Identification and Application of Emerging Biomarkers in Treatment of Non-Small-Cell Lung Cancer: Systematic Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Protocol
2.2. Research Question
2.3. Eligibility Criteria
2.3.1. Inclusion Criteria
- Manuscripts published between 2020 and 2024;
- Studies including predictive biomarkers;
- Studies including diagnostic biomarkers;
- Studies including monitoring biomarkers;
- Clinical follow-up studies with biomarkers for oncological treatments (predictive biomarkers);
- Full-access articles;
- Articles in English;
- Clinical trials.
2.3.2. Exclusion Criteria
- Articles in pre-print mode or letters to the editor;
- Studies with biomarkers not specific to NSCLC;
- Studies published >5 years ago;
- Studies with inconclusive experimental designs;
- Studies of selected population analysis;
- Quality of life studies;
- Studies on treatment maintenance strategies and palliative care management.
2.4. Data Sources and Search Strategy
2.5. Selection and Data Extraction
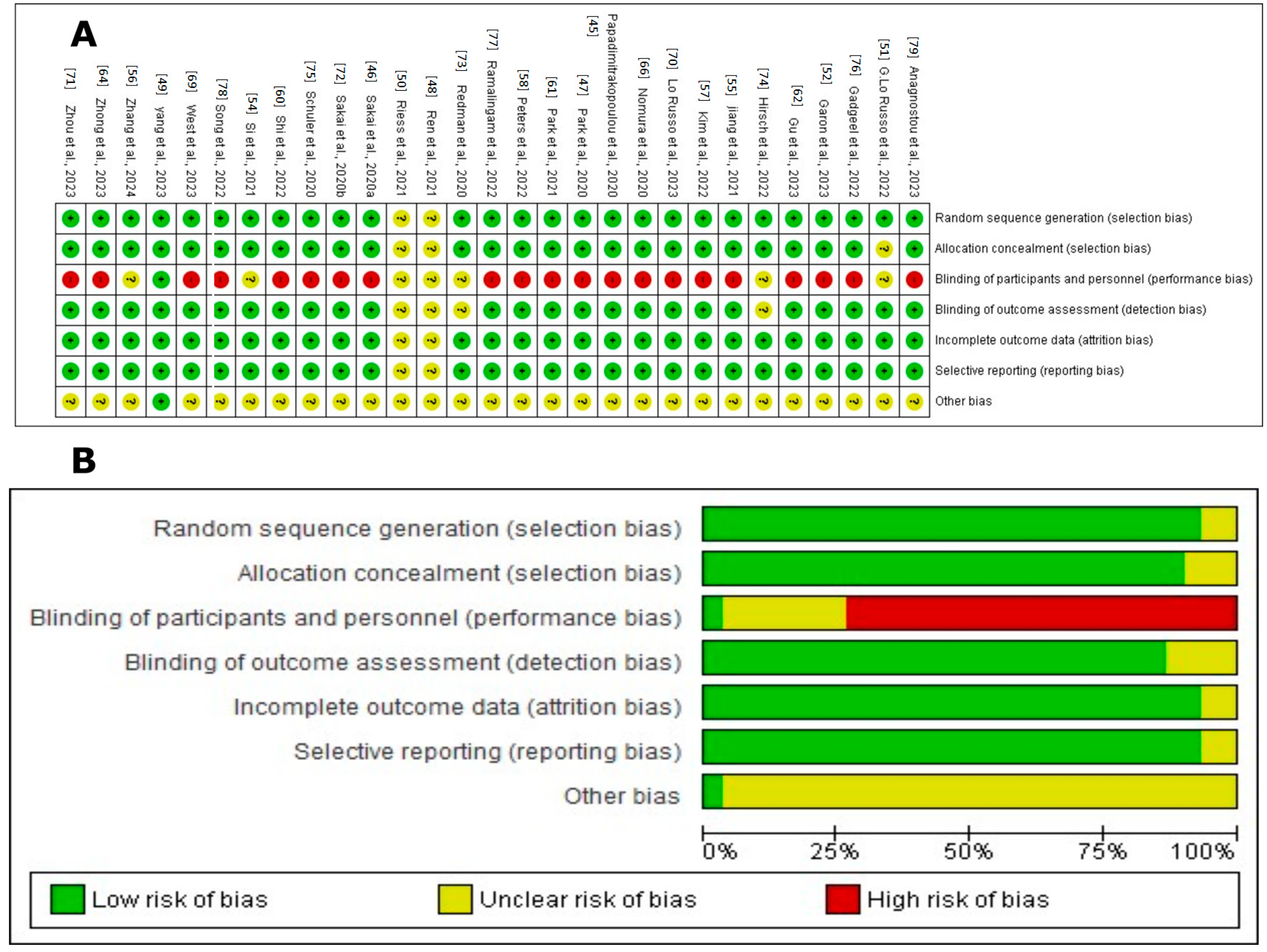
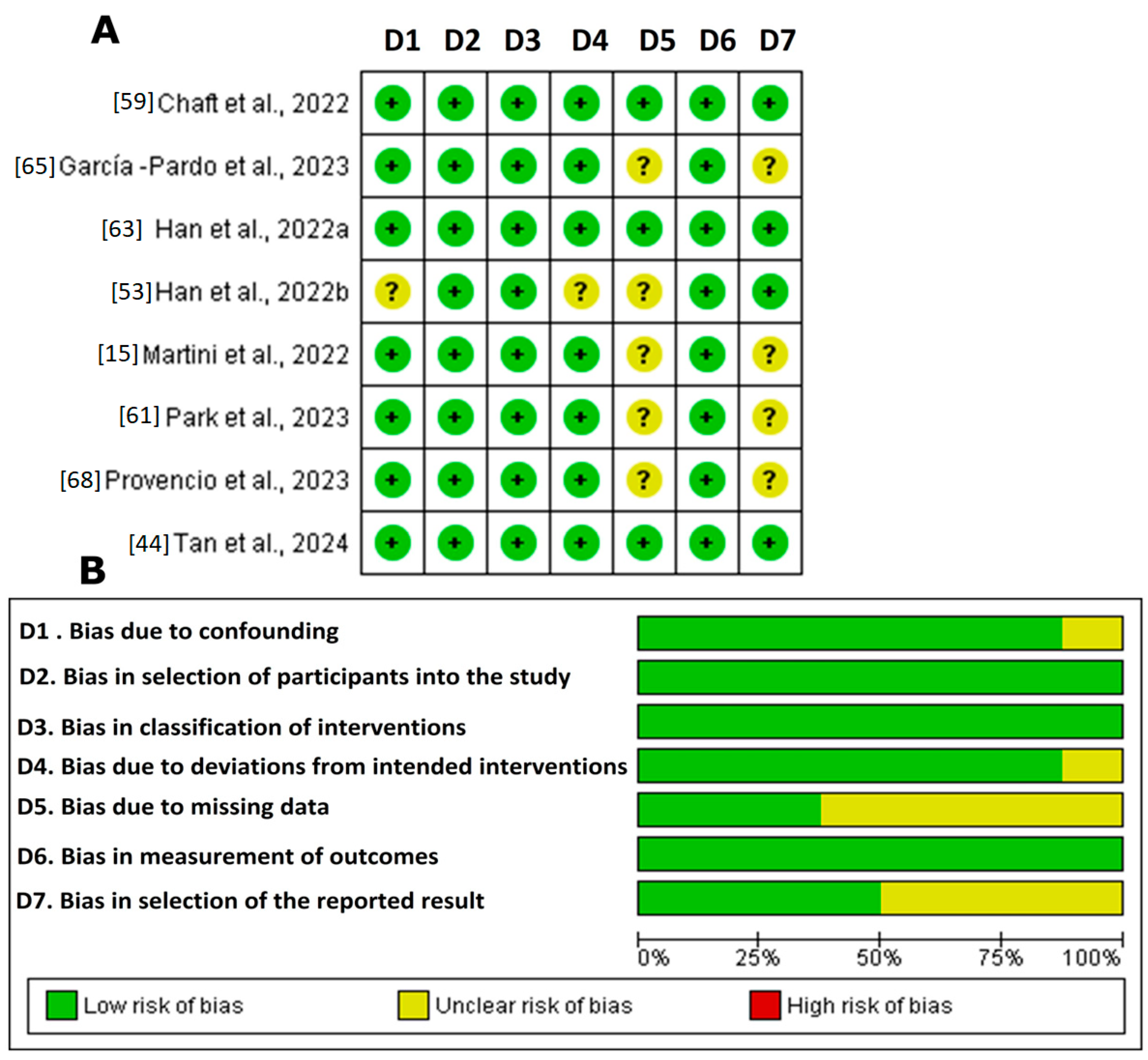
2.6. Risk of Bias Assessment
2.7. Ethical Considerations
3. Results
3.1. Characteristics of the Included Studies
3.2. Findings of the Studies
| Author and Publication Year | Country | Type of Design | Biological Sample | Number of Participants | Age of Participants | Sex of Participants | Biomarker Evaluated |
|---|---|---|---|---|---|---|---|
| Ren et al., 2022 [48] | China | Randomized Clinical Trial | Peripheral blood ctDNA samples | 389 | 18 to 75 years. | Men and women participated, but most patients were men | ctDNA dynamics |
| Yang et al., 2023 [49] | China | Randomized Clinical Trial | Peripheral blood ctDNA samples | 248 | Median age 54.0 years (brigatinib group), 53.0 years (alectinib group) | 54% female (brigatinib), 55% female (alectinib) | ALK fusions in plasma ctDNA |
| Riess et al., 2021 [50] | USA | Randomized Clinical Trial | Peripheral blood ctDNA samples | 11 | 60 (51–70) | 27% male, 73% female | EGFR exon 20 insertions |
| Lo Russo et al., 2022 [51] | USA | Randomized Clinical Trial | Peripheral blood ctDNA samples | 65 | Median age was 70.9 years (Q1–Q3: 63.7–77.1 years) | 21 (32.3%) were female and the rest were male | 36 immunobiomarkers like CD14, CD15, CD16, CD33, CD56, CD19, CD3, HLA-DR |
| Garon et al., 2023 [52] | USA, Japan | Randomized Clinical Trial | Peripheral blood ctDNA samples | 449 | NS | NS | The biomarkers evaluated were EGFR and co-occurring/treatment-emergent (TE) genomic alterations in ctDNA |
| Han et al., 2023 [53] | China | Non-Randomized Clinical Trial | Peripheral blood ctDNA samples | 40 | 18 to 75 years | NS | bTMB through ctDNA profiling |
| Si et al., 2021 [54] | USA | Randomized Clinical Trial | Peripheral blood ctDNA samples | 809 | NS | NS | bTMB |
| Jiang et al., 2021 [55] | China | Randomized Clinical Trial | Tumor biopsies for whole-exome and transcriptome sequencing and plasma samples for ctDNA analysis | 40 | 18 to 75 years old | Included both male (19, 47.5%) and female (21, 52.5%) patients | PD-L1, TMB, CD8+ TIL density, DSPP |
| Zhang et al., 2024 [56] | China | Randomized Clinical Trial | Peripheral blood ctDNA samples | 47 | Median age was 65 years (range: 52–76) | Mostly male (45/46, 97.8%); females (1/46, 2.2%) | ctDNA dynamics, BRCA2, BRINP3, FBXW7, KIT, RB1 |
| Tan et al., 2024 [44] | Australia | Non-Randomized Clinical Trial | Peripheral blood ctDNA samples | 47 | Median age was 60 years (range: 32–86) | 62% female, 38% male | EGFR T790M, EGFR exon 19 deletion, L858R mutation |
| Kim et al., 2022 [57] | USA | Randomized Clinical Trial | Peripheral blood ctDNA samples | 152 | NS | NS | bTMB |
| Peters et al., 2022 [58] | Multinational | Randomized Clinical Trial | Peripheral blood ctDNA samples | 471 | NS | NS | bTMB |
| Chaft et al., 2022 [59] | USA | Non-Randomized Clinical Trial | Tumor samples | 181 | Median age of 65 years (range: 37–83) | 93 females (51%) | Major pathological response (MPR), PD-L1 tumor proportion score (TPS) |
| Shi et al., 2022 [60] | China | Randomized Clinical Trial | Peripheral blood ctDNA samples | 290 | 18 to 75 years | Majority male (93.8% sintilimab, 90.4% docetaxel) | PD-L1, OVOL2, CTCF via tissue/blood sequencing |
| Papadimitrakopoulou et al., 2020 [45] | Multinational | Randomized Clinical Trial | Peripheral blood ctDNA samples | NS | NS | NS | EGFR mutations |
| Sakai et al., 2021a [46] | Japan | Randomized Clinical Trial | Formalin-fixed paraffin-embedded tumor tissue | 389 | NS | NS | Tumor mutation burden (TMB) |
| Park et al., 2023 [61] | South Korea | Non-Randomized Clinical Trial | Peripheral blood ctDNA samples | 100 | NS | 84 male, 16 female | bTMB, cfDNA concentration, hVAF, VAFSD |
| Park et al., 2021 [47] | South Korea | Randomized Clinical Trial | Peripheral blood ctDNA samples | 19 | Median age of 70 years (range: 32–84) | 13 females (68%) and 6 males (32%) | Activating EGFR mutations in ctDNA and tumor DNA |
| Gu et al., 2023 [62] | China | Randomized Clinical Trial | Peripheral blood ctDNA samples | 92 | Median age 65 years | 34% male and 66% female | EGFR mutations, ctDNA for minimal residual disease (MRD) |
| Han et al., 2022 [63] | China | Non-Randomized Clinical Trial | Peripheral blood ctDNA samples | 33 | Median age 56 years (range: 31–71) | 26 males (78.79%) and 7 females (21.21%) | ctDNA analysis with 448-gene panel for short-term dynamics |
| Zhong et al., 2023 [64] | China | Randomized Clinical Trial | Peripheral blood ctDNA samples | 69 | Median age was 58 years (range: 33–76 years) | 38 males (55.1%) and 31 females (44.9%) | EGFR mutation detection via ctDNA |
| García-Pardo et al., 2023 [65] | Canada | Non-Randomized Clinical Trial | Peripheral blood ctDNA samples | 150 | Median age at diagnosis 68 years (range: 33–91 years) | 80 men (53%), 70 women (47%) | ctDNA genotyping |
| Nomura et al., 2020 [66] | Japan | Randomized Clinical Trial | Peripheral blood ctDNA samples | 216 | NS | Both males and females (NS) | ctDNA (Guardant360® ancillary study) |
| Martini et al., 2022 [67] | Italy | Non-Randomized Clinical Trial | Basal fecal samples and peripheral blood samples for ctDNA analysis | 14 | NS | NS | Gut microbiota species and ctDNA RAS/BRAF WT MSS disease |
| Provencio et al., 2022 [68] | Spain | Non-Randomized Clinical Trial | Peripheral blood samples for ctDNA analysis and formalin-fixed paraffin-embedded tissue samples | 46 | Median age was 67.6 years | Majority male (66.5%) | ctDNA analysis for prognosis and predictive value |
| West et al., 2022 [69] | Multinational study | Randomized Clinical Trial | Peripheral blood ctDNA samples | 920 | NS | Both males and females included, exact distribution not provided | KRAS, STK11, KEAP1, TP53 mutations |
| Lo Russo et al., 2023 [70] | Italy | Randomized Clinical Trial | Blood and stool samples for circulating immune profiling and gut bacterial taxonomic abundance analysis | 65 | Median age was 70 years, with a range of 47–87 years | 44 men (68%) and 21 women (32%) | Immune circulating cell subsets and gene expression levels |
| Zhou et al., 2023 [71] | Multinational study | Randomized Clinical Trial | Formalin-fixed paraffin-embedded tumor tissue for PD-L1 expression assessment | NS | NS | Both males and females included, but the exact distribution is not provided | PD-L1 expression on tumor cells |
| Sakai et al., 2021b [72] | Japan | Randomized Clinical Trial | Peripheral blood ctDNA samples | 52 | Median age 67 (range: 37–82 years) | 17 male (32.7%), 35 female (67.3%) | EGFR genomic alterations including the T790M mutation |
| Redman et al., 2020 [73] | USA | Randomized Clinical Trial | Formalin-fixed paraffin-embedded tumor specimens for genomic DNA extraction | 1864 | NS | Both males and females included | Multiple biomarkers defined by the FoundationOne® NGS assay |
| Hirsch et al., 2022 [74] | USA | Randomized Clinical Trial | Formalin-fixed paraffin-embedded tumor specimens | 1313 | NS | Both males and females included | EGFR copy number and protein expression |
| Schuler et al., 2020 [75] | Multinational study | Randomized Clinical Trial | Tumor samples | 55 | Median age was 60 years | Both males (60%) and females (40%) | MET dysregulation |
| Gadgeel et al., 2022 [76] | Multinational study | Randomized Clinical Trial | Tumor samples | 577 | NS | Both males and females included | PD-L1 expression |
| Ramalingam et al., 2021 [77] | Multinational study | Randomized Clinical Trial | Formalin-fixed paraffin-embedded tumor samples | 970 | NS | Both males (82%) and females included | 52-gene expression histology classifier (LP52) |
| Song et al., 2022 [78] | China | Randomized Clinical Trial | Peripheral blood ctDNA samples | 78 | Median age 62 years | Both male (47.4%) and female (52.6%) participants | HER2 mutations |
| Anagnostou et al., 2023 [79] | Multinational study | Randomized Clinical Trial | Peripheral blood ctDNA samples | 50 | NS | Both males and females included | ctDNA dynamics |
| Park et al., 2021 [80] | South Korea | Randomized Clinical Trial | Peripheral blood ctDNA samples | 21 | Mean age of 68.5 years | 17 females and 4 males | EGFR exon 19 deletions, exon 21 point mutations (ctDNA) |
3.3. Risk of Bias Assessment
4. Discussion
4.1. Most Significant Findings
4.2. Quality of Evidence
4.3. Future Directions and Clinical Implications
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NSCLC | Non-Small-Cell Lung Cancer |
| SCLC | Small Cell Lung Cancer |
| ctDNA | Circulating Tumor DNA |
| miRNA | microRNA |
| bTMB | Blood Tumor Mutational Burden |
| NGS | Next-Generation Sequencing |
| EGFR | Epidermal Growth Factor Receptor |
| ALK | Anaplastic Lymphoma Kinase |
| ROS1 | ROS Proto-Oncogene 1, Receptor Tyrosine Kinase |
| KRAS | Kirsten Rat Sarcoma Viral Oncogene Homolog |
| PD-1 | Programmed Death-1 |
| PD-L1 | Programmed Death Ligand 1 |
| CT | Computed Tomography |
| MRI | Magnetic Resonance Imaging |
| PET | Positron Emission Tomography |
| FISH | Fluorescence In Situ Hybridization |
| PCR | Polymerase Chain Reaction |
| IHC | Immunohistochemistry |
| TKIs | Tyrosine Kinase Inhibitors |
| EMT | Epithelial–Mesenchymal Transition |
| MET | Mesenchymal Epithelial Transition |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| VEGF | Vascular Endothelial Growth Factor |
| FDA | U.S. Food and Drug Administration |
| NIH | National Institutes of Health |
| ICBs | Immune Checkpoint Inhibitors |
| NTRK | Neurotrophic Tyrosine Kinase Receptor |
| TNM | Tumor, Node, Metastasis |
| TME | Tumor Microenvironment |
| CEA | Carcinoembryonic Antigen |
| CA125 | Carbohydrate Antigen 125 |
| STK11 | Serine/Threonine Kinase 11 |
| DDR2 | Discoidin Domain Receptor 2 |
| RET | Rearranged during Transfection |
| JAK-STAT | Janus Kinase Signal Transducer and Activator of Transcription |
| RAS/MAPK | Rat Sarcoma/Mitogen-Activated Protein Kinase |
| PI3K/AKT/mTOR | Phosphoinositide 3-Kinase/Protein Kinase B/Mammalian Target of Rapamycin |
| MEK/ERK | Mitogen-Activated Protein Kinase Kinase/Extracellular Signal-Regulated Kinase |
| BRAF | B-Raf Proto-Oncogene, Serine/Threonine Kinase |
| TUBB3 | Tubulin Beta-3 |
| TGF-β | Transforming Growth Factor Beta |
| LAG-3 | Lymphocyte Activation Gene-3 |
| MDS | Multidimensional Scaling |
| UMAP | Uniform Manifold Approximation and Projection |
| GEO | Gene Expression Omnibus |
| TCGA | The Cancer Genome Atlas |
| AMP | Association for Molecular Pathology |
| CAP | College of American Pathologists |
| IASLC | International Association for the Study of Lung Cancer |
| ASCO | American Society of Clinical Oncology |
| ESMO | European Society for Medical Oncology |
| NCCN | National Comprehensive Cancer Network |
| FGFR1 | Fibroblast Growth Factor Receptor 1 |
| TIM-3 | T-Cell Immunoglobulin and Mucin Domain Containing-3 |
| TMB | Tumor Mutational Burden |
| OPN | Osteopontin |
| DNA | Deoxyribonucleic Acid |
| SNPs | Single-Nucleotide Polymorphisms |
| FDA-NIH | FDA-NIH Biomarker Working Group |
| NKX2-1 | Thyroid Transcription Factor-1 |
| TTF-1 | Thyroid Transcription Factor-1 |
| NAPSA | Napsin A |
| CYFRA 21-1 | Cytokeratin 19 Fragment |
| SCCA | Squamous Cell Carcinoma Antigen |
| m6A | N6-Methyladenine |
| SFTA2 | Surfactant Protein A2 |
| KIAA1522 | KIAA1522 Protein |
| PFS | Progression-Free Survival |
| NLR | Neutrophil-to-Lymphocyte Ratio |
| RCTs | Randomized Clinical Trials |
| MPR | Major Pathological Response |
| TPS | Tumor Proportion Score |
| MDR | Minimal Residual Disease |
| hVAF | High Variant Allele Frequency |
| VAFSD | Variant Allele Frequency Standard Deviation |
| RECIST | Response Evaluation Criteria In Solid Tumors |
| ORR | Objective Response Rate |
| BRCA2 | Breast Cancer 2, Early Onset |
| DSPP | Dentin Sialophosphoprotein |
| CTCF | CCCTC-Binding Factor |
| OVOL2 | Ovo-Like Transcriptional Repressor 2 |
| FFPE | Formalin-Fixed Paraffin-Embedded |
| ABCP | Atezolizumab Bevacizumab Carboplatin Paclitaxel |
| BCP | Bevacizumab Carboplatin Paclitaxel |
| KEAP1 | Kelch-Like ECH-Associated Protein |
| TP53 | Tumor Protein p53 |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| ddPCR | Droplet Digital Polymerase Chain Reaction |
| LP52 | 52-Gene Expression Histology Classifier |
| BRAFV600E | BRAF Gene Mutation V600E |
| ASOs | Antisense Oligonucleotides |
| siRNAs | Small Interfering RNAs |
Appendix A
References
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non–Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Gridelli, C.; Rossi, A.; Carbone, D.P.; Guarize, J.; Karachaliou, N.; Mok, T.; Petrella, F.; Spaggiari, L.; Rosell, R. Non-Small-Cell Lung Cancer. Nat. Rev. Dis. Primers 2015, 1, 15009. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and Prospects of Early Detection in Lung Cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef]
- Polanco, D.; Pinilla, L.; Gracia-Lavedan, E.; Mas, A.; Bertran, S.; Fierro, G.; Seminario, A.; Gómez, S.; Barbé, F. Prognostic Value of Symptoms at Lung Cancer Diagnosis: A Three-Year Observational Study. J. Thorac. Dis. 2021, 13, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Alduais, Y.; Zhang, H.; Fan, F.; Chen, J.; Chen, B. Non-Small Cell Lung Cancer (NSCLC): A Review of Risk Factors, Diagnosis, and Treatment. Medicine 2023, 102, e32899. [Google Scholar] [CrossRef] [PubMed]
- Petrella, F.; Rizzo, S.; Attili, I.; Passaro, A.; Zilli, T.; Martucci, F.; Bonomo, L.; Del Grande, F.; Casiraghi, M.; De Marinis, F.; et al. Stage III Non-Small-Cell Lung Cancer: An Overview of Treatment Options. Curr. Oncol. 2023, 30, 3160–3175. [Google Scholar] [CrossRef] [PubMed]
- Rodak, O.; Peris-Díaz, M.D.; Olbromski, M.; Podhorska-Okołów, M.; Dzięgiel, P. Current Landscape of Non-Small Cell Lung Cancer: Epidemiology, Histological Classification, Targeted Therapies, and Immunotherapy. Cancers 2021, 13, 4705. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, Y.; Wen, Y.; Zhou, C. Non-small Cell Lung Cancer in China. Cancer Commun. 2022, 42, 937–970. [Google Scholar] [CrossRef] [PubMed]
- Cardona, A.F.; Mejía, S.A.; Viola, L.; Chamorro, D.F.; Rojas, L.; Ruíz-Patiño, A.; Serna, A.; Martínez, S.; Muñoz, Á.; Rodríguez, J.; et al. Lung Cancer in Colombia. J. Thorac. Oncol. 2022, 17, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.-K. Non-Small-Cell Lung Cancers: A Heterogeneous Set of Diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Senosain, M.-F.; Massion, P.P. Intratumor Heterogeneity in Early Lung Adenocarcinoma. Front. Oncol. 2020, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- Flury, D.V.; Minervini, F.; Kocher, G.J. Heterogeneity of Stage IIIA Non-Small Cell Lung Cancer—Different Tumours, Different Nodal Status, Different Treatment, Different Prognosis: A Narrative Review. Curr. Chall Thorac. Surg. 2022, 4, 13. [Google Scholar] [CrossRef]
- Palumbo, A.; De Oliveira Meireles Da Costa, N.; Bonamino, M.H.; Ribeiro Pinto, L.F.; Nasciutti, L.E. Genetic Instability in the Tumor Microenvironment: A New Look at an Old Neighbor. Mol. Cancer 2015, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Siveke, J.T. Fibroblast-Activating Protein: Targeting the Roots of the Tumor Microenvironment. J. Nucl. Med. 2018, 59, 1412–1414. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Dai, Y. Tumor Microenvironment and Therapeutic Response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Bevere, M.; Masetto, F.; Carazzolo, M.E.; Bettega, A.; Gkountakos, A.; Scarpa, A.; Simbolo, M. An Overview of Circulating Biomarkers in Neuroendocrine Neoplasms: A Clinical Guide. Diagnostics 2023, 13, 2820. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D. Identification of Driver Mutations in Lung Cancer: First Step in Personalized Cancer. Targ. Oncol. 2013, 8, 3–14. [Google Scholar] [CrossRef]
- Zografos, E.; Dimitrakopoulos, F.-I.; Koutras, A. Prognostic Value of Circulating Tumor DNA (ctDNA) in Oncogene-Driven NSCLC: Current Knowledge and Future Perspectives. Cancers 2022, 14, 4954. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, F.; Rachiglio, A.M.; Iannaccone, A.; Chicchinelli, N.; Normanno, N. Circulating Tumor Cells and ctDNA in NSCLC. In Oncogenomics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 465–475. ISBN 978-0-12-811785-9. [Google Scholar]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Latini, A.; Borgiani, P.; Novelli, G.; Ciccacci, C. miRNAs in Drug Response Variability: Potential Utility as Biomarkers for Personalized Medicine. Pharmacogenomics 2019, 20, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Han, J.; Cui, Y.; Fan, K.; Zhou, X. Circulating microRNA-21 as Noninvasive Predictive Biomarker for Response in Cancer Immunotherapy. Med. Hypotheses 2013, 81, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, X.; Liu, C.; Zhang, X.; Wu, Y.; Diao, M.; Tan, S.; Huang, S.; Cheng, Y.; You, T. MicroRNA-21 as a Diagnostic and Prognostic Biomarker of Lung Cancer: A Systematic Review and Meta-Analysis. Biosci. Rep. 2022, 42, BSR20211653. [Google Scholar] [CrossRef] [PubMed]
- Mithoowani, H.; Febbraro, M. Non-Small-Cell Lung Cancer in 2022: A Review for General Practitioners in Oncology. Curr. Oncol. 2022, 29, 1828–1839. [Google Scholar] [CrossRef] [PubMed]
- Qian, F.; Yang, W.; Chen, Q.; Zhang, X.; Han, B. Screening for Early Stage Lung Cancer and Its Correlation with Lung Nodule Detection. J. Thorac. Dis. 2018, 10, S846–S859. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Buttitta, F.; D’Angelo, E. Adjuvant Osimertinib Treatment in Patients with Early Stage NSCLC (IB-IIIA): Pathological Pathway Adaptations. Oncotarget 2022, 13, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Sabari, J.K.; Santini, F.; Bergagnini, I.; Lai, W.V.; Arbour, K.C.; Drilon, A. Changing the Therapeutic Landscape in Non-Small Cell Lung Cancers: The Evolution of Comprehensive Molecular Profiling Improves Access to Therapy. Curr. Oncol. Rep. 2017, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Coco, S.; Truini, A.; Vanni, I.; Dal Bello, M.; Alama, A.; Rijavec, E.; Genova, C.; Barletta, G.; Sini, C.; Burrafato, G.; et al. Next Generation Sequencing in Non-Small Cell Lung Cancer: New Avenues Toward the Personalized Medicine. CDT 2015, 16, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dey, M.K.; Devireddy, R.; Gartia, M.R. Biomarkers in Cancer Detection, Diagnosis, and Prognosis. Sensors 2023, 24, 37. [Google Scholar] [CrossRef] [PubMed]
- Liberini, V.; Mariniello, A.; Righi, L.; Capozza, M.; Delcuratolo, M.D.; Terreno, E.; Farsad, M.; Volante, M.; Novello, S.; Deandreis, D. NSCLC Biomarkers to Predict Response to Immunotherapy with Checkpoint Inhibitors (ICI): From the Cells to In Vivo Images. Cancers 2021, 13, 4543. [Google Scholar] [CrossRef] [PubMed]
- Pretelli, G.; Spagnolo, C.C.; Ciappina, G.; Santarpia, M.; Pasello, G. Overview on Therapeutic Options in Uncommon EGFR Mutant Non-Small Cell Lung Cancer (NSCLC): New Lights for an Unmet Medical Need. Int. J. Mol. Sci. 2023, 24, 8878. [Google Scholar] [CrossRef] [PubMed]
- Simarro, J.; Pérez-Simó, G.; Mancheño, N.; Ansotegui, E.; Muñoz-Núñez, C.F.; Gómez-Codina, J.; Juan, Ó.; Palanca, S. Impact of Molecular Testing Using Next-Generation Sequencing in the Clinical Management of Patients with Non-Small Cell Lung Cancer in a Public Healthcare Hospital. Cancers 2023, 15, 1705. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tam, K.Y. Combination Strategies Using EGFR-TKi in NSCLC Therapy: Learning from the Gap between Pre-Clinical Results and Clinical Outcomes. Int. J. Biol. Sci. 2018, 14, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Melosky, B.; Kambartel, K.; Häntschel, M.; Bennetts, M.; Nickens, D.J.; Brinkmann, J.; Kayser, A.; Moran, M.; Cappuzzo, F. Worldwide Prevalence of Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer: A Meta-Analysis. Mol. Diagn. Ther. 2022, 26, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Araghi, M.; Mannani, R.; Heidarnejad Maleki, A.; Hamidi, A.; Rostami, S.; Safa, S.H.; Faramarzi, F.; Khorasani, S.; Alimohammadi, M.; Tahmasebi, S.; et al. Recent Advances in Non-Small Cell Lung Cancer Targeted Therapy; an Update Review. Cancer Cell Int. 2023, 23, 162. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lin, Z. Non-Small Cell Lung Cancer Targeted Therapy: Drugs and Mechanisms of Drug Resistance. Int. J. Mol. Sci. 2022, 23, 15056. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, J.; Qin, C.; Yan, H.; Liu, T.; Hu, H.; Tang, S.; Tang, S.; Zhou, H. Biomarker-Targeted Therapies in Non–Small Cell Lung Cancer: Current Status and Perspectives. Cells 2022, 11, 3200. [Google Scholar] [CrossRef] [PubMed]
- Saller, J.J.; Boyle, T.A. Molecular Pathology of Lung Cancer. Cold Spring Harb. Perspect. Med. 2022, 12, a037812. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Saifi, M.; Batra, U.; Suryavanshi, M.; Gupta, K. Incidence of ROS1-Rearranged Non-Small-Cell Lung Carcinoma in India and Efficacy of Crizotinib in Lung Adenocarcinoma Patients. LCTT 2020, 11, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS. Med. 2009, 15, 264–269. [Google Scholar] [CrossRef]
- Cochrane Cochrane Handbook for Systematic Reviews of Interventions. Available online: https://training.cochrane.org/handbook (accessed on 9 June 2023).
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A Tool for Assessing Risk of Bias in Non-Randomised Studies of Interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef] [PubMed]
- Haddaway, N.R.; Page, M.J.; Pritchard, C.C.; McGuinness, L.A. PRISMA2020: An R Package and Shiny App for Producing PRISMA 2020-Compliant Flow Diagrams, with Interactivity for Optimised Digital Transparency and Open Synthesis. Campbell Syst. Rev. 2022, 18, e1230. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Brown, C.; Mersiades, A.; Lee, C.K.; John, T.; Kao, S.; Newnham, G.; O’Byrne, K.; Parakh, S.; Bray, V.; et al. A Phase II Trial of Alternating Osimertinib and Gefitinib Therapy in Advanced EGFR-T790M Positive Non-Small Cell Lung Cancer: OSCILLATE. Nat. Commun. 2024, 15, 1823. [Google Scholar] [CrossRef] [PubMed]
- Papadimitrakopoulou, V.A.; Han, J.; Ahn, M.; Ramalingam, S.S.; Delmonte, A.; Hsia, T.; Laskin, J.; Kim, S.; He, Y.; Tsai, C.; et al. Epidermal Growth Factor Receptor Mutation Analysis in Tissue and Plasma from the AURA3 Trial: Osimertinib versus Platinum-pemetrexed for T790M Mutation-positive Advanced Non–Small Cell Lung Cancer. Cancer 2020, 126, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Tsuboi, M.; Kenmotsu, H.; Yamanaka, T.; Takahashi, T.; Goto, K.; Daga, H.; Ohira, T.; Ueno, T.; Aoki, T.; et al. Tumor Mutation Burden as a Biomarker for Lung Cancer Patients Treated with Pemetrexed and Cisplatin (the JIPANG-TR). Cancer Sci. 2021, 112, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-K.; Cho, H.-J.; Choi, Y.-D.; Oh, I.-J.; Kim, Y.-C. A Phase II Trial of Osimertinib as the First-Line Treatment of Non–Small Cell Lung Cancer Harboring Activating EGFR Mutations in Circulating Tumor DNA: LiquidLung-O-Cohort 1. Cancer Res. Treat. 2021, 53, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Chen, J.; Xu, X.; Jiang, T.; Cheng, Y.; Chen, G.; Pan, Y.; Fang, Y.; Wang, Q.; Huang, Y.; et al. Camrelizumab Plus Carboplatin and Paclitaxel as First-Line Treatment for Advanced Squamous NSCLC (CameL-Sq): A Phase 3 Trial. J. Thorac. Oncol. 2022, 17, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.-H.; Liu, G.; Lu, S.; He, J.; Burotto, M.; Ahn, M.-J.; Kim, D.-W.; Liu, X.; Zhao, Y.; Vincent, S.; et al. Brigatinib Versus Alectinib in ALK-Positive NSCLC After Disease Progression on Crizotinib: Results of Phase 3 ALTA-3 Trial. J. Thorac. Oncol. 2023, 18, 1743–1755. [Google Scholar] [CrossRef]
- Riess, J.W.; Reckamp, K.L.; Frankel, P.; Longmate, J.; Kelly, K.A.; Gandara, D.R.; Weipert, C.M.; Raymond, V.M.; Keer, H.N.; Mack, P.C.; et al. Erlotinib and Onalespib Lactate Focused on EGFR Exon 20 Insertion Non-Small Cell Lung Cancer (NSCLC): A California Cancer Consortium Phase I/II Trial (NCI 9878). Clin. Lung Cancer 2021, 22, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Lo Russo, G.; Sgambelluri, F.; Prelaj, A.; Galli, F.; Manglaviti, S.; Bottiglieri, A.; Di Mauro, R.M.; Ferrara, R.; Galli, G.; Signorelli, D.; et al. PEOPLE (NCT03447678), a First-Line Phase II Pembrolizumab Trial, in Negative and Low PD-L1 Advanced NSCLC: Clinical Outcomes and Association with Circulating Immune Biomarkers. ESMO Open 2022, 7, 100645. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Reck, M.; Nishio, K.; Heymach, J.V.; Nishio, M.; Novello, S.; Paz-Ares, L.; Popat, S.; Aix, S.P.; Graham, H.; et al. Ramucirumab plus Erlotinib versus Placebo plus Erlotinib in Previously Untreated EGFR-Mutated Metastatic Non-Small-Cell Lung Cancer (RELAY): Exploratory Analysis of next-Generation Sequencing Results. ESMO Open 2023, 8, 101580. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Guo, J.; Tang, X.; Zhu, H.; Zhu, D.; Zhang, X.; Meng, X.; Hua, Y.; Wang, Z.; Zhang, Y.; et al. Efficacy and Safety of Sintilimab plus Docetaxel in Patients with Previously Treated Advanced Non-Small Cell Lung Cancer: A Prospective, Single-Arm, Phase II Study in China. J. Cancer Res. Clin. Oncol. 2023, 149, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Kuziora, M.; Quinn, K.J.; Helman, E.; Ye, J.; Liu, F.; Scheuring, U.; Peters, S.; Rizvi, N.A.; Brohawn, P.Z.; et al. A Blood-Based Assay for Assessment of Tumor Mutational Burden in First-Line Metastatic NSCLC Treatment: Results from the MYSTIC Study. Clin. Cancer Res. 2021, 27, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Wang, P.; Zhang, J.; Zhao, Y.; Zhou, J.; Fan, Y.; Shu, Y.; Liu, X.; Zhang, H.; He, J.; et al. Toripalimab plus Chemotherapy as Second-Line Treatment in Previously EGFR-TKI Treated Patients with EGFR-Mutant-Advanced NSCLC: A Multicenter Phase-II Trial. Sig. Transduct. Target Ther. 2021, 6, 355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, G.; Niu, Y.; Zhang, G.; Ji, Y.; Yan, X.; Zhang, X.; Wang, Q.; Jing, X.; Wang, J.; et al. Sintilimab with Two Cycles of Chemotherapy for the Treatment of Advanced Squamous Non-Small Cell Lung Cancer: A Phase 2 Clinical Trial. Nat. Commun. 2024, 15, 1512. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S.; Velcheti, V.; Mekhail, T.; Yun, C.; Shagan, S.M.; Hu, S.; Chae, Y.K.; Leal, T.A.; Dowell, J.E.; Tsai, M.L.; et al. Blood-Based Tumor Mutational Burden as a Biomarker for Atezolizumab in Non-Small Cell Lung Cancer: The Phase 2 B-F1RST Trial. Nat. Med. 2022, 28, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Dziadziuszko, R.; Morabito, A.; Felip, E.; Gadgeel, S.M.; Cheema, P.; Cobo, M.; Andric, Z.; Barrios, C.H.; Yamaguchi, M.; et al. Atezolizumab versus Chemotherapy in Advanced or Metastatic NSCLC with High Blood-Based Tumor Mutational Burden: Primary Analysis of BFAST Cohort C Randomized Phase 3 Trial. Nat. Med. 2022, 28, 1831–1839. [Google Scholar] [CrossRef] [PubMed]
- Chaft, J.E.; Oezkan, F.; Kris, M.G.; Bunn, P.A.; Wistuba, I.I.; Kwiatkowski, D.J.; Owen, D.H.; Tang, Y.; Johnson, B.E.; Lee, J.M.; et al. Neoadjuvant Atezolizumab for Resectable Non-Small Cell Lung Cancer: An Open-Label, Single-Arm Phase II Trial. Nat. Med. 2022, 28, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wu, L.; Yu, X.; Xing, P.; Wang, Y.; Zhou, J.; Wang, A.; Shi, J.; Hu, Y.; Wang, Z.; et al. Sintilimab versus Docetaxel as Second-line Treatment in Advanced or Metastatic Squamous Non-small-cell Lung Cancer: An Open-label, Randomized Controlled Phase 3 Trial (ORIENT-3). Cancer Commun. 2022, 42, 1314–1330. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-K.; Jun, H.R.; Oh, H.-J.; Lee, J.-Y.; Cho, H.-J.; Kim, Y.-C.; Lee, J.E.; Yoon, S.H.; Choi, C.M.; Lee, J.C.; et al. Evaluation of Blood Tumor Mutation Burden for the Efficacy of Second-Line Atezolizumab Treatment in Non-Small Cell Lung Cancer: BUDDY Trial. Cells 2023, 12, 1246. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zhang, H.; Lu, Y.; Li, M.; Yang, S.; Liang, J.; Ye, Z.; Li, Z.; He, M.; Shi, X.; et al. EGFR-TKI Combined with Pemetrexed versus EGFR-TKI Monotherapy in Advanced EGFR-Mutated NSCLC: A Prospective, Randomized, Exploratory Study. Cancer Res. Treat. 2023, 55, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Tang, X.; Zhu, H.; Zhu, D.; Zhang, X.; Meng, X.; Hua, Y.; Wang, Z.; Zhang, Y.; Huang, W.; et al. Short-Term Dynamics of Circulating Tumor DNA Predicting Efficacy of Sintilimab plus Docetaxel in Second-Line Treatment of Advanced NSCLC: Biomarker Analysis from a Single-Arm, Phase 2 Trial. J. Immunother. Cancer 2022, 10, e004952. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Zhang, X.; Tian, P.; Chu, T.; Guo, Q.; Yu, X.; Yu, Z.; Li, Y.; Chen, L.; Liu, J.; et al. Tislelizumab plus Chemotherapy for Patients with EGFR -Mutated Non-Squamous Non-Small Cell Lung Cancer Who Progressed on EGFR Tyrosine Kinase Inhibitor Therapy. J. Immunother. Cancer 2023, 11, e006887. [Google Scholar] [CrossRef]
- García-Pardo, M.; Czarnecka-Kujawa, K.; Law, J.H.; Salvarrey, A.M.; Fernandes, R.; Fan, Z.J.; Waddell, T.K.; Yasufuku, K.; Liu, G.; Donahoe, L.L.; et al. Association of Circulating Tumor DNA Testing Before Tissue Diagnosis with Time to Treatment Among Patients With Suspected Advanced Lung Cancer: The ACCELERATE Nonrandomized Clinical Trial. JAMA Netw. Open 2023, 6, e2325332. [Google Scholar] [CrossRef] [PubMed]
- Nomura, S.; Goto, Y.; Mizutani, T.; Kataoka, T.; Kawai, S.; Okuma, Y.; Murakami, H.; Tanaka, K.; Ohe, Y. A Randomized Phase III Study Comparing Continuation and Discontinuation of PD-1 Pathway Inhibitors for Patients with Advanced Non-Small-Cell Lung Cancer (JCOG1701, SAVE Study). Jpn. J. Clin. Oncol. 2020, 50, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Martini, G.; Ciardiello, D.; Dallio, M.; Famiglietti, V.; Esposito, L.; Corte, C.M.D.; Napolitano, S.; Fasano, M.; Gravina, A.G.; Romano, M.; et al. Gut Microbiota Correlates with Antitumor Activity in Patients with mCRC and NSCLC Treated with Cetuximab plus Avelumab. Int. J. Cancer 2022, 151, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Provencio, M.; Serna-Blasco, R.; Nadal, E.; Insa, A.; García-Campelo, M.R.; Casal Rubio, J.; Dómine, M.; Majem, M.; Rodríguez-Abreu, D.; Martínez-Martí, A.; et al. Overall Survival and Biomarker Analysis of Neoadjuvant Nivolumab Plus Chemotherapy in Operable Stage IIIA Non–Small-Cell Lung Cancer (NADIM Phase II Trial). JCO 2022, 40, 2924–2933. [Google Scholar] [CrossRef] [PubMed]
- West, H.J.; McCleland, M.; Cappuzzo, F.; Reck, M.; Mok, T.S.; Jotte, R.M.; Nishio, M.; Kim, E.; Morris, S.; Zou, W.; et al. Clinical Efficacy of Atezolizumab plus Bevacizumab and Chemotherapy in KRAS- Mutated Non-Small Cell Lung Cancer with STK11, KEAP1, or TP53 Comutations: Subgroup Results from the Phase III IMpower150 Trial. J. Immunother. Cancer 2022, 10, e003027. [Google Scholar] [CrossRef] [PubMed]
- Lo Russo, G.; Prelaj, A.; Dolezal, J.; Beninato, T.; Agnelli, L.; Triulzi, T.; Fabbri, A.; Lorenzini, D.; Ferrara, R.; Brambilla, M.; et al. PEOPLE (NTC03447678), a Phase II Trial to Test Pembrolizumab as First-Line Treatment in Patients with Advanced NSCLC with PD-L1 <50%: A Multiomics Analysis. J. Immunother. Cancer 2023, 11, e006833. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Srivastava, M.K.; Xu, H.; Felip, E.; Wakelee, H.; Altorki, N.; Reck, M.; Liersch, R.; Kryzhanivska, A.; Oizumi, S.; et al. Comparison of SP263 and 22C3 Immunohistochemistry PD-L1 Assays for Clinical Efficacy of Adjuvant Atezolizumab in Non-Small Cell Lung Cancer: Results from the Randomized Phase III IMpower010 Trial. J. Immunother. Cancer 2023, 11, e007047. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Takahama, T.; Shimokawa, M.; Azuma, K.; Takeda, M.; Kato, T.; Daga, H.; Okamoto, I.; Akamatsu, H.; Teraoka, S.; et al. Predicting Osimertinib-treatment Outcomes through EGFR Mutant-fraction Monitoring in the Circulating Tumor DNA of EGFR T790M-positive Patients with Non-small Cell Lung Cancer (WJOG8815L). Mol. Oncol. 2021, 15, 126–137. [Google Scholar] [CrossRef]
- Redman, M.W.; Papadimitrakopoulou, V.A.; Minichiello, K.; Hirsch, F.R.; Mack, P.C.; Schwartz, L.H.; Vokes, E.; Ramalingam, S.; Leighl, N.; Bradley, J.; et al. Biomarker-Driven Therapies for Previously Treated Squamous Non-Small-Cell Lung Cancer (Lung-MAP SWOG S1400): A Biomarker-Driven Master Protocol. Lancet Oncol. 2020, 21, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Redman, M.W.; Moon, J.; Agustoni, F.; Herbst, R.S.; Semrad, T.J.; Varella-Garcia, M.; Rivard, C.J.; Kelly, K.; Gandara, D.R.; et al. EGFR High Copy Number Together With High EGFR Protein Expression Predicts Improved Outcome for Cetuximab-Based Therapy in Squamous Cell Lung Cancer: Analysis From SWOG S0819, a Phase III Trial of Chemotherapy With or Without Cetuximab in Advanced NSCLC. Clin. Lung Cancer 2022, 23, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.; Berardi, R.; Lim, W.-T.; De Jonge, M.; Bauer, T.M.; Azaro, A.; Gottfried, M.; Han, J.-Y.; Lee, D.H.; Wollner, M.; et al. Molecular Correlates of Response to Capmatinib in Advanced Non-Small-Cell Lung Cancer: Clinical and Biomarker Results from a Phase I Trial. Ann. Oncol. 2020, 31, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Gadgeel, S.; Hirsch, F.R.; Kerr, K.; Barlesi, F.; Park, K.; Rittmeyer, A.; Zou, W.; Bhatia, N.; Koeppen, H.; Paul, S.M.; et al. Comparison of SP142 and 22C3 Immunohistochemistry PD-L1 Assays for Clinical Efficacy of Atezolizumab in Non–Small Cell Lung Cancer: Results From the Randomized OAK Trial. Clin. Lung Cancer 2022, 23, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Novello, S.; Guclu, S.Z.; Bentsion, D.; Zvirbule, Z.; Szilasi, M.; Bernabe, R.; Syrigos, K.; Byers, L.A.; Clingan, P.; et al. Veliparib in Combination With Platinum-Based Chemotherapy for First-Line Treatment of Advanced Squamous Cell Lung Cancer: A Randomized, Multicenter Phase III Study. JCO 2021, 39, 3633–3644. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Li, Y.; Chen, S.; Ying, S.; Xu, S.; Huang, J.; Wu, D.; Lv, D.; Bei, T.; Liu, S.; et al. Efficacy and Safety of Pyrotinib in Advanced Lung Adenocarcinoma with HER2 Mutations: A Multicenter, Single-Arm, Phase II Trial. BMC Med. 2022, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Anagnostou, V.; Ho, C.; Nicholas, G.; Juergens, R.A.; Sacher, A.; Fung, A.S.; Wheatley-Price, P.; Laurie, S.A.; Levy, B.; Brahmer, J.R.; et al. ctDNA Response after Pembrolizumab in Non-Small Cell Lung Cancer: Phase 2 Adaptive Trial Results. Nat. Med. 2023, 29, 2559–2569. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, S.; Lee, J.C.; Choi, C.; Lee, S.Y.; Jang, T.; Oh, I.; Kim, Y. Phase II OPEN-LABEL Multicenter Study to Assess the Antitumor Activity of Afatinib in Lung Cancer Patients with Activating Epidermal Growth Factor Receptor Mutation from Circulating Tumor DNA: LIQUID-LUNG-A. Thoracic. Cancer 2021, 12, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Qian, J.; Xu, M.-D.; Gu, K.; Qian, F.-F.; Lu, J.; Zhang, X.-Y.; Wang, H.-M.; Yan, B.; Zhang, B.; et al. Prognostic and Predictive Value of Blood Tumor Mutational Burden in Patients With Lung Cancer Treated With Docetaxel. J. Natl. Compr. Cancer Netw. 2020, 18, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Willis, C.; Bauer, H.; Au, T.H.; Menon, J.; Unni, S.; Tran, D.; Rivers, Z.; Akerley, W.; Schabath, M.B.; Badin, F.; et al. Real-World Survival Analysis by Tumor Mutational Burden in Non-Small Cell Lung Cancer: A Multisite U.S. Study. Oncotarget 2022, 13, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yao, M.; Yang, Y. Higher Tumor Mutation Burden Was a Predictor for Better Outcome for NSCLC Patients Treated with PD-1 Antibodies: A Systematic Review and Meta-Analysis. SLAS Technol. 2021, 26, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, J.; Chen, L.; Guo, L.; Ye, M.; Wu, Y.; Ji, Q. Plasma miR-32 Levels in Non-Small Cell Lung Cancer Patients Receiving Platinum-Based Chemotherapy Can Predict the Effectiveness and Prognosis of Chemotherapy. Medicine 2019, 98, e17335. [Google Scholar] [CrossRef] [PubMed]
- Simeonidis, S.; Koutsilieri, S.; Vozikis, A.; Cooper, D.N.; Mitropoulou, C.; Patrinos, G.P. Application of Economic Evaluation to Assess Feasibility for Reimbursement of Genomic Testing as Part of Personalized Medicine Interventions. Front. Pharmacol. 2019, 10, 830. [Google Scholar] [CrossRef] [PubMed]
- Fagery, M.; Khorshidi, H.A.; Wong, S.Q.; Vu, M.; IJzerman, M. Health Economic Evidence and Modeling Challenges for Liquid Biopsy Assays in Cancer Management: A Systematic Literature Review. PharmacoEconomics 2023, 41, 1229–1248. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.M.; Leighl, N.B. Economic Impact of Tissue Testing and Treatments of Metastatic NSCLC in the Era of Personalized Medicine. Front. Oncol. 2014, 4, 258. [Google Scholar] [CrossRef] [PubMed]
- Hofmarcher, T.; Malmberg, C.; Lindgren, P. A Global Analysis of the Value of Precision Medicine in Oncology—The Case of Non-Small Cell Lung Cancer. Front. Med. 2023, 10, 1119506. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Anothaisintawee, T.; Butani, D.; Wang, Y.; Zemlyanska, Y.; Wong, C.B.N.; Virabhak, S.; Hrishikesh, M.A.; Teerawattananon, Y. Assessing the Cost-Effectiveness of Precision Medicine: Protocol for a Systematic Review and Meta-Analysis. BMJ Open 2022, 12, e057537. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef] [PubMed]
- Postel, M.; Roosen, A.; Laurent-Puig, P.; Taly, V.; Wang-Renault, S.-F. Droplet-Based Digital PCR and next Generation Sequencing for Monitoring Circulating Tumor DNA: A Cancer Diagnostic Perspective. Expert Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar] [CrossRef]
- Valpione, S.; Campana, L. Detection of Circulating Tumor DNA (ctDNA) by Digital Droplet Polymerase Chain Reaction (Dd-PCR) in Liquid Biopsies. Methods Enzymol. 2019, 629, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Van Den Broek, D.; Denis, M.G.; Hofman, P.; Hubank, M.; Mouliere, F.; Paz-Ares, L.; Schuuring, E.; Sültmann, H.; Vainer, G.; et al. Recommendations for a Practical Implementation of Circulating Tumor DNA Mutation Testing in Metastatic Non-Small-Cell Lung Cancer. ESMO Open 2022, 7, 100399. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-G.; Zhu, W.-Y.; Huang, Y.-Y.; Ma, L.-N.; Zhou, S.-Q.; Wang, Y.-K.; Zeng, F.; Zhou, J.-H.; Zhang, Y.-K. High Expression of Serum miR-21 and Tumor miR-200c Associated with Poor Prognosis in Patients with Lung Cancer. Med. Oncol. 2012, 29, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Cao, Y.; He, Z.; He, J.; Hu, C.; Duan, H.; Jiang, J. Serum Levels of miR-19b and miR-146a as Prognostic Biomarkers for Non-Small Cell Lung Cancer. Tohoku J. Exp. Med. 2014, 232, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yin, Y.; Liu, X.; Xi, X.; Xue, W.; Qu, Y. Non-Small Cell Lung Cancer Associated microRNA Expression Signature: Integrated Bioinformatics Analysis, Validation and Clinical Significance. Oncotarget 2017, 8, 24564–24578. [Google Scholar] [CrossRef] [PubMed]
- Schuurbiers, M.; Huang, Z.; Saelee, S.; Javey, M.; de Visser, L.; van den Broek, D.; van den Heuvel, M.; Lovejoy, A.F.; Monkhorst, K.; Klass, D. Biological and Technical Factors in the Assessment of Blood-Based Tumor Mutational Burden (bTMB) in Patients with NSCLC. J. Immunother. Cancer 2022, 10, e004064. [Google Scholar] [CrossRef] [PubMed]
- Raiber-Moreau, E.-A.; Portella, G.; Butler, M.G.; Clement, O.; Konigshofer, Y.; Hadfield, J. Development and Validation of Blood Tumor Mutational Burden Reference Standards. Genes Chromosomes Cancer 2023, 62, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, T.R.; Taube, J.M. PD-L1 and Emerging Biomarkers in Immune Checkpoint Blockade Therapy. Cancer J. 2018, 24, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Domblides, C.; Ruppert, A.-M.; Antoine, M.; Wislez, M. Immunotherapy’s New Challenge: Identification of Predictive Biomarkers for Tumor Response. Transl. Cancer Res. 2017, 6, S306–S308. [Google Scholar] [CrossRef]
- Goh, K.Y.; Cheng, T.Y.D.; Tham, S.C.; Lim, D.W.-T. Circulating Biomarkers for Prediction of Immunotherapy Response in NSCLC. Biomedicines 2023, 11, 508. [Google Scholar] [CrossRef]
- Cao, W.; Tang, Q.; Zeng, J.; Jin, X.; Zu, L.; Xu, S. A Review of Biomarkers and Their Clinical Impact in Resected Early-Stage Non-Small-Cell Lung Cancer. Cancers 2023, 15, 4561. [Google Scholar] [CrossRef]
- Tostes, K.; Siqueira, A.P.; Reis, R.M.; Leal, L.F.; Arantes, L.M.R.B. Biomarkers for Immune Checkpoint Inhibitor Response in NSCLC: Current Developments and Applicability. Int. J. Mol. Sci. 2023, 24, 11887. [Google Scholar] [CrossRef] [PubMed]
- Leidinger, P.; Backes, C.; Blatt, M.; Keller, A.; Huwer, H.; Lepper, P.; Bals, R.; Meese, E. The Blood-Borne miRNA Signature of Lung Cancer Patients Is Independent of Histology but Influenced by Metastases. Mol. Cancer 2014, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Luo, J.; Wu, S.; Si, H.; Gao, C.; Xu, W.; Abdullah, S.E.; Higgs, B.W.; Dennis, P.A.; Van Der Heijden, M.S.; et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov. 2020, 10, 1842–1853. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, D.B.; Bhalla, S.; Beasley, M.B.; Sholl, L.M.; Kerr, K.M.; Gnjatic, S.; Wistuba, I.I.; Rimm, D.L.; Tsao, M.S.; Hirsch, F.R. PD-L1 as a Biomarker of Response to Immune-Checkpoint Inhibitors. Nat. Rev. Clin. Oncol. 2021, 18, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Niu, X.; Cheng, Y.; Zhang, Y.; Xia, L.; Xia, W.; Lu, S. Exosomal PD-L1 Predicts Response with Immunotherapy in NSCLC Patients. Clin. Exp. Immunol. 2022, 208, 316–322. [Google Scholar] [CrossRef]
- Massafra, M.; Passalacqua, M.I.; Gebbia, V.; Macrì, P.; Lazzari, C.; Gregorc, V.; Buda, C.; Altavilla, G.; Santarpia, M. Immunotherapeutic Advances for NSCLC. BTT 2021, 15, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Incorvaia, L.; Fanale, D.; Badalamenti, G.; Barraco, N.; Bono, M.; Corsini, L.R.; Galvano, A.; Gristina, V.; Listì, A.; Vieni, S.; et al. Programmed Death Ligand 1 (PD-L1) as a Predictive Biomarker for Pembrolizumab Therapy in Patients with Advanced Non-Small-Cell Lung Cancer (NSCLC). Adv. Ther. 2019, 36, 2600–2617. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Pulliam, S.; Karki, N.R.; Khan, J.; Jogezai, S.; Sultan, S.; Muhammad, L.; Khan, M.; Jamil, N.; Waheed, A.; et al. PD-L1 Over-Expression Varies in Different Subtypes of Lung Cancer: Will This Affect Future Therapies? Clin. Pract. 2022, 12, 653–671. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Kwong, L.N. Diagnostic and Therapeutic Applications of miRNA-Based Strategies to Cancer Immunotherapy. Cancer Metastasis Rev. 2018, 37, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Wang, Z.; Lu, Z.; Xia, J.; Xie, Z.; Jiao, M.; Liu, R.; Chu, Y. MicroRNAs: Immune Modulators in Cancer Immunotherapy. Immunother. Adv. 2021, 1, ltab006. [Google Scholar] [CrossRef]
- Sirker, M.; Liang, W.; Zhang, L.; Fimmers, R.; Rothschild, M.A.; Gomes, I.; Schneider, P.M. Impact of Using Validated or Standard Reference Genes for miRNA qPCR Data Normalization in Cell Type Identification. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e199–e201. [Google Scholar] [CrossRef]
- Nuzziello, N.; Liguori, M. Seeking a Standardized Normalization Method for the Quantification of microRNA Expression. Muscle Nerve 2019, 59, E39. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, F.E.C.; Guevara-Montoya, M.C.; Serna-Ramirez, V.; Liscano, Y. Neuroinflammation and Schizophrenia: New Therapeutic Strategies through Psychobiotics, Nanotechnology, and Artificial Intelligence (AI). JPM 2024, 14, 391. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, F.E.C.; Lizcano Martinez, S.; Liscano, Y. Effectiveness of Psychobiotics in the Treatment of Psychiatric and Cognitive Disorders: A Systematic Review of Randomized Clinical Trials. Nutrients 2024, 16, 1352. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome Is Associated with Favorable Responses to Anti–Programmed Death 1 Immunotherapy in Chinese Patients with NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ferrero, M.; Dong, N.; D’Auria, G.; Reyes-Prieto, M.; Herreros-Pomares, A.; Calabuig-Fariñas, S.; Duréndez, E.; Aparisi, F.; Blasco, A.; et al. Analysis of the Gut Microbiota: An Emerging Source of Biomarkers for Immune Checkpoint Blockade Therapy in Non-Small Cell Lung Cancer. Cancers 2021, 13, 2514. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G.; Iannone, L.F.; Bersanelli, M.; Mini, E.; Catalano, M. The Gut Microbiome and Efficacy of Cancer Immunotherapy. Pharmacol. Ther. 2022, 231, 107973. [Google Scholar] [CrossRef]
- Katayama, Y.; Yamada, T.; Shimamoto, T.; Iwasaku, M.; Kaneko, Y.; Uchino, J.; Takayama, K. The Role of the Gut Microbiome on the Efficacy of Immune Checkpoint Inhibitors in Japanese Responder Patients with Advanced Non-Small Cell Lung Cancer. Transl. Lung. Cancer Res. 2019, 8, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.; Ng, T.L. A Narrative Review from Gut to Lungs: Non-Small Cell Lung Cancer and the Gastrointestinal Microbiome. Transl. Lung. Cancer Res. 2023, 12, 909–926. [Google Scholar] [CrossRef] [PubMed]
- Casciaro, M.; Salvo, E.D.; Pioggia, G.; Gangemi, S. Microbiota and microRNAs in Lung Diseases: Mutual Influence and Role Insights. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 13000–13008. [Google Scholar] [PubMed]
- Chrysostomou, D.; Roberts, L.A.; Marchesi, J.R.; Kinross, J.M. Gut Microbiota Modulation of Efficacy and Toxicity of Cancer Chemotherapy and Immunotherapy. Gastroenterology 2023, 164, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M. Training the Microbiota to Increase Immune Checkpoint Blockade and to Reduce Toxicity. Eur. J. Immunol. 2023, 53, 2250183. [Google Scholar] [CrossRef] [PubMed]
- Ryu, G.; Kim, H.; Koh, A. Approaching Precision Medicine by Tailoring the Microbiota. Mamm. Genome 2021, 32, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Musolino, C.; Tonacci, A.; Pioggia, G.; Gangemi, S. Interactions between the MicroRNAs and Microbiota in Cancer Development: Roles and Therapeutic Opportunities. Cancers 2020, 12, 805. [Google Scholar] [CrossRef] [PubMed]
- Purkayastha, K.; Dhar, R.; Pethusamy, K.; Srivastava, T.; Shankar, A.; Rath, G.K.; Karmakar, S. The Issues and Challenges with Cancer Biomarkers. J. Cancer Res. Ther. 2023, 19, S20–S35. [Google Scholar] [CrossRef]
- Truesdell, J.; Miller, V.A.; Fabrizio, D. Approach to Evaluating Tumor Mutational Burden in Routine Clinical Practice. Transl. Lung. Cancer Res. 2018, 7, 678–681. [Google Scholar] [CrossRef] [PubMed]
- Restrepo, J.C.; Dueñas, D.; Corredor, Z.; Liscano, Y. Advances in Genomic Data and Biomarkers: Revolutionizing NSCLC Diagnosis and Treatment. Cancers 2023, 15, 3474. [Google Scholar] [CrossRef] [PubMed]
- Van Den Broek, D.; Hiltermann, T.J.N.; Biesma, B.; Dinjens, W.N.M.; ‘T Hart, N.A.; Hinrichs, J.W.J.; Leers, M.P.G.; Monkhorst, K.; Van Oosterhout, M.; Scharnhorst, V.; et al. Implementation of Novel Molecular Biomarkers for Non-Small Cell Lung Cancer in the Netherlands: How to Deal with Increasing Complexity. Front. Oncol. 2020, 9, 1521. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. The Challenges of Evaluating Predictive Biomarkers Using Small Biopsy Tissue Samples and Liquid Biopsies from Non-Small Cell Lung Cancer Patients. J. Thorac. Dis. 2019, 11, S57–S64. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, J.; Das, B.; Shin, S.; Chen, A. Challenges and Future Directions in the Management of Tumor Mutational Burden-High (TMB-H) Advanced Solid Malignancies. Cancers 2023, 15, 5841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, J.; Wang, G.; He, X.; Mi, Y.; Cao, Y.; Yu, X. Predictive Efficacy of Blood-Based Tumor Mutation Burden Assay for Immune Checkpoint Inhibitors Therapy in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 795933. [Google Scholar] [CrossRef] [PubMed]
- Arghiani, N.; Shah, K. Modulating microRNAs in Cancer: Next-Generation Therapies. Cancer Biol. Med. 2021, 19, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, K.; Benhamou, R.I. Targeting MicroRNAs with Small Molecules. ncRNA 2024, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Pimienta, D.A.; Cruz Mosquera, F.E.; Palacios Velasco, I.; Giraldo Rodas, M.; Oñate-Garzón, J.; Liscano, Y. Specific Focus on Antifungal Peptides against Azole Resistant Aspergillus Fumigatus: Current Status, Challenges, and Future Perspectives. J. Fungi 2022, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Wan, J.; Zhou, L.; Ma, W.; Yang, Y.; Luo, W.; Yu, Z.; Wang, H. Stimuli-responsive Nanotherapeutics for Precision Drug Delivery and Cancer Therapy. WIREs Nanomed. Nanobiotechnol. 2019, 11, e1527. [Google Scholar] [CrossRef] [PubMed]
- Holder, J.E.; Ferguson, C.; Oliveira, E.; Lodeiro, C.; Trim, C.M.; Byrne, L.J.; Bertolo, E.; Wilson, C.M. The Use of Nanoparticles for Targeted Drug Delivery in Non-Small Cell Lung Cancer. Front. Oncol. 2023, 13, 1154318. [Google Scholar] [CrossRef] [PubMed]
- Macarrón Palacios, A.; Korus, P.; Wilkens, B.G.C.; Heshmatpour, N.; Patnaik, S.R. Revolutionizing in Vivo Therapy with CRISPR/Cas Genome Editing: Breakthroughs, Opportunities and Challenges. Front. Genome Ed. 2024, 6, 1342193. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Ma, Q.; Yin, W.; Ma, X.; He, Z. CRISPR/Cas9 Gene-Editing in Cancer Immunotherapy: Promoting the Present Revolution in Cancer Therapy and Exploring More. Front. Cell Dev. Biol. 2021, 9, 674467. [Google Scholar] [CrossRef] [PubMed]
- Yahya, S.; Abdelmenym Mohamed, S.I.; Yahya, S. Gene Editing: A Powerful Tool for Cancer Immunotherapy. Biointerface Res. Appl. Chem. 2022, 13, 98. [Google Scholar] [CrossRef]
- Mandair, D.; Reis-Filho, J.S.; Ashworth, A. Biological Insights and Novel Biomarker Discovery through Deep Learning Approaches in Breast Cancer Histopathology. NPJ. Breast. Cancer 2023, 9, 21. [Google Scholar] [CrossRef]

| Author and Publication Year | Biomarker Evaluated | Method of Biomarker Analysis | Cut-Off-Value | Predictive and Prognostic Value | Overall Survival (OS) and Progression-Free Survival (PFS) | Hazard Ratios (HRs) and 95% Confidence Intervals (CIs) for Tumor Size | Treatment | Statistical Methods Used for Biomarker Evaluation | Conclusions |
|---|---|---|---|---|---|---|---|---|---|
| Ren et al., 2022 [48] | ctDNA dynamics | Dynamic monitoring | ctDNA clearance after two cycles | Predicts camrelizumab plus chemotherapy efficacy | Prolonged OS and PFS | OS HR = 0.55 | Camrelizumab + carboplatin and paclitaxel | Kaplan–Meier, Cox models | ctDNA dynamics useful in predicting treatment efficacy in advanced squamous NSCLC |
| Yang et al., 2023 [49] | ALK fusions in ctDNA | NGS of plasma ctDNA | NS | ALK fusion detectability related to prognosis | PFS similar for brigatinib (19.3 mo) and alectinib (19.2 mo) | PFS HR = 0.97 (95% CI: 0.66–1.42) | Brigatinib vs. alectinib | Kaplan–Meier, Cox regression | No superiority of brigatinib over alectinib in ALK-positive NSCLC previously treated with crizotinib |
| Riess et al., 2021 [50] | EGFR exon 20 insertions | Next-generation sequencing (NGS) | NS | Not predictive | PFS: 5.4 months | NS | Erlotinib and onalespib | Standard 3 + 3 dose escalation, RECIST 1.1 | Limited activity of erlotinib and onalespib in EGFR exon 20 insertion NSCLC |
| Lo Russo et al., 2022 [51] | 36 immunobiomarkers like CD14, CD16 | Multiparametric flow cytometry | NS | Improved PFS and OS | CD14 = 0.018 OS significant improvements | NS | Pembrolizumab | Orthogonal component clustering, multivariable Cox regression | Predictive outcomes in PD-L1-low NSCLC with pembrolizumab |
| Garon et al., 2023 [52] | EGFR, TE genomic alterations in ctDNA | NGS of ctDNA | NS | Baseline EGFR alterations indicate shorter PFS | Median PFS: 12.7 mo (with aEGFR), 22 mo (without) | HR for PFS = 1.87 (95% CI: 1.42–2.51) | Ramucirumab + erlotinib | Kaplan–Meier | Baseline EGFR mutations linked to shorter PFS, predictive of ramucirumab + erlotinib efficacy |
| Han et al., 2023 [53] | bTMB | NGS with 448 gene panel | ≥1.72 mutations/Mb | Predictive of sintilimab + docetaxel efficacy | Median PFS: 5.8 mo; Median OS: 12.6 mo | NS | Docetaxel + sintilimab | Kaplan–Meier, Clopper–Pearson | Sintilimab + docetaxel improves PFS, OS in advanced NSCLC |
| Si et al., 2021 [54] | bTMB | GuardantOMNI ctDNA assay | ≥20 mutations/Mb | Predictive of benefit with durvalumab + tremelimumab vs. chemotherapy | NS | NS | Durvalumab ± tremelimumab or chemotherapy | Cox models, minimal p-value cross-validation | High bTMB predicts clinical benefit with durvalumab + tremelimumab vs. chemotherapy |
| Jiang et al., 2021 [55] | PD-L1, TMB, CD8+ TIL density, DSPP mutation | IHC and whole-exome sequencing | PD-L1 ≥ 1% expression | DSPP mutation associated with longer PFS | Median PFS: 7.0 mo; Median OS: 23.5 mo | NS | Toripalimab + carboplatin and pemetrexed | Kaplan–Meier | DSPP mutation suggested as potential biomarker for toripalimab efficacy in second-line treatment |
| Zhang et al., 2024 [56] | ctDNA dynamics, BRCA2, BRINP3, FBXW7, KIT, RB1 | Comprehensive NGS ctDNA profiling | Clearance after 2 cycles | BRCA2, others shorten PFS; clearance associated with longer PFS | Median PFS: 11.4 months, OS: 27.2 months | NS | Sintilimab, nab-paclitaxel, carboplatin | Kaplan–Meier, hazard ratios | Effective first-line treatment with promising PFS; genetic markers as predictive values |
| Tan et al., 2024 [44] | EGFR T790M, EGFRm | Droplet digital PCR, targeted sequencing | NS | Decrease in/clearance of EGFRm and T790M predicts longer PFS/OS | Median PFS: 9.4 months, OS: 26 months | NS | Alternating osimertinib and gefitinib | Kaplan–Meier, Cox regression | Feasible therapy with profound impact on clonal dynamics, though primary endpoint unmet |
| Kim et al., 2022 [57] | bTMB | Foundation Medicine bTMB assay | ≥16 mutations/Mb | Higher ORR, possibly longer OS for bTMB ≥ 16 | Longer OS for bTMB ≥ 16, PFS and OS not significant at cut-off | NS | First-line atezolizumab monotherapy | Kaplan–Meier, log-rank, Cox model | bTMB ≥ 16 linked to higher ORR; further studies needed for bTMB as a standalone predictive marker |
| Peters et al., 2022 [58] | bTMB | Foundation Medicine bTMB CTA | ≥16 for analysis | bTMB ≥ 16 did not meet primary endpoint for PFS benefit | No significant difference in PFS or OS | HR: 0.77 (95% CI: 0.59–1.00, p = 0.053) | Atezolizumab vs. chemotherapy | Kaplan–Meier, Cox model | bTMB ≥ 16 not supported as standalone predictive marker; potential for optimization |
| Chaft et al., 2022 [59] | MPR, PD-L1 TPS | Blood immunophenotype, exome sequencing | MPR ≤ 10% viable cells, PD-L1 high TPS | High PD-L1 TPS predicts MPR | 3-year DFS: 72%, OS: 80% | DFS HR = 0.373, OS HR = 0.273 | Neoadjuvant atezolizumab monotherapy | Kaplan–Meier | Neoadjuvant atezolizumab met primary endpoint of MPR; further validation needed |
| Shi et al., 2022. [60] | PD-L1, OVOL2, CTCF | Deep sequencing | NS | OVOL2 high = better PFS, CTCF high = worse PFS | Median OS significantly longer in sintilimab arm | NS | Sintilimab vs. docetaxel | Cox regression | Sintilimab improved survival, response rates; potential new treatment option for advanced NSCLC |
| Papadimitrakopoulou et al., 2020 [45] | EGFR T790M, EGFRm | Cobas, ddPCR, NGS | T790M-negative predicts longer PFS | T790M status predictive | Median PFS: 12.5 vs. 8.3 months (osimertinib) | NS | Osimertinib, platinum-pemetrexed | Kaplan–Meier | T790M-negative plasma predicts longer PFS |
| Sakai et al., 2021a [46] | TMB | Targeted deep sequencing | ≥12–16 mut/Mb | High TMB predicts better RFS | TMB ≥ 12 mut/Mb: improved RFS | HR = 0.477 | Pemetrexed/cisplatin vs. vinorelbine/cisplatin | Kaplan–Meier, Cox model | High TMB beneficial for pemetrexed/cisplatin in NS-NSCLC |
| Park et al., 2023 [61] | bTMB, cfDNA, hVAF, VAFSD | CT-ULTRA, targeted NGS | bTMB ≥ 11.5 mut/Mb at baseline | cfDNA, bTMB dynamics predictive | Median OS: 13.1 months, PFS: 2.1 months | NS | 1200 mg of atezolizumab every three weeks | Kaplan–Meier, Cox model | Baseline biomarkers predict atezolizumab efficacy |
| Park et al., 2021 [47] | Activating EGFR mutations in ctDNA and DNA | Mutyper and Cobas v2 assays | NS | ctDNA sensitivity for EGFR mutations | Median PFS: 11.1 months | NS | Osimertinib 80 mg daily | Kaplan–Meier | ctDNA assays effective in detecting EGFR mutations |
| Gu et al., 2023 [62] | EGFR mutations, ctDNA MRD | NGS | NS | MRD predicts therapeutic efficacy | PFS longer with TKI + pemetrexed in altered genes | NS | EGFR-TKI monotherapy or combined with pemetrexed | Kaplan–Meier, log-rank tests | ctDNA MRD as a biomarker for therapy efficacy in NSCLC |
| Han et al., 2022 [63] | ctDNA dynamics | NGS with 448-gene panel | ≥2 mutations positive, ≤1 mutation negative | ctDNA clearance at 6 weeks predictive | NS | NS | Sintilimab + docetaxel, maintenance with sintilimab | Kaplan–Meier, Cox regression | ctDNA dynamics predict sintilimab efficacy |
| Zhong et al., 2023 [64] | EGFR mutations, ctDNA mutation detection | NGS targeting 425 genes | Positive ctDNA at baseline predicts shorter PFS | ctDNA as a predictive biomarker | NS | NS | Tislelizumab + carboplatin and nab-paclitaxel | Kaplan–Meier, Cox regression | ctDNA predicts progression in tislelizumab therapy |
| García-Pardo et al., 2023 [65] | ctDNA genotyping | Plasma ctDNA testing with NGS before diagnosis | NS | Early ctDNA testing accelerates treatment initiation | NS | NS | Advanced nonsquamous NSCLC treatments | Standard genotyping comparison | Early ctDNA genotyping shortens treatment initiation |
| Nomura et al., 2020 [66] | ctDNA (Guardant360®) | Guardant360® | NS | Non-inferiority of discontinuing PD-1 inhibitors | NS | NS | Continue or discontinue PD-1 inhibitors | Kaplan–Meier, Cox model, non-inferiority test | Study supports possible discontinuation of PD-1 inhibitors without affecting survival |
| Martini et al., 2022 [67] | Gut microbiota, ctDNA RAS/BRAF WT MSS | 16S rRNA sequencing, targeted NGS | NS | Certain gut bacteria species linked to longer PFS | NS | NS | Cetuximab + avelumab combination therapy | Kendall Tau-b, Kaplan–Meier, log-rank | Gut microbiota as potential biomarker for cetuximab + avelumab efficacy |
| Provencio et al., 2022 [68] | ctDNA for prognosis and predictive value | NGS of FFPE and plasma samples | MAF ≥ 1% at baseline | Low ctDNA levels associated with better survival outcomes | 3-year OS: 81.9% (ITT), 91.0% (per protocol) | HR for PFS: 0.20, OS: 0.07 | Neoadjuvant paclitaxel, carboplatin, and nivolumab | Kaplan–Meier, Cox regression, competing risk | ctDNA levels predict success in chemoimmunotherapy |
| West et al., 2022 [69] | KRAS, STK11, KEAP1, TP53 mutations | Blood-based ctDNA sequencing | NS | KRAS, STK11, KEAP1 mutations affect treatment efficacy | mKRAS: improved OS and PFS with ABCP vs. BCP | OS: HR = 0.50 (0.34–0.72); PFS: HR = 0.42 (0.29–0.61) | Atezolizumab, bevacizumab, carboplatin, paclitaxel | Kaplan–Meier, Cox model | Improved outcomes in mKRAS with ABCP; ctDNA levels after treatment correlate with PFS and OS |
| Lo Russo et al., 2023 [70] | Immune cell subsets, gene expression levels | Flow cytometry, gene expression, metagenomic sequencing | NS | Immune profiles correlate with PFS | Median PFS: 2.9 months | NS | Pembrolizumab as first-line treatment | Sequential Cox regression, Benjamini–Hochberg, LASSO | Multiomic markers predict PFS; NK cells at baseline may determine pembrolizumab benefit |
| Zhou et al., 2023 [71] | PD-L1 on tumor cells | SP263 and 22C3 assays | NS | High concordance between SP263 and 22C3 assays | NS | NS | Adjuvant atezolizumab vs. supportive care | Concordance assessment, survival evaluation | SP263 and 22C3 assays effectively predict adjuvant atezolizumab benefit in early-stage NSCLC |
| Sakai et al., 2021b [72] | EGFR T790M mutation | Cobas v2, ddPCR, deep sequencing | NS | Monitors EGFR T790M for treatment response | NS | NS | Osimertinib administration | Univariate regression, Cox model | Effective monitoring of EGFR T790M with ctDNA enhances osimertinib treatment decisions |
| Redman et al., 2020 [73] | Multiple biomarkers | FoundationOne® NGS assay | NS | Multiple biomarkers assessed for targeted therapies | Median OS varied by treatment arm, no PFS provided | NS | Targeted therapies, immunotherapies in sqNSCLC | Kaplan–Meier, Cox regression, other methods | The study validates the use of molecularly targeted therapies in genomically defined sqNSCLC subgroups |
| Hirsch et al., 2022 [74] | EGFR copy number, protein expression | EGFR FISH, IHC | TC ≥ 50% or IC ≥ 10% for SP142 and TPS ≥ 50% for 22C3 | High EGFR copy number and protein expression predictive | Improved OS in SCC with cetuximab addition (12.6 vs. 4.6 months) | HR: 0.32 (0.18–0.59), p = 0.0002 | Chemotherapy with or without cetuximab | Cox model for OS and PFS | EGFR FISH and IHC are predictive of cetuximab benefit in SCC, independently of KRAS status |
| Schuler et al., 2020 [75] | MET dysregulation | Immunohistochemistry, FISH, NGS | NS | Capmatinib’s antitumor activity based on MET dysregulation | NS | NS | Capmatinib administration | Safety and activity assessment | Capmatinib shows promise in NSCLC with MET dependency, particularly with high MET GCN or METex14 mutations |
| Gadgeel et al., 2022 [76] | PD-L1 expression | SP142 and 22C3 IHC assays | TC ≥ 50% or IC ≥ 10% for SP142 and TPS ≥ 50% for 22C3 | Atezolizumab improves survival over docetaxel | Atezolizumab benefits across all PD-L1 subgroups, especially high PD-L1 | OS and PFS: HR varied by assay, greater in high PD-L1 groups | Atezolizumab or docetaxel | Analysis in PD-L1 subgroups, assay selection | SP142 and 22C3 assays effectively predict atezolizumab efficacy in metastatic NSCLC across PD-L1 thresholds |
| Ramalingam et al., 2021 [77] | LP52 gene expression | Whole-transcriptome sequencing | NS | Veliparib’s efficacy in sqNSCLC | No significant OS benefit in smokers; slight benefit in general population | OS: HR = 0.853 (0.747 to 0.974), PFS not different | Veliparib or placebo with carboplatin and paclitaxel | Kaplan–Meier, log-rank test, biomarker analysis | Veliparib shows a marginal OS benefit; LP52 may help to identify responsive patients |
| Song et al., 2022 [78] | HER2 mutations | NGS | NS | Pyrotinib’s efficacy based on HER2 mutation types | 6-month PFS rate: 49.5%, median PFS: 5.6 months, OS: 10.5 months | NS | Pyrotinib treatment | Kaplan–Meier, Cox regression, Fisher’s exact test | Pyrotinib effective in HER2-mutant NSCLC; potential for ctDNA to aid in disease monitoring |
| Anagnostou et al., 2023 [79] | ctDNA dynamics | Serial quantitative ctDNA assessments | Maximal mutant allele fraction clearance at cycle 3 | ctDNA clearance after two cycles predictive | Median PFS significantly longer with molecular response, OS not reached vs. 7.23 months | NS | Pembrolizumab treatment | Kaplan–Meier, exploratory biomarker analyses | ctDNA dynamics correlate with pembrolizumab efficacy, guiding treatment adjustments |
| Park et al., 2021 [80] | Activating EGFRm | EGFRm ctDNA analysis using PANA Mutype | NS | Efficacy of afatinib in EGFRm-positive lung cancer | Median PFS: 12.0 months, OS data immature | NS | Afatinib 40 mg daily | Mann–Whitney U, Pearson’s χ2, Fisher’s exact, Kaplan–Meier | Afatinib shows favorable ORR and PFS in treatment-naïve patients with detectable EGFRm in ctDNA |
| Biomarker | Role of Biomarker | Main Findings | Clinical Implications | Authors | Treatment Response | Heterogeneity in NSCLC | Clinical and Demographic Characteristics |
|---|---|---|---|---|---|---|---|
| ctDNA | Tumor burden and disease progression | - Detection correlated with tumor burden and disease progression. - Reduction in levels during treatment indicative of better PFS and OS. | Non-invasive biomarker for monitoring of response and adjustment of therapeutic strategies. | Ren et al., 2022 [48]; Yang et al., 2023 [49]; Garon et al., 2023 [52]; Zhang et al., 2024 [56]; Tan et al., 2024 [44] | Predictive of response to camrelizumab, carboplatin, paclitaxel, osimertinib, sintilimab, nab-paclitaxel, gefitinib | Varies with mutation type (EGFR, ALK), tumor stage, distribution of histological subtypes, and frequency of co-alterations. | Predominantly men, history of heavy smoking, ECOG 1, stage IV disease. |
| PD-L1 | Tumor progression and prognosis | High levels associated with better response to ICIs and longer survival. | Crucial in selecting patients for immunotherapy. | Jiang et al., 2021 [55]; Zhou et al., 2023 [71]; Gadgeel et al., 2022 [76] | Predictive of response to atezolizumab, pembrolizumab, toripalimab | Varies with expression levels, tumor microenvironment, distribution of histological subtypes, and frequency of co-alterations. | Predominantly men, history of heavy smoking, ECOG 1, stage IV disease. |
| miRNA * | Response to immunotherapy and survival outcomes | miRNA-21 and miRNA-155 correlated with response to immunotherapy and survival outcomes. Plasma miR-32 levels correlated with chemotherapy response and prognosis. | Patient stratification and treatment personalization based on molecular profiles. | Xu et al., 2019 [84] | Predictive of chemotherapy efficacy and prognosis with platinum-based chemotherapy | Varies with miRNA type, interaction with other molecular pathways, and frequency of co-alterations. | Patients aged 45–78, predominantly men, 81.4% smokers, ECOG 1–2, stage II–IV disease. |
| bTMB | Tumor progression and prognosis | High bTMB associated with better outcomes in combined immunotherapy and chemotherapy. | Accurate measurement predicts immunotherapy efficacy and guides treatment selection. | Han et al., 2023 [53]; Si et al., 2021 [54]; Kim et al., 2022 [57]; Peters et al., 2022 [58] | Predictive of response to sintilimab + docetaxel, durvalumab + tremelimumab, atezolizumab | Varies with mutation burden, specific gene mutations, distribution of histological subtypes, and frequency of co-alterations. | Predominantly men, history of heavy smoking, ECOG 1, stage IV disease. |
| Variable | ctDNA [91,92,93] | miRNA [94,95,96] | bTMB [54,81,97,98] | Immunological Markers [20,99,100] |
|---|---|---|---|---|
| Type of Biomarker | Genetic (circulating DNA) | Genetic (non-coding RNA) | Genetic (mutational burden) | Protein (immune proteins) |
| Detection Method | NGS, digital PCR | Real-time PCR, microarrays | NGS, digital PCR | IHC, flow cytometry |
| Clinical Utility | Diagnosis, prognosis, monitoring | Prognosis, monitoring | Prognostic, predictive | Diagnostic, predictive |
| Prognostic and Predictive Aspects | High sensitivity for early detection | Correlates with immunotherapy response | Predicts response to specific immunotherapies | Expression correlated with survival and response |
| Variability Factors | Influenced by tumor burden, detection techniques | Influenced by sample conditions | Requires standardization in measurement | Sensitive to detection methods and immune status |
| Advantages | Non-invasive, high sensitivity | Non-invasive, easily quantifiable | Information on tumor heterogeneity | Directly related to mechanisms of action of therapies |
| Limitations | Cost, need for sequencing | Inter- and intra-individual variability | Influenced by technical and biological factors | Requires validation for specific interpretation |
| Cost-Effectiveness | Moderate–high | Low–moderate | High due to sequencing technologies | Moderate, depends on the marker and method |
| Usage Recommendations | Widely recommended in clinical guidelines | In research, some clinical applications | Recommended in specific contexts | Emerging use, supported by recent studies |
| Recent Innovations | Advances in digital PCR technology | New miRNAs associated with NSCLC | Improvements in accuracy and cost of NGS | New predictive markers of response to PD-1/PD-L1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Restrepo, J.C.; Martínez Guevara, D.; Pareja López, A.; Montenegro Palacios, J.F.; Liscano, Y. Identification and Application of Emerging Biomarkers in Treatment of Non-Small-Cell Lung Cancer: Systematic Review. Cancers 2024, 16, 2338. https://doi.org/10.3390/cancers16132338
Restrepo JC, Martínez Guevara D, Pareja López A, Montenegro Palacios JF, Liscano Y. Identification and Application of Emerging Biomarkers in Treatment of Non-Small-Cell Lung Cancer: Systematic Review. Cancers. 2024; 16(13):2338. https://doi.org/10.3390/cancers16132338
Chicago/Turabian StyleRestrepo, Juan Carlos, Darly Martínez Guevara, Andrés Pareja López, John Fernando Montenegro Palacios, and Yamil Liscano. 2024. "Identification and Application of Emerging Biomarkers in Treatment of Non-Small-Cell Lung Cancer: Systematic Review" Cancers 16, no. 13: 2338. https://doi.org/10.3390/cancers16132338






