Chromogenic LMO2 mRNA ISH Expression Correlates with LMO2 Protein and Gene Expression and Captures Their Survival Impact in Diffuse Large B-Cell Lymphoma, NOS
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection
2.2. Immunohistochemistry
2.3. Fluorescence and Chromogenic In Situ Hybridization (FISH and CISH)
2.4. Gene Expression Analysis
2.5. Statistical Analysis
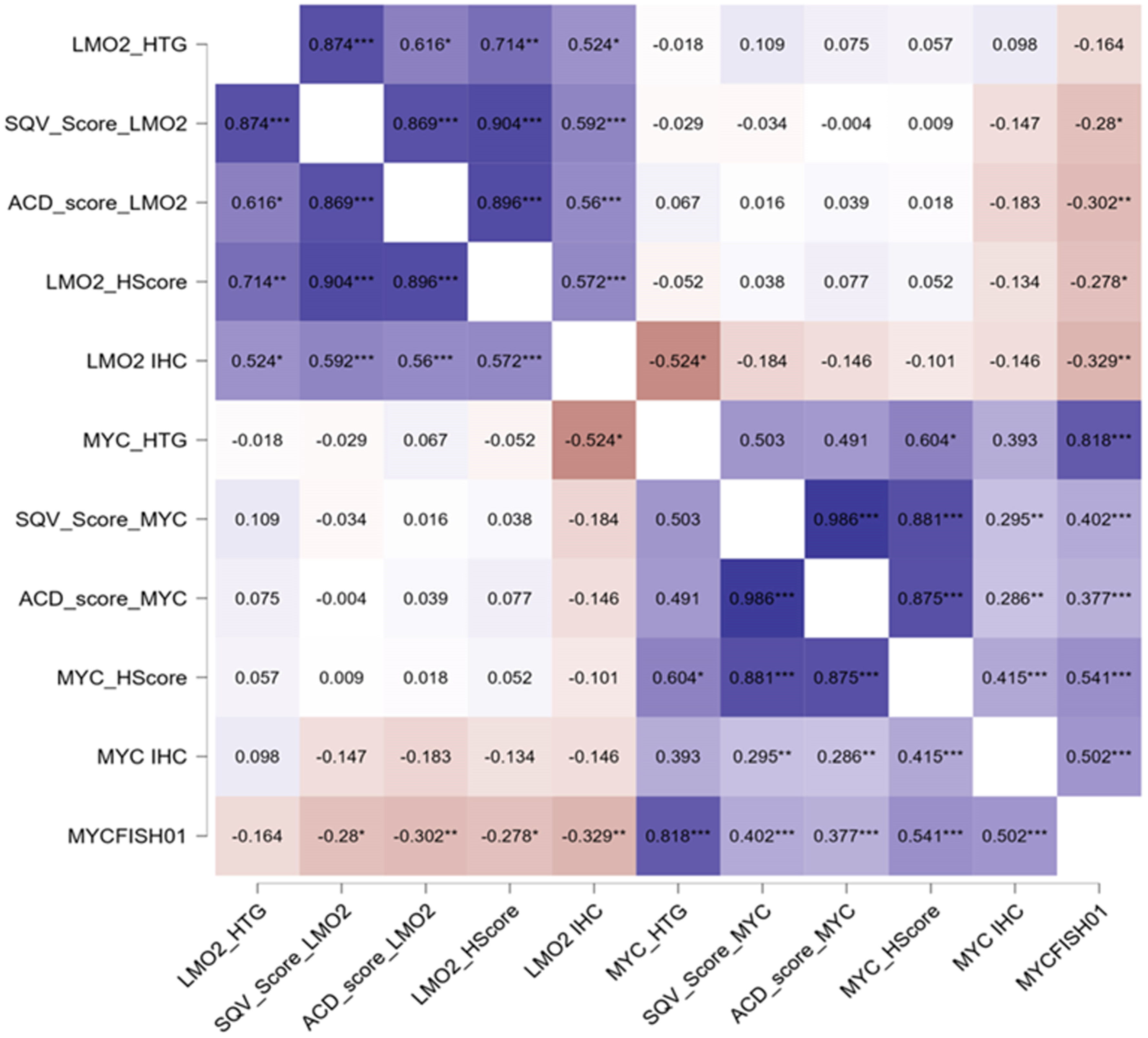
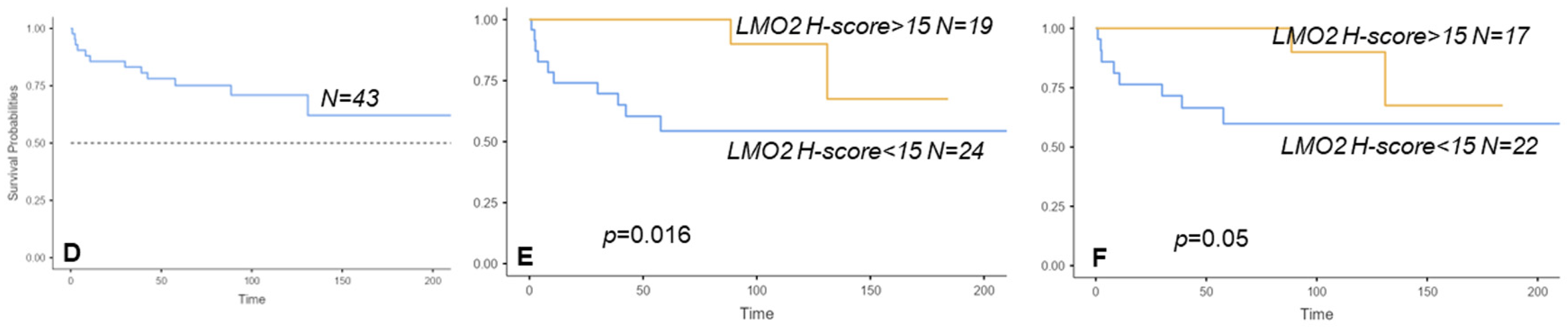
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Royer-Pokora, B.; Loos, U.; Ludwig, W.D. TTG-2, a New Gene Encoding a Cysteine-Rich Protein with the LIM Motif, Is Overexpressed in Acute T-Cell Leukaemia with the t(11;14)(P13;Q11). Oncogene 1991, 6, 1887–1893. [Google Scholar] [PubMed]
- Boehm, T.; Foroni, L.; Kaneko, Y.; Perutz, M.F.; Rabbitts, T.H. The Rhombotin Family of Cysteine-Rich LIM-Domain Oncogenes: Distinct Members Are Involved in T-Cell Translocations to Human Chromosomes 11p15 and 11p13. Proc. Natl. Acad. Sci. USA 1991, 88, 4367–4371. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Pannell, R.; Forster, A.; Rabbitts, T.H. The Oncogenic LIM-Only Transcription Factor Lmo2 Regulates Angiogenesis but Not Vasculogenesis in Mice. Proc. Natl. Acad. Sci. USA 2000, 97, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct Types of Diffuse Large B-Cell Lymphoma Identified by Gene Expression Profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Wright, G.; Dave, S.S.; Xiao, W.; Powell, J.; Zhao, H.; Xu, W.; Tan, B.; Goldschmidt, N.; Iqbal, J.; et al. Stromal Gene Signatures in Large-B-Cell Lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The Use of Molecular Profiling to Predict Survival after Chemotherapy for Diffuse Large-B-Cell Lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- Lossos, I.S.; Czerwinski, D.K.; Alizadeh, A.A.; Wechser, M.A.; Tibshirani, R.; Botstein, D.; Levy, R. Prediction of Survival in Diffuse Large-B-Cell Lymphoma Based on the Expression of Six Genes. N. Engl. J. Med. 2004, 350, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, R.; Chen, J.; Tibshirani, R.; Johnson, N.A.; Sehn, L.H.; Natkunam, Y.; Briones, J.; Advani, R.; Connors, J.M.; Byrne, G.E.; et al. Paraffin-Based 6-Gene Model Predicts Outcome in Diffuse Large B-Cell Lymphoma Patients Treated with R-CHOP. Blood 2008, 111, 5509–5514. [Google Scholar] [CrossRef] [PubMed]
- Natkunam, Y.; Farinha, P.; Hsi, E.D.; Hans, C.P.; Tibshirani, R.; Sehn, L.H.; Connors, J.M.; Gratzinger, D.; Rosado, M.; Zhao, S.; et al. LMO2 Protein Expression Predicts Survival in Patients with Diffuse Large B-Cell Lymphoma Treated with Anthracycline-Based Chemotherapy with and without Rituximab. J. Clin. Oncol. 2008, 26, 447–454. [Google Scholar] [CrossRef]
- Vazquez, I.; Papaleo, N.; Garcia, E.; Salido, M.; Salar, A.; Hernandez, S.; Calvo, X.; Colomo, L. Clinical Interest of LMO2 Testing for the Diagnosis of Aggressive Large B-Cell Lymphomas. Cancers 2020, 12, 884. [Google Scholar] [CrossRef]
- Vazquez, I.; Papaleo, N.; Lop, J.; Puiggros, A.; Sanchez-Gonzalez, B.; Diez-Feijoo, R.; Gimeno, E.; Andrade-Campos, M.; Salar, A.; Espinet, B.; et al. Lack of Expression of LMO2 Clone SP51 Identifies MYC Rearrangements in Aggressive Large B-Cell Lymphomas. Virchows Arch. 2021, 479, 1073–1078. [Google Scholar] [CrossRef]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A Report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- De Jong, D.; Rosenwald, A.; Chhanabhai, M.; Gaulard, P.; Klapper, W.; Lee, A.; Sander, B.; Thorns, C.; Campo, E.; Molina, T.; et al. Immunohistochemical Prognostic Markers in Diffuse Large B-Cell Lymphoma: Validation of Tissue Microarray as a Prerequisite for Broad Clinical Applications—A Study from the Lunenburg Lymphoma Biomarker Consortium. J. Clin. Oncol. 2007, 25, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Muller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the Molecular Classification of Diffuse Large B-Cell Lymphoma by Immunohistochemistry Using a Tissue Microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Ventura, R.A.; Martin-Subero, J.I.; Jones, M.; McParland, J.; Gesk, S.; Mason, D.Y.; Siebert, R. FISH Analysis for the Detection of Lymphoma-Associated Chromosomal Abnormalities in Routine Paraffin-Embedded Tissue. J. Mol. Diagn. 2006, 8, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A Novel in Situ RNA Analysis Platform for Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Sola, D.; Victora, G.D.; Ying, C.Y.; Phan, R.T.; Saito, M.; Nussenzweig, M.C.; Dalla-Favera, R. The Proto-Oncogene MYC Is Required for Selection in the Germinal Center and Cyclic Reentry. Nat. Immunol. 2012, 13, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Calado, D.P.; Sasaki, Y.; Godinho, S.A.; Pellerin, A.; Köchert, K.; Sleckman, B.P.; De Alborán, I.M.; Janz, M.; Rodig, S.; Rajewsky, K. The Cell-Cycle Regulator c-Myc Is Essential for the Formation and Maintenance of Germinal Centers. Nat. Immunol. 2012, 13, 1092–1100. [Google Scholar] [CrossRef]
- Valentino, C.; Kendrick, S.; Johnson, N.; Gascoyne, R.; Chan, W.C.; Weisenburger, D.; Braziel, R.; Cook, J.R.; Tubbs, R.; Campo, E.; et al. Colorimetric in Situ Hybridization Identifies MYC Gene Signal Clusters Correlating with Increased Copy Number, MRNA, and Protein in Diffuse Large b-Cell Lymphoma. Am. J. Clin. Pathol. 2013, 139, 242–254. [Google Scholar] [CrossRef]
- Stasik, C.J.; Nitta, H.; Zhang, W.; Mosher, C.H.; Cook, J.R.; Tubbs, R.R.; Unger, J.M.; Brooks, T.A.; Persky, D.O.; Wilkinson, S.T.; et al. Increased MYC Gene Copy Number Correlates with Increased MRNA Levels in Diffuse Large B-Cell Lymphoma. Haematologica 2010, 95, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Colomo, L.; Vazquez, I.; Papaleo, N.; Espinet, B.; Ferrer, A.; Franco, C.; Comerma, L.; Hernandez, S.; Calvo, X.; Salar, A.; et al. LMO2-Negative Expression Predicts the Presence of MYC Translocations in Aggressive B-Cell Lymphomas. Am. J. Surg. Pathol. 2017, 41, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.; Verdun, R.E.; Lossos, I.S. Low LIM-Domain Only 2 (LMO2) Expression in Aggressive B Cell Lymphoma Correlates with MYC and MYC/BCL2 Rearrangements, Especially in Germinal Center Cell-Type Tumors. Leuk. Lymphoma 2021, 62, 2547–2550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bian, T.; Zhang, Y.; Zheng, Y.; Zhang, J.; Zhou, X.; Xie, J. A Combination of LMO2 Negative and CD38 Positive Is Useful for the Diagnosis of Burkitt Lymphoma. Diagn. Pathol. 2019, 14, 100. [Google Scholar] [CrossRef] [PubMed]
- Massoni-Badosa, R.; Aguilar-Fernández, S.; Nieto, J.C.; Soler-Vila, P.; Elosua-Bayes, M.; Marchese, D.; Kulis, M.; Vilas-Zornoza, A.; Bühler, M.M.; Rashmi, S.; et al. An Atlas of Cells in the Human Tonsil. Immunity 2024, 57, 379–399.e18. [Google Scholar] [CrossRef] [PubMed]
- Milpied, P.; Cervera-Marzal, I.; Mollichella, M.L.; Tesson, B.; Brisou, G.; Traverse-Glehen, A.; Salles, G.; Spinelli, L.; Nadel, B. Human Germinal Center Transcriptional Programs Are De-Synchronized in B Cell Lymphoma. Nat. Immunol. 2018, 19, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; May, W.S.; Gao, F.; Flagg, T.; Deng, X. Bcl2 Suppresses DNA Repair by Enhancing C-Myc Transcriptional Activity. J. Biol. Chem. 2006, 281, 14446–14456. [Google Scholar] [CrossRef] [PubMed]
- Parvin, S.; Ramirez-Labrada, A.; Aumann, S.; Lu, X.Q.; Weich, N.; Santiago, G.; Cortizas, E.M.; Sharabi, E.; Zhang, Y.; Sanchez-Garcia, I.; et al. LMO2 Confers Synthetic Lethality to PARP Inhibition in DLBCL. Cancer Cell 2019, 36, 237–249.e6. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, N.; Climent, F.; Tapia, G.; Luizaga, L.; Azcarate, J.; Bosch-Schips, J.; Muñoz-Marmol, A.M.; Salido, M.; Lome-Maldonado, C.; Vazquez, I.; et al. Round-Robin Testing for LMO2 and MYC as Immunohistochemical Markers to Screen MYC Rearrangements in Aggressive Large B-Cell Lymphoma. Virchows Arch. 2023. [Google Scholar] [CrossRef]
- Ott, G.; Rosenwald, A.; Campo, E. Review Article Understanding MYC -Driven Aggressive B-Cell Lymphomas: Pathogenesis and Classification. J. Am. Soc. Hematol. 2016, 122, 575–583. [Google Scholar] [CrossRef]
- Alduaij, W.; Collinge, B.; Ben-Neriah, S.; Jiang, A.; Hilton, L.K.; Boyle, M.; Meissner, B.; Chong, L.; Miyata-Takata, T.; Slack, G.W.; et al. Molecular Determinants of Clinical Outcomes in a Real-World Diffuse Large B-Cell Lymphoma Population. Blood 2023, 141, 2493–2507. [Google Scholar] [CrossRef] [PubMed]
- Latchmansingh, K.A.; Wang, X.; Verdun, R.E.; Marques-Piubelli, M.L.; Vega, F.; You, M.J.; Chapman, J.; Lossos, I.S. LMO2 Expression Is Frequent in T-Lymphoblastic Leukemia and Correlates with Survival, Regardless of T-Cell Stage. Mod. Pathol. 2022, 35, 1220–1226. [Google Scholar] [CrossRef] [PubMed]



| All Cases | tDLBCL from MZL | tDLBCL from FL | DLBCL-NOS | HGBCL DH/TH | HGBCL-NOS | BL | |
|---|---|---|---|---|---|---|---|
| Number of cases | 82 | 1 | 12 | 56 | 9 | 2 | 2 |
| Age | 67 (38–93) | 63 | 65 (47–80) | 67 (38–88) | 73 (59–93) | 53 (53–54) | 43 (38–49) |
| Sex (male:female) | 44:38 | 1:0 | 7:5 | 28:28 | 6:3 | 1:1 | 1:1 |
| Nodal | 58 (71%) | 1 (100%) | 12 (100%) | 36 (64%) | 7 (78%) | 2 (100%) | 2 (100%) |
| IPI * | |||||||
| Low | 22/77 (29%) | 0/1 (0) | 3/10 (30%) | 19/54 (35%) | 0/8 (0) | 0/2 (0) | 0/2 (0) |
| Low-intermediate | 16/77 (21%) | 0/1 (0) | 4/10 (40%) | 11/54 (20%) | 0/8 (0) | 1/2 (50%) | 0/2 (0) |
| High-intermediate | 24/77 (31%) | 1/1 (100%) | 3/10 (30%) | 13/54 (24%) | 4/8 (50%) | 1/2 (50%) | 2/2 (100%) |
| High | 15/77 (19%) | 0/1 (0) | 0/10 (0) | 11/54 (20%) | 4/8 (50%) | 0/2 (0) | 0/2 (0) |
| Immunohistochemistry | |||||||
| CD10 | 43 (52%) | 0 | 8 (67%) | 25 (45%) | 6 (67%) | 2 (100%) | 2 (100%) |
| BCL-6 | 76 (93%) | 1 (100%) | 11 (92%) | 52 (93%) | 8 (89%) | 2 (100%) | 2 (100%) |
| MUM1/IRF4 | 52 (63%) | 1 (100%) | 4 (33%) | 43 (78%) | 2 (22%) | 2 (100%) | 0 |
| MYC | 34 (41%) | 0 | 5 (42%) | 17 (30%) | 8 (89%) | 2 (100%) | 2 (100%) |
| LMO2 | 48 (58%) | 1 (100%) | 10 (83%) | 34 (61%) | 3 (33%) | 0 | 0 |
| BCL-2 | 66 (80%) | 1 (100%) | 11 (92%) | 45 (80%) | 8 (89%) | 1 (50%) | 0 |
| COO | |||||||
| GCB-like | 49 (60%) | 0 | 9 (75%) | 29 (52%) | 8 (89%) | 2 (100%) | 2 (100%) |
| Non-GCB-like | 33 (40%) | 1 (100%) | 3 (25%) | 27 (48%) | 1 (11%) | 0 | 0 |
| FISH | |||||||
| MYC rearranged | 20/82 (24%) | 0/1 (0) | 5/12 (42%) | 2/56 (4%) | 9/9 (100%) | 2/2 (100%) | 2/2 (100%) |
| BCL-2 rearranged * | 20/68 (24%) | 0/1 (0) | 8/12 (67%) | 5/42 (12%) | 7/9 (78%) | 0/2 (0) | 0/2 (0) |
| BCL-6 rearranged * | 17/61 (28%) | 0/1 (0) | 5/11 (45%) | 9/36 (25%) | 3/9 (33%) | 0/2 (0) | 0/2 (0) |
| LMO2 H-Score | MYC H-Score | |
|---|---|---|
| Control tonsil | ||
| GC dark zone | 115 (110, 120) | 40 (30, 40) |
| GC light zone | 260 (250, 275) | 120 (115, 125) |
| Mantle/marginal zones | 30 (30, 40) | 50 (30, 50) |
| Interfollicular areas | 30 (20, 40) | 50 (40, 55) |
| All cases (N = 82) | 15 (5, 90) | 30 (8.75, 90) |
| tDLBCL from MZL (N = 1) | 110 | 140 |
| tDLBCL from FL (N = 12) | 97.5 (42.5, 127.5) | 75 (56.25, 162.5) |
| DLBCL-NOS (N = 56) | 15 (5, 72.5) | 25 (5, 47.5) |
| GCB-like (N = 29) | 40 (7.5, 82.5) | 20 (5, 52.5) |
| Non-GCB-like (N = 27) | 10 (5, 50) | 30 (5, 50) |
| HGBCL DH/TH (N = 9) | 5 (5, 12.5) | 100 (32.5, 177) |
| HGBCL-NOS (N = 2) | 5 | 190 |
| BL (N = 2) | 7.5 | 155 |
| Diagnosis | MYC Rearranged (N = 20) | MYC Non-Rearranged (N = 62) |
|---|---|---|
| tDLBCL | 5/20 (25%) | 8/62 (13%) |
| DLBCL-NOS | 2/20 (10%) | 54/62 (87%) |
| HGBCL-NOS | 2/20 (10%) | - |
| HGBCL DH/TH | 9/20 (45%) | - |
| BL | 2/20 (10%) | - |
| Nodal/Extranodal | 14 (70%)/6 (30%) | 44 (71%)/18 (29%) |
| CD10 expression | 14/20 (70%) | 29/62 (47%) |
| GCB-like/Non-GCB-like * | 16 (80%)/4 (20%) | 33 (53%)/29 (47%) |
| LMO2 GEP, median (Q1;Q3) | 1301 (638; 2848.5) | 1679.5 (1056; 5207.75) |
| LMO2 H-score, median (Q1;Q3) | 5 (5; 37.5) | 35 (5; 102.5) |
| LMO2 protein expression | 6/20 (30%) | 42/62 (68%) |
| MYC GEP, median (Q1;Q3) | 5171 (3638; 6879) | 1025.5 (804.25; 1688) |
| MYC H-score, median (Q1;Q3) | 157.5 (72.5; 190) | 30 (5; 56.25) |
| MYC protein expression | 17/20 (85%) | 17/62 (27%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaleo, N.; Molina-Alvarez, A.; Onieva, R.; Fuertes, D.; Sanchez-Gonzalez, B.; Riera, X.; Lopez-Segura, D.; Lome-Maldonado, C.; Ara-Mancebo, X.; Yelamos, J.; et al. Chromogenic LMO2 mRNA ISH Expression Correlates with LMO2 Protein and Gene Expression and Captures Their Survival Impact in Diffuse Large B-Cell Lymphoma, NOS. Cancers 2024, 16, 2378. https://doi.org/10.3390/cancers16132378
Papaleo N, Molina-Alvarez A, Onieva R, Fuertes D, Sanchez-Gonzalez B, Riera X, Lopez-Segura D, Lome-Maldonado C, Ara-Mancebo X, Yelamos J, et al. Chromogenic LMO2 mRNA ISH Expression Correlates with LMO2 Protein and Gene Expression and Captures Their Survival Impact in Diffuse Large B-Cell Lymphoma, NOS. Cancers. 2024; 16(13):2378. https://doi.org/10.3390/cancers16132378
Chicago/Turabian StylePapaleo, Natalia, Andrea Molina-Alvarez, Ricard Onieva, Diana Fuertes, Blanca Sanchez-Gonzalez, Xenia Riera, David Lopez-Segura, Carmen Lome-Maldonado, Xavier Ara-Mancebo, Jose Yelamos, and et al. 2024. "Chromogenic LMO2 mRNA ISH Expression Correlates with LMO2 Protein and Gene Expression and Captures Their Survival Impact in Diffuse Large B-Cell Lymphoma, NOS" Cancers 16, no. 13: 2378. https://doi.org/10.3390/cancers16132378
APA StylePapaleo, N., Molina-Alvarez, A., Onieva, R., Fuertes, D., Sanchez-Gonzalez, B., Riera, X., Lopez-Segura, D., Lome-Maldonado, C., Ara-Mancebo, X., Yelamos, J., Salido, M., Vazquez, I., Calvo, X., & Colomo, L. (2024). Chromogenic LMO2 mRNA ISH Expression Correlates with LMO2 Protein and Gene Expression and Captures Their Survival Impact in Diffuse Large B-Cell Lymphoma, NOS. Cancers, 16(13), 2378. https://doi.org/10.3390/cancers16132378








