Auranofin and Glutathione-Depleting Combination Unveils Synergistic Lethality in Glioblastoma Cells with Aberrant Epidermal Growth Factor Receptor Expression. Cancers, 16(13), 2319. https://doi.org/10.3390/cancers16132319







