Serial Changes of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Treated with Atezolizumab Plus Bevacizumab
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Protocol and Participants
2.2. Etiology of Liver Diseases
2.3. Treatment Protocol
2.4. Evaluation of the Treatment Response
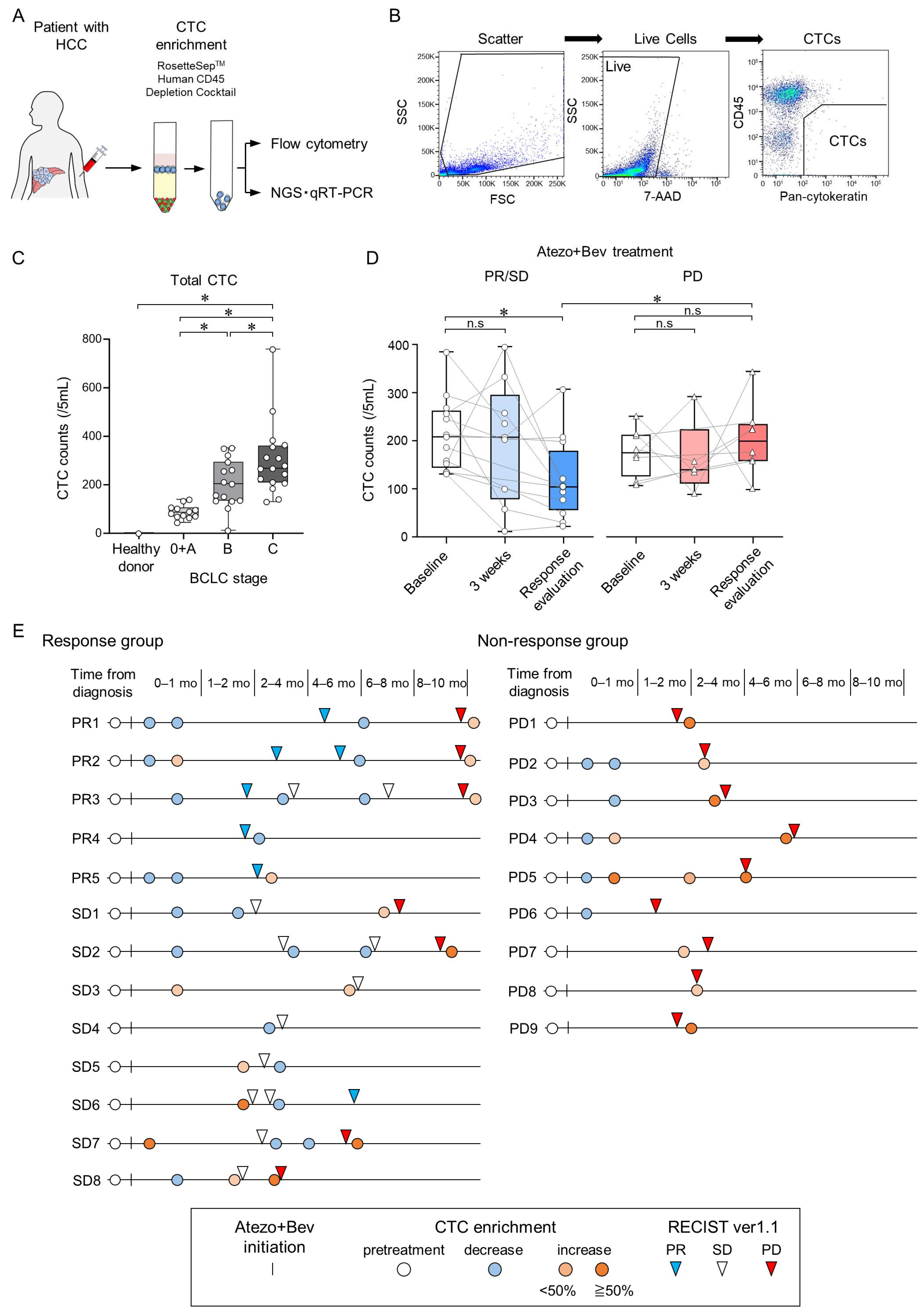
2.5. Enrichment of CTCs and RNA Extraction
2.6. Flow-Cytometric Analysis
2.7. Next Generation Sequencing
2.8. RNA Extraction and Nested PCR
2.9. Statistical Analyses
3. Results
3.1. Changes in CTC Counts during Atezo+Bev Treatment Reflect the Treatment Response in Patients with Unresectable HCC
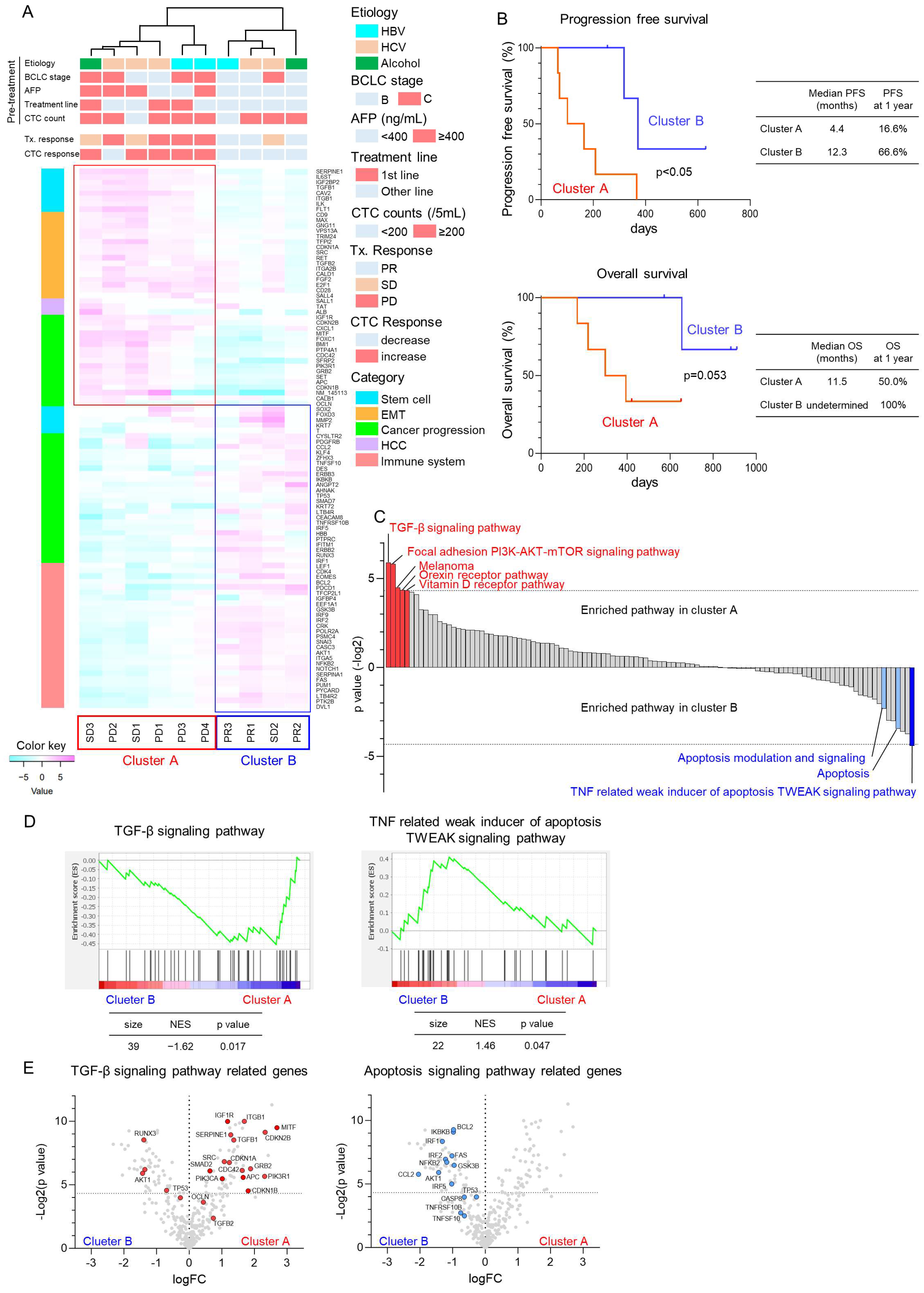
3.2. Molecular Changes of CTCs Using NGS in Patients with HCC Treated with Atezo+Bev
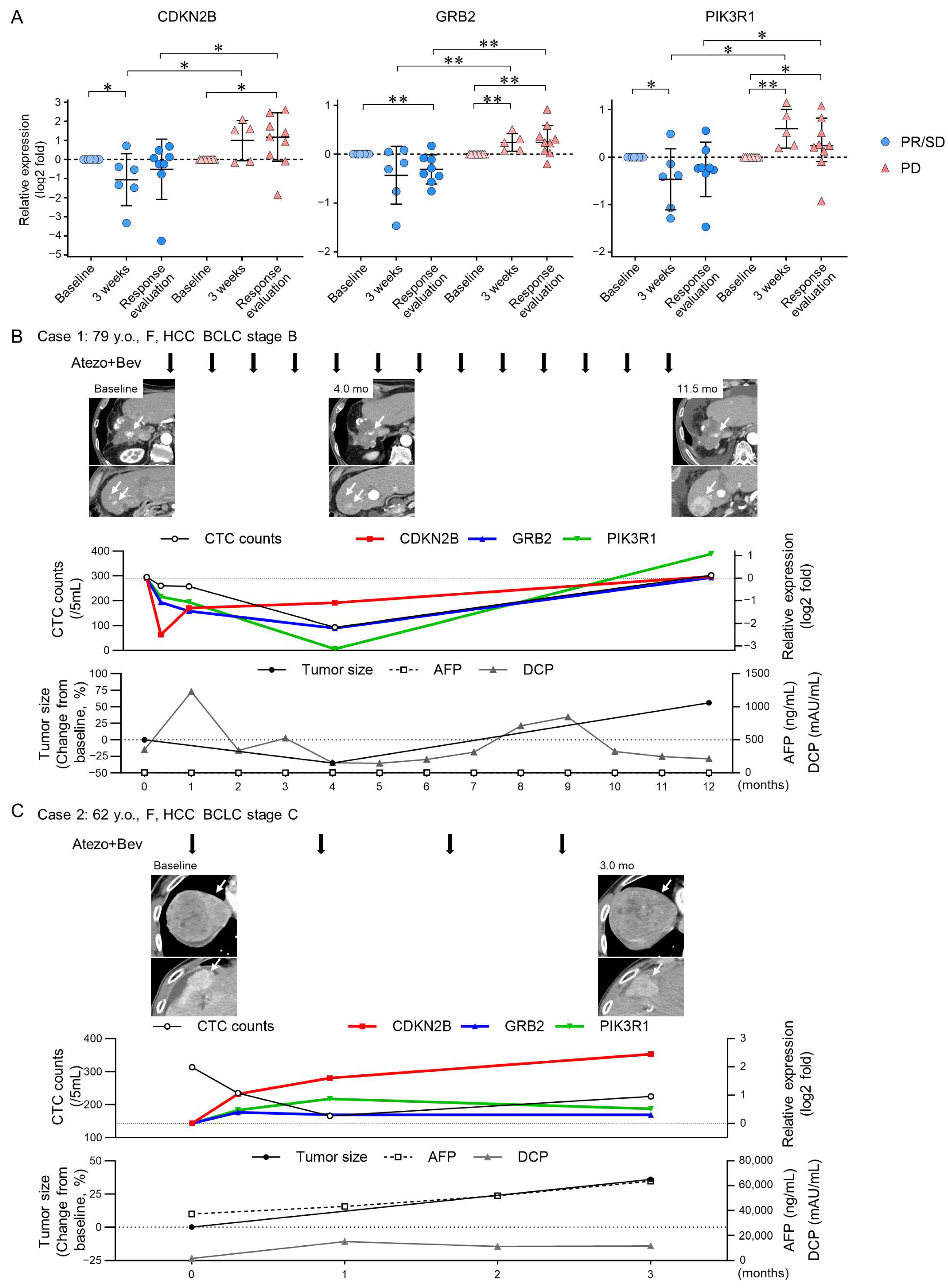
3.3. TGF-β Signaling-Related Gene Expression in CTCs Reflect Early Treatment Response to Atezo+Bev
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Abbas, A.R.; de Galarreta, M.R.; Guan, Y.; Lu, S.; Koeppen, H.; Zhang, W.; Hsu, C.H.; He, A.R.; Ryoo, B.Y.; et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat. Med. 2022, 28, 1599–1611. [Google Scholar] [CrossRef] [PubMed]
- Greten, T.F.; Villanueva, A.; Korangy, F.; Ruf, B.; Yarchoan, M.; Ma, L.; Ma, L.; Ruppin, E.; Wang, X.W. Biomarkers for immunotherapy of hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2023, 20, 780–798. [Google Scholar] [CrossRef]
- Ring, A.; Nguyen-Sträuli, B.D.; Wicki, A.; Aceto, N. Biology, vulnerabilities and clinical applications of circulating tumour cells. Nat. Rev. Cancer 2023, 23, 95–111. [Google Scholar] [CrossRef]
- Bates, M.; Mohamed, B.M.; Ward, M.P.; Kelly, T.E.; O’Connor, R.; Malone, V.; Brooks, R.; Brooks, D.; Selemidis, S.; Martin, C.; et al. Circulating tumour cells: The Good, the Bad and the Ugly. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188863. [Google Scholar] [CrossRef] [PubMed]
- Winograd, P.; Hou, S.; Court, C.M.; Lee, Y.T.; Chen, P.J.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Teng, P.C.; Wang, J.J.; et al. Hepatocellular carcinoma-circulating tumor cells expressing PD-L1 are prognostic and potentially associated with response to checkpoint inhibitors. Hepatol. Commun. 2020, 4, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Ofuji, K.; Hiramatsu, K.; Nosaka, T.; Naito, T.; Matsuda, H.; Endo, K.; Higuchi, M.; Ohtani, M.; Nemoto, T.; et al. Circulating tumor cells detected with a microcavity array predict clinical outcome in hepatocellular carcinoma. Cancer Med. 2021, 10, 2300–2309. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, A.; Philouze, P.; Lauret, A.; Alphonse, G.; Malesys, C.; Ardail, D.; Payen, L.; Céruse, P.; Wozny, A.S.; Rodriguez-Lafrasse, C. Circulating tumor cell detection during neoadjuvant chemotherapy to predict early response in locally advanced oropharyngeal cancers: A Prospective Pilot Study. J. Pers. Med. 2022, 12, 445. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Shimizu, T.; Tsujino, I.; Obana, Y.; Seki, T.; Fuchinoue, F.; Ohni, S.; Oinuma, T.; Kusumi, Y.; Yamada, T.; et al. Semi-nested real-time reverse transcription polymerase chain reaction methods for the successful quantitation of cytokeratin mRNA expression levels for the subtyping of non-small-cell lung carcinoma using paraffin-embedded and microdissected lung biopsy specimens. Acta Histochem. Cytochem. 2013, 46, 85–96. [Google Scholar]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J. Oncol. 2010, 2010, 617421. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Forgues, M.; Wang, W.; Kim, J.W.; Ye, Q.; Jia, H.; Budhu, A.; Zanetti, K.A.; Chen, Y.; Qin, L.X.; et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008, 68, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Teng, P.C.; Chen, P.J.; Posadas, E.; Tseng, H.R.; Lu, S.C.; Yang, J.D. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology 2021, 73, 422–436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef] [PubMed]
- Boral, D.; Vishnoi, M.; Liu, H.N.; Yin, W.; Sprouse, M.L.; Scamardo, A.; Hong, D.S.; Tan, T.Z.; Thiery, J.P.; Chang, J.C.; et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017, 8, 196. [Google Scholar] [CrossRef]
- Schulze, K.; Gasch, C.; Staufer, K.; Nashan, B.; Lohse, A.W.; Pantel, K.; Riethdorf, S.; Wege, H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int. J. Cancer 2013, 133, 2165–2171. [Google Scholar] [CrossRef]
- Sun, Y.F.; Xu, Y.; Yang, X.R.; Guo, W.; Zhang, X.; Qiu, S.J.; Shi, R.Y.; Hu, B.; Zhou, J.; Fan, J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013, 57, 1458–1468. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.T.; Bardia, A.; Spring, L.M.; Giobbie-Hurder, A.; Kalinich, M.; Dubash, T.; Sundaresan, T.; Hong, X.; LiCausi, J.A.; Ho, U.; et al. A digital RNA signature of circulating tumor cells predicting early therapeutic response in localized and metastatic breast cancer. Cancer Discov. 2018, 8, 1286–1299. [Google Scholar] [CrossRef]
- Espejo-Cruz, M.L.; González-Rubio, S.; Zamora-Olaya, J.; Amado-Torres, V.; Alejandre, R.; Sánchez-Frías, M.; Ciria, R.; De la Mata, M.; Rodríguez-Perálvarez, M.; Ferrín, G. Circulating tumor cells in hepatocellular carcinoma: A comprehensive review and critical appraisal. Int. J. Mol. Sci. 2021, 22, 13073. [Google Scholar] [CrossRef]
- Qi, L.N.; Xiang, B.D.; Wu, F.X.; Ye, J.Z.; Zhong, J.H.; Wang, Y.Y.; Chen, Y.Y.; Chen, Z.S.; Ma, L.; Chen, J.; et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 2018, 78, 4731–4744. [Google Scholar] [CrossRef]
- Akhurst, R.J.; Hata, A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 2012, 11, 790–811. [Google Scholar] [CrossRef]
- Gulley, J.L.; Schlom, J.; Barcellos-Hoff, M.H.; Wang, X.J.; Seoane, J.; Audhuy, F.; Lan, Y.; Dussault, I.; Moustakas, A. Dual inhibition of TGF-β and PD-L1: A novel approach to cancer treatment. Mol. Oncol. 2022, 16, 2117–2134. [Google Scholar] [CrossRef]
- Lawrence, R.; Watters, M.; Davies, C.R.; Pantel, K.; Lu, Y.J. Circulating tumour cells for early detection of clinically relevant cancer. Nat. Rev. Clin. Oncol. 2023, 20, 487–500. [Google Scholar] [CrossRef]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Bao, S.; Jiang, X.; Jin, S.; Tu, P.; Lu, J. TGF-β1 induces immune escape by enhancing PD-1 and CTLA-4 expression on T lymphocytes in hepatocellular carcinoma. Front. Oncol. 2021, 11, 694145. [Google Scholar] [CrossRef]
- Devan, A.R.; Pavithran, K.; Nair, B.; Murali, M.; Nath, L.R. Deciphering the role of transforming growth factor-beta 1 as a diagnostic-prognostic-therapeutic candidate against hepatocellular carcinoma. World J. Gastroenterol. 2022, 28, 5250–5264. [Google Scholar] [CrossRef]
- Sia, D.; Jiao, Y.; Martinez-Quetglas, I.; Kuchuk, O.; Villacorta-Martin, C.; Castro de Moura, M.; Putra, J.; Camprecios, G.; Bassaganyas, L.; Akers, N.; et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017, 153, 812–826. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Deng, J.; Zhou, M.; Liao, T.; Kuang, W.; Xia, H.; Yin, Z.; Tan, Q.; Li, Y.; Song, S.; Zhou, E.; et al. Targeting cancer cell ferroptosis to reverse immune checkpoint inhibitor therapy resistance. Front. Cell Dev. Biol. 2022, 10, 818453. [Google Scholar] [CrossRef]
- Sun, Q.; Hong, Z.; Zhang, C.; Wang, L.; Han, Z.; Ma, D. Immune checkpoint therapy for solid tumours: Clinical dilemmas and future trends. Signal Transduct. Target. Ther. 2023, 8, 320. [Google Scholar] [CrossRef] [PubMed]
- Fehm, T.; Becker, S.; Becker-Pergola, G.; Sotlar, K.; Gebauer, G.; Dürr-Störzer, S.; Neubauer, H.; Wallwiener, D.; Solomayer, E.F. Presence of apoptotic and nonapoptotic disseminated tumor cells reflects the response to neoadjuvant systemic therapy in breast cancer. Breast Cancer Res. 2006, 8, R60. [Google Scholar] [CrossRef] [PubMed]
| Target | Application | Target Species | Host Species | Clone | Company | Catalogue No. |
|---|---|---|---|---|---|---|
| CD45 | FCM | Human | Mouse | 2D1 | BioLegend | 368516 |
| pan-Cytokeratin | FCM | Human | Mouse | C-11 | Cayman Chemical | 10478 |
| Gene | Species | Dye | Company | Catalogue No. |
|---|---|---|---|---|
| CDKN2B | Human | FAM | ThermoFisher Scientific | Hs00793225_m1 |
| GRB2 | Human | FAM | ThermoFisher Scientific | Hs00157817_m1 |
| PIK3R1 | Human | FAM | ThermoFisher Scientific | Hs00933163_m1 |
| HPRT1 | Human | FAM | ThermoFisher Scientific | Hs02800695_m1 |
| Characteristics | HCC (n = 44) | Healthy Donors (n = 10) |
|---|---|---|
| Age, median (IQR), years | 75 (69–80) | 71 (56–77) |
| Sex, male/female, n | 31/13 | 7/3 |
| Etiology, HBV/HCV/NBNC, n | 3/15/26 | — |
| PLT, ×109/L, median (IQR) | 151 (119–215) | 252 (121–270) |
| PT, INR, median (IQR) | 1.07 (0.99–1.26) | 0.98 (0.88–1.00) |
| ALB, g/dL, median (IQR) | 3.5 (3.1–3.8) | 4.1 (3.8–4.4) |
| T-bil, g/dL, median (IQR) | 0.8 (0.6–1.2) | 0.9 (0.6–1.1) |
| ALT, IU/L, median (IQR) | 25 (15–34) | 22 (12–22) |
| Child-Pugh class, A/B/C, n | 33/10/1 | |
| AFP, ng/mL, median (IQR) | 12.2 (4.1–117) | — |
| DCP, mAU/mL, median (IQR) | 146 (24–1356) | — |
| Maximum tumor size, cm, median (IQR) | 3.0 (1.6–6.7) | — |
| Number of tumors, 1/2/3+, n | 15/4/25 | — |
| Vascular invasion, absent/present, n | 35/9 | — |
| Extrahepatic metastasis, n | ||
| None | 36 | — |
| Lymph node | 3 | — |
| Bone | 1 | — |
| Lung | 1 | — |
| Lung, Bone | 2 | — |
| Lymph node, Bone, Adrenal gland | 1 | — |
| BCLC stage, 0/A/B/C, n | 6/6/16/16 | — |
| Stem Cell Potency | ||||||||
| BMI1 | COL2A1 | FGF5 | GRIN1 | ISL1 | MYF5 | PDX1 | SEMA3A | TERT |
| CALB1 | COMMD3 | FLT1 | GSX2 | JARID2 | MYOD1 | PECAM1 | SERPINA1 | TFCP2L1 |
| CD34 | CRABP2 | FOXA2 | HAND1 | KIT | NANOG | PODXL | SERPINB3 | THY1 |
| CD9 | CRK | FOXC1 | HBB | KLF4 | NEUROD1 | POU5F1 | SET | TRIM24 |
| CDH5 | CTLA4 | FOXD3 | HBZ | LAMB1 | NEUROG1 | PROM1 | SFRP2 | WT1 |
| CDK2 | CTNNB1 | GABRB3 | HESX1 | LAMC1 | NODAL | PTEN | SKIL | XIST |
| CDK4 | DDX4 | GAL | HGF | LEFTY1 | NOG | PTF1A | SMARCAD1 | ZFHX3 |
| CDKN1A | DES | GATA4 | HNF4A | LEFTY2 | NPPA | RAF1 | SOX17 | ZFP42 |
| CDKN2A | DNMT3B | GATA6 | HOXB1 | LHX5 | NR5A2 | REST | STAT1 | ZIC3 |
| CDKN2B | E2F1 | GBX2 | IAPP | LIFR | NR6A1 | RFX4 | STAT3 | |
| CDX2 | ENO2 | GCM1 | IFITM1 | LIN28A | OLIG2 | RIF1 | SYCP3 | |
| CDYL | EOMES | GDF3 | IFITM2 | MEIS1 | ONECUT1 | RNF112 | SYP | |
| CGB | EPCAM | GFAP | IGF2BP2 | MNX1 | OTX1 | RUNX2 | TAT | |
| COL1A1 | ERBB2 | GJA9 | IL6ST | MUC1 | PAX4 | SALL1 | TCF7L1 | |
| COL1A2 | ESX1 | GRB7 | ILK | MYC | PAX6 | SALL4 | TDGF1 | |
| Epithelial–mesenchymal transition | ||||||||
| AHNAK | COL3A1 | FGFR3 | IL1RN | KRT18 | MMP3 | RELA | SRC | TSPAN13 |
| AKT1 | COL5A2 | FN1 | INS | KRT19 | MMP9 | RGS2 | STEAP1 | TWIST1 |
| AKT2 | CYCS | FOXC2 | IPO8 | KRT4 | MSN | RHOA | TCF3 | VCAN |
| ANGPT2 | DESI1 | FYN | ITGA5 | KRT5 | MST1R | SERPINE1 | TCF4 | VIM |
| AXL | DSC2 | FZD7 | ITGAV | KRT7 | NES | SMAD2 | TFPI2 | VPS13A |
| BMP1 | DSP | GEMIN2 | ITGB1 | KRT72 | NOTCH1 | SMAD4 | TGFA | WNT11 |
| BMP7 | EGFR | GNG11 | JAG1 | KRT8 | NUDT13 | SMAD7 | TGFB1 | WNT5A |
| BRAF | ELK1 | GRB2 | JUN | LAMA1 | OCLN | SNAI1 | TGFB2 | WNT5B |
| CALD1 | ERBB3 | GSC | KRAS | LEF1 | PDGFRB | SNAI2 | TGFB3 | ZEB1 |
| CAMK2N1 | ESR1 | GSK3B | KRT1 | MAP1B | PLEK2 | SNAI3 | TGFBR1 | ZEB2 |
| CAV2 | F11R | HMBS | KRT1 | MAX | PTK2 | SOX10 | TGFBR2 | |
| CDC42 | FADD | IGF1 | KRT10 | MDM2 | PTP4A1 | SOX2 | TIMP1 | |
| CDH1 | FGFR1 | IGF1R | KRT13 | MITF | RAC1 | SPARC | TMEFF1 | |
| CDH2 | FGFR2 | IGFBP4 | KRT14 | MMP2 | RB1 | SPP1 | TMEM132A | |
| HCC progression | ||||||||
| ABCC5 | BRIX1 | CYSLTR1 | FGFBP1 | KDR | MRPL19 | POLR2A | RET | VEGFA |
| ABL1 | CASC3 | CYSLTR2 | FGFR4 | LRRC23 | MUC16 | POP4 | RPL37A | WNT1 |
| ACTC1 | CASP8 | DVL1 | FOS | LTB4R | NFKB1 | PSAT1 | RUNX3 | XIAP |
| AFP | CASP9 | EEF1A1 | FZD1 | LTB4R2 | NFKB2 | PSMC4 | SHC1 | YAP1 |
| ALB | CCND1 | EEF1A2 | GADD45A | MAP2K1 | NFKBIA | PTGS1 | SOCS3 | |
| ALOX5 | CCND2 | EIF2B1 | GPC3 | MAP3K5 | NRAS | PTGS2 | SOS1 | |
| APC | CCND3 | ELF1 | HRAS | MAPK1 | OAS2 | PTK2B | T | |
| ASGR1 | CDKN1B | FGF19 | IFI44L | MAPK14 | PDCD1 | PUM1 | TBXT | |
| BAX | CEACAM5 | FGF2 | ITGA2B | MAPK3 | PES1 | PYCARD | TNFRSF10B | |
| BCAR1 | CEACAM8 | FGF23 | ITGB3 | MAPK8 | PIK3CA | RASSF1 | TNFSF10 | |
| BID | CELA1 | FGF4 | KAT6A | MET | PIK3R1 | RELN | TP53 | |
| Cancer immunotherapy | ||||||||
| APOBEC3G | CCL2 | CCNE1 | CD8A | CXCL9 | IRF1 | IRF5 | PTPRC | |
| BCL2 | CCL3 | CD274 | CHUK | FAS | IRF2 | IRF6 | TLR4 | |
| BCL2L1 | CCL4 | CD28 | CX3CL1 | FASLG | IRF3 | IRF7 | ||
| BCL2L11 | CCL5 | CD4 | CXCL1 | IKBKB | IRF4 | IRF9 | ||
| APOBEC3G | CCL2 | CCNE1 | CD8A | CXCL9 | IRF1 | IRF5 | PTPRC | |
| Characteristics | Atezolizumab + Bevacizumab (n = 22) |
|---|---|
| Age, median (IQR), years | 69 (62–79) |
| Sex, male/female, n | 17/5 |
| Etiology, HBV/HCV/NBNC, n | 3/8/11 |
| PLT, ×109/L, median (IQR) | 154 (110–204) |
| PT, INR, median (IQR) | 1.13 (0.98–1.24) |
| ALB, g/dL, median (IQR) | 3.5 (3.0–3.9) |
| T-bil, g/dL, median (IQR) | 1.0 (0.6–1.4) |
| ALT, IU/L, median (IQR) | 26 (16–38) |
| Child–Pugh class, A/B, n | 17/5 |
| AFP, ng/mL, median (IQR) | 16.5 (4.4–404) |
| DCP, mAU/mL, median (IQR) | 261 (24–1452) |
| Maximum tumor size, cm, median (IQR) | 3.8 (2.0–7.2) |
| Number of tumors, 1/2/3+, n | 4/1/17 |
| Vascular invasion, absent/present, n | 15/7 |
| Extrahepatic metastasis, n | |
| None | 16 |
| Lymph node | 2 |
| Bone | 1 |
| Lung | 1 |
| Lymph node, Lung | 1 |
| Lymph node, Bone, Adrenal gland | 1 |
| BCLC stage, A/B/C, n | 1/10/11 |
| Prior systemic therapy, n | |
| None | 13 |
| Sorafenib | 1 |
| Lenvatinib | 6 |
| HAIC | 1 |
| Lenvatinib, HAIC | 1 |
| Observation period, median, days | 305 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murata, Y.; Nosaka, T.; Akazawa, Y.; Tanaka, T.; Takahashi, K.; Naito, T.; Matsuda, H.; Ohtani, M.; Nakamoto, Y. Serial Changes of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Treated with Atezolizumab Plus Bevacizumab. Cancers 2024, 16, 2410. https://doi.org/10.3390/cancers16132410
Murata Y, Nosaka T, Akazawa Y, Tanaka T, Takahashi K, Naito T, Matsuda H, Ohtani M, Nakamoto Y. Serial Changes of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Treated with Atezolizumab Plus Bevacizumab. Cancers. 2024; 16(13):2410. https://doi.org/10.3390/cancers16132410
Chicago/Turabian StyleMurata, Yosuke, Takuto Nosaka, Yu Akazawa, Tomoko Tanaka, Kazuto Takahashi, Tatsushi Naito, Hidetaka Matsuda, Masahiro Ohtani, and Yasunari Nakamoto. 2024. "Serial Changes of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Treated with Atezolizumab Plus Bevacizumab" Cancers 16, no. 13: 2410. https://doi.org/10.3390/cancers16132410
APA StyleMurata, Y., Nosaka, T., Akazawa, Y., Tanaka, T., Takahashi, K., Naito, T., Matsuda, H., Ohtani, M., & Nakamoto, Y. (2024). Serial Changes of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Treated with Atezolizumab Plus Bevacizumab. Cancers, 16(13), 2410. https://doi.org/10.3390/cancers16132410






