Canine Cancer Diagnostics by X-ray Diffraction of Claws
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
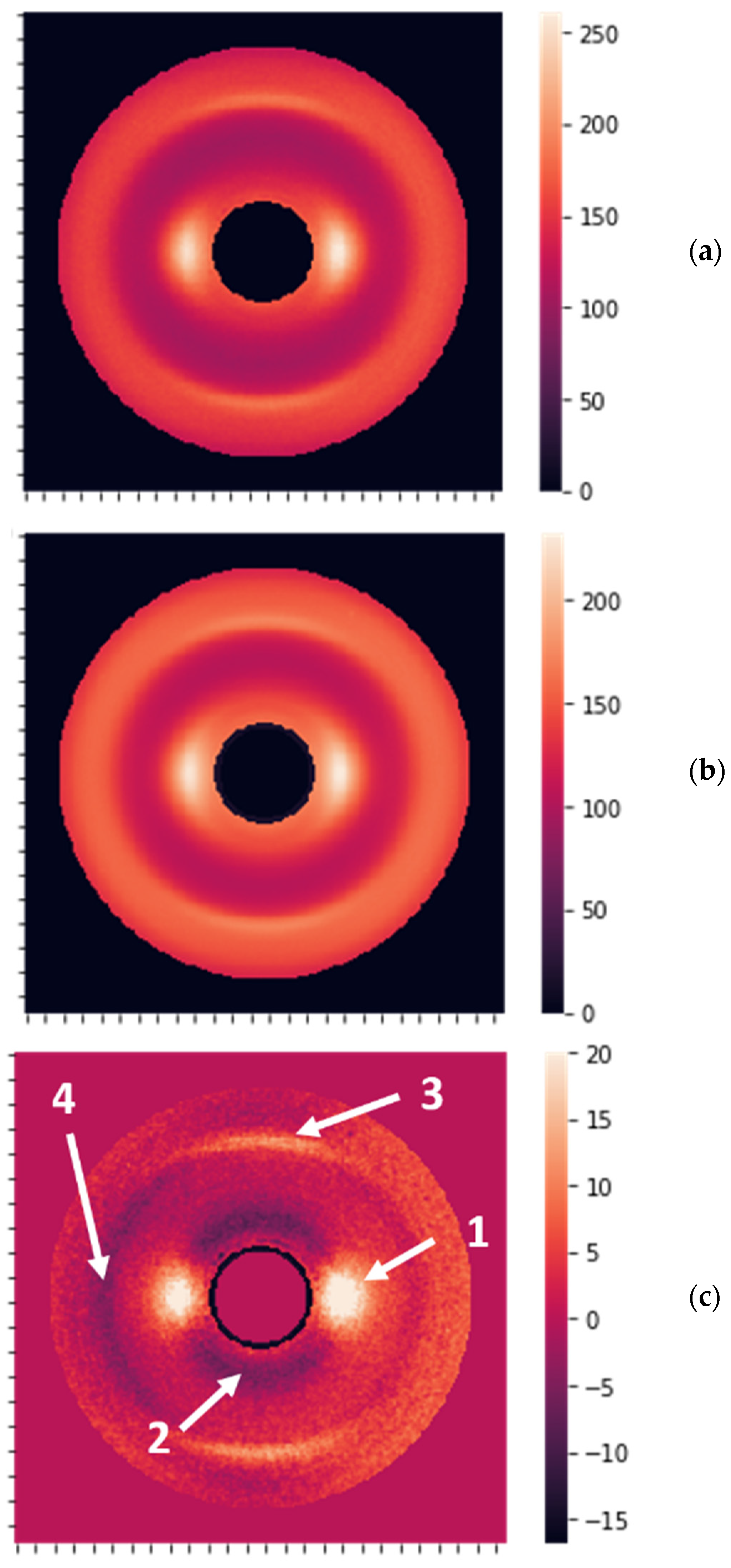
2.1. Sample Preparation and XRD Measurements
2.2. Data Preprocessing
- Calibration of raw data using silver behenate (AgBH): The sample-to-detector distance may vary from batch to batch and, thus, generally requires calibration to unify scale. The image was rescaled during calibration to adjust the q-range to the same predefined standard value. The data used in the current study had a calibration spread within 3%, and calibration of these particular data was not performed.
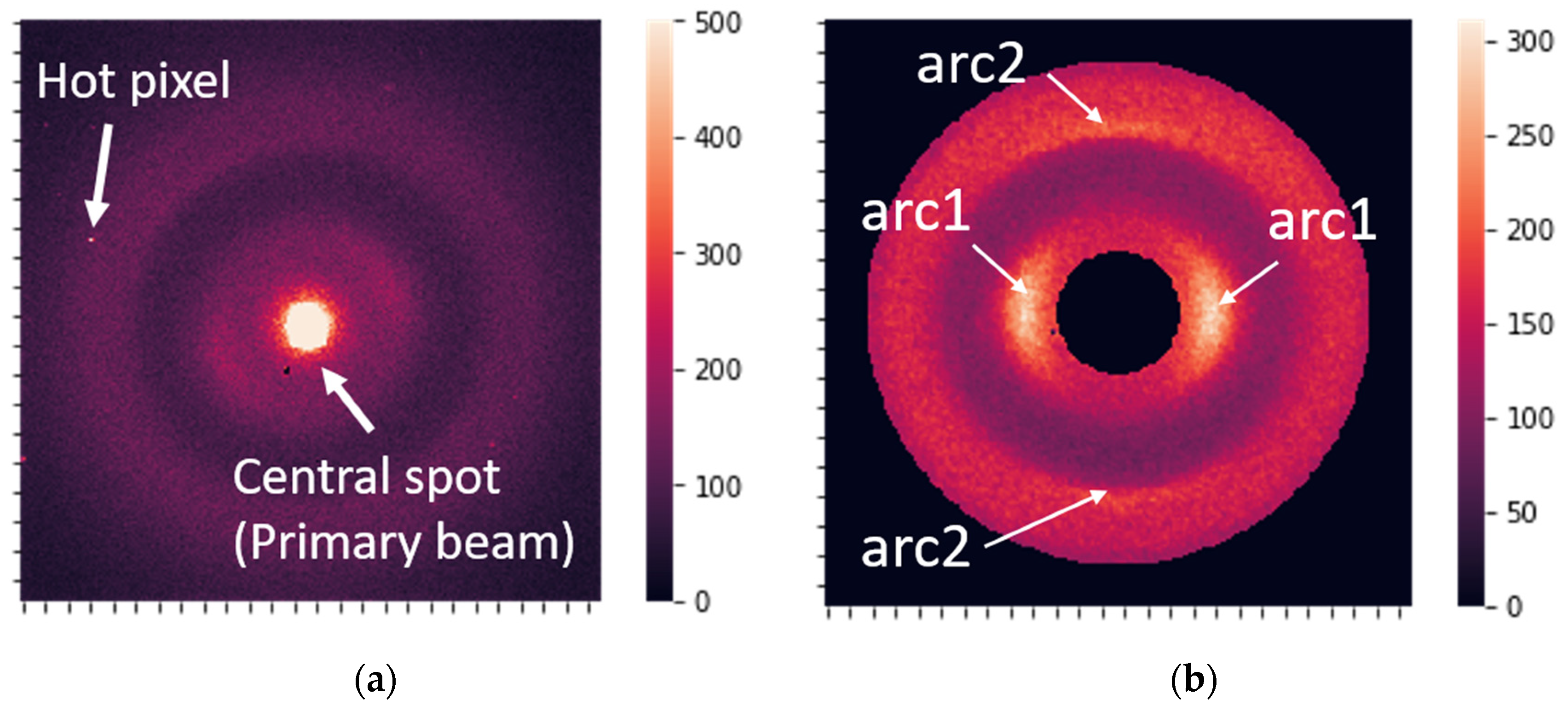
- Centering, cropping the central spot, and rotation of the images (CR preprocessing). The data were also cropped to a circular shape to make them symmetric.
- Hot-spot and hot-pixel removal: Detecting hot pixels and substituting them with the average intensity value over the circle with the corresponding radius.
- Standardizing the diffracted beam’s total intensity, i.e., the integral signal of CR-preprocessed images. The total intensity of the preprocessed images was adjusted to 5 mln counts. Typically, integral intensity is in the range of 2–10 mln counts for unnormalized images.
2.3. Extraction of Features
2.4. Data Analysis
- Using mean Fourier coefficients per patient. In this case, we reduced data for training and, as a result, lost information. The results obtained for mean coefficients were not stable.
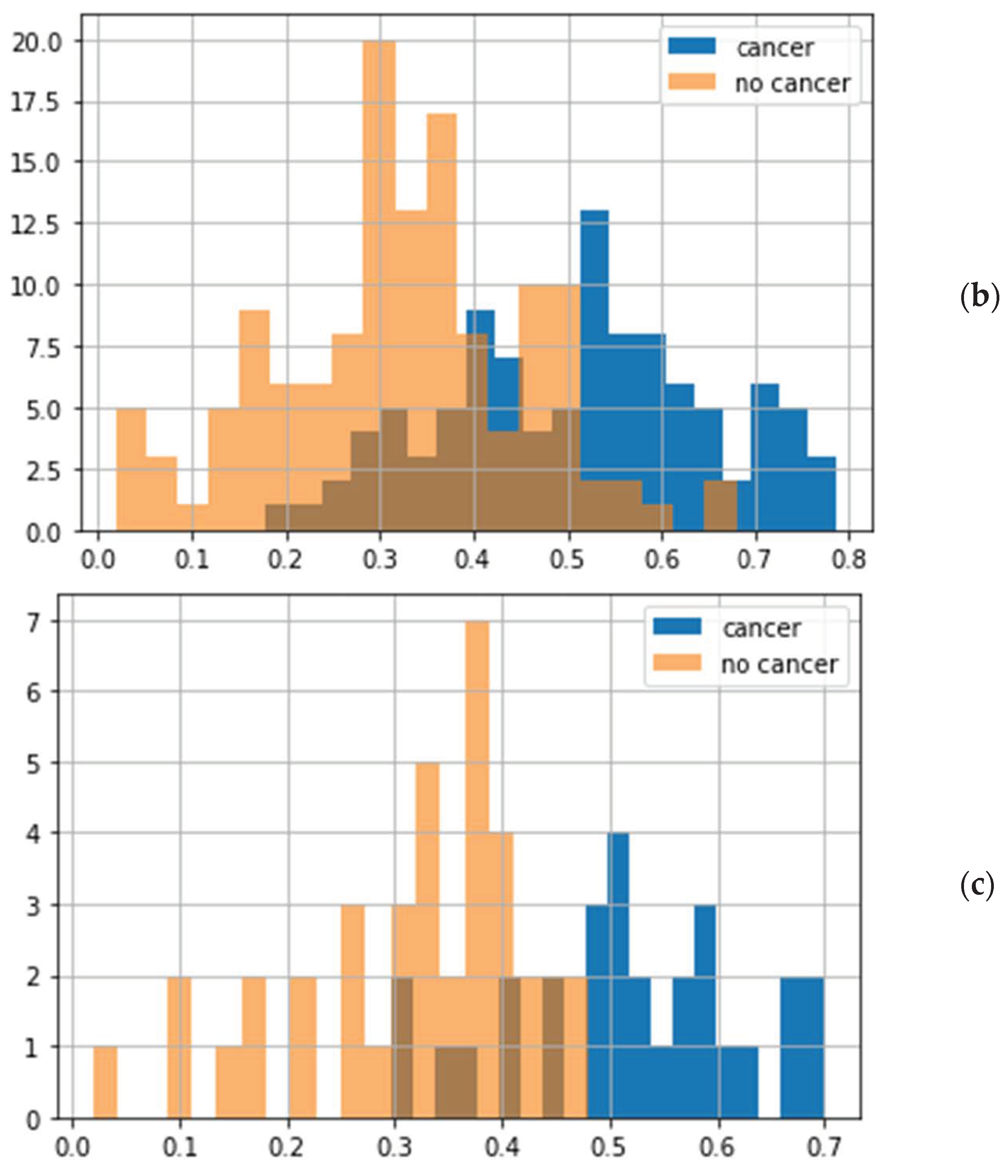
- At first, samples were classified by a supervised model, i.e., diagnosis of “cancer”/“no cancer” was predicted, and then the transfer from samples to patients was performed by using the rule “if N samples per patient are classified as cancerous, then the patient has cancer”. N can vary between 1 and 4 for patients with 4 samples. The best results were obtained for models with N = 2 or 3. The disadvantage of this method is that the same number of samples per patient is required, which was not the case due to the rejection of some images after quality control.
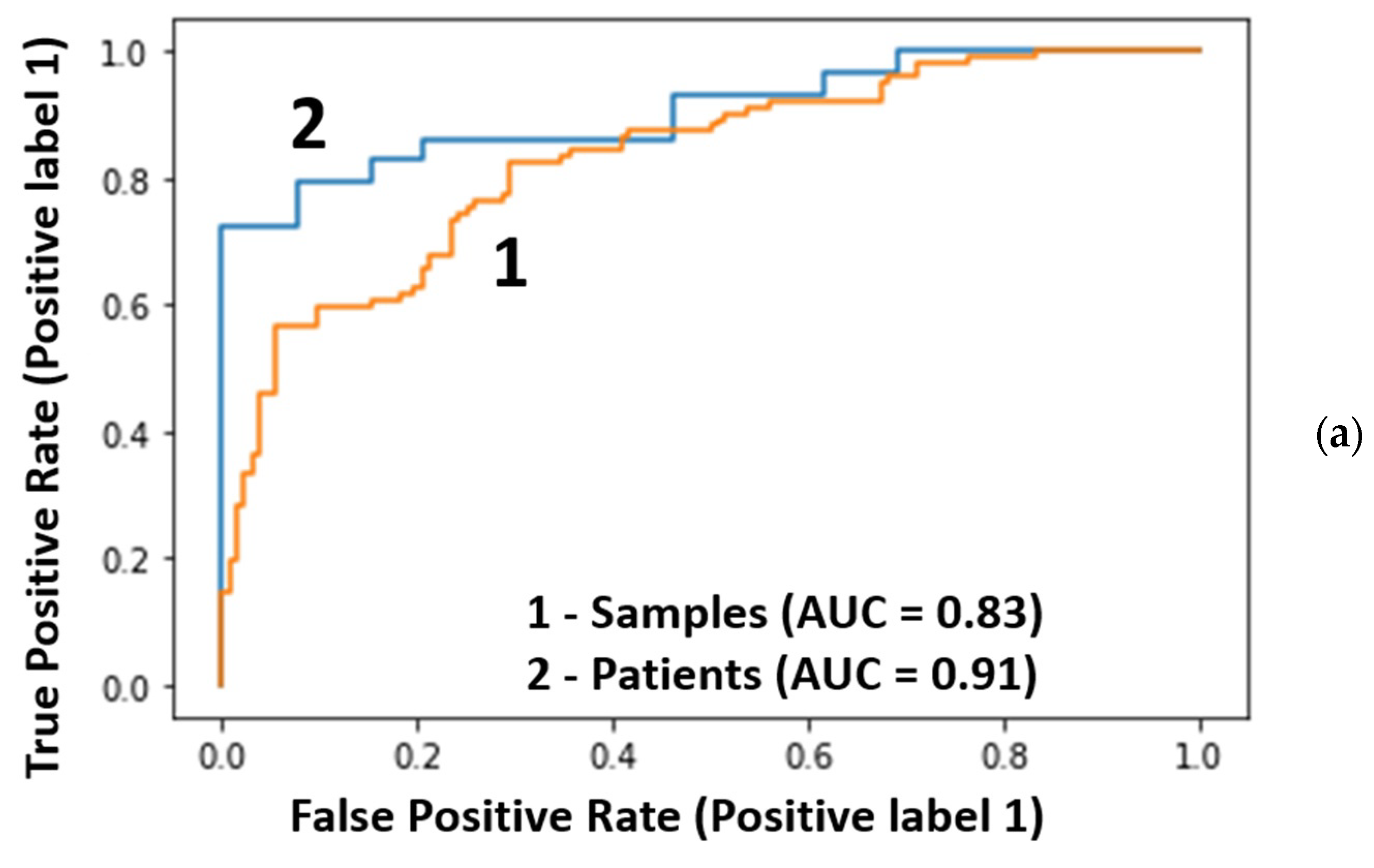
- Transfer from samples to patients by averaging predicted cancer probabilities. This means that for each sample from the testing group, the cancer probability was calculated using a supervised model (random forest classifier) optimized by training samples. Then, the final classification was performed based on the probabilities averaged for each patient. A different number of samples per patient is not a problem for this model.Method 3 was the most successful and provided the most reliable metrics.
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gardner, H.L.; Fenger, J.M.; London, C.A. Dogs as a Model for Cancer. Annu. Rev. Anim. Biosci. 2016, 4, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Thamm, D.H. Canine Cancer: Strategies in Experimental Therapeutics. Front. Oncol. 2019, 9, 1257. [Google Scholar] [CrossRef] [PubMed]
- Kehl, A.; Aupperle-Lellbach, H.; de Brot, S.; van der Weyden, L. Review of Molecular Technologies for Investigating Canine Cancer. Animals 2024, 14, 769. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Cho, J.-Y. Comparative oncology: Overcoming human cancer through companion animal studies. Exp. Mol. Med. 2023, 55, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.futuremarketinsights.com/reports/cancer-diagnostics-market (accessed on 20 February 2024).
- Pulumati, A.; Pulumati, A.; Dwarakanath, B.S.; Verma, A.; Papineni, R.V.L. Technological advancements in cancer diagnostics: Improvements and limitations. Cancer Rep. 2023, 6, e1764. [Google Scholar] [CrossRef] [PubMed]
- Goyal, L.; Hingmire, S.; Parikh, P.M. Newer Diagnostic Methods in Oncology. Med. J. Arm. Forces India 2006, 62, 162–168. [Google Scholar] [CrossRef]
- Hemminki, K.; Liu, H.; Heminki, A.; Sundquist, J. Power and limits of modern cancer diagnostics: Cancer of unknown primary. Ann. Oncol. 2012, 23, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Nover, A.B.; Jagtap, S.; Anjum, W.; Yegingil, H.; Shih, W.Y.; Shih, W.-H.; Brooks, A.D. Modern Breast Cancer Detection: A Technological Review. Int. J. Biomed. Imaging 2009, 2009, 902326. [Google Scholar] [CrossRef] [PubMed]
- Evers, D.; Hendriks, B.; Lucassen, G.; Ruers, T. Optical spectroscopy: Current advances and future applications in cancer diagnostics and therapy. Future Oncol. 2012, 8, 307–320. [Google Scholar] [CrossRef]
- Kim, J.A.; Wales, D.J.; Yang, G.-Z. Optical spectroscopy for in vivo medical diagnosis—A review of the state of the art and future perspectives. Prog. Biomed. Eng. 2020, 2, 042001. [Google Scholar] [CrossRef]
- Shin, H.; Choi, B.H.; Shim, O.; Kim, J.; Park, Y.; Cho, S.K.; Kim, H.K.; Choi, Y. Single test-based diagnosis of multiple cancer types using Exosome-SERS-AI for early stage cancers. Nat. Commun. 2023, 14, 1644. [Google Scholar] [CrossRef] [PubMed]
- Eyden, B. Electron microscopy in the diagnosis of tumours. Curr. Diag. Pathol. 2002, 8, 216–224. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Gao, X.; Chen, Y.; Liu, T. Nanotechnology in cancer diagnosis: Progress, challenges and opportunities. J. Hematol. Oncol. 2019, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Crouse, D.T. X-ray Diffraction and the Discovery of the Structure of DNA. A Tutorial and Historical Account of James Watson and Francis Crick’s Use of X-ray Diffraction in Their Discovery of the Double Helix Structure of DNA. J. Chem. Educ. 2007, 84, 803. [Google Scholar] [CrossRef]
- Mayer, C. X-ray Diffraction in Biology: How Can We See DNA and Proteins in Three Dimensions? In X-ray Scattering; Ares, A.E., Ed.; InTech: Nappanee, IN, USA, 2017. [Google Scholar] [CrossRef]
- Gawas, U.B.; Mandrekar, V.K.; Majik, M.S. Structural analysis of proteins using X-ray diffraction technique. In Advances in Biological Science Research; Meena, S.N., Naik, M.M., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 69–84. [Google Scholar] [CrossRef]
- Macarthur, I. Structure of α-KERATIN. Nature 1943, 152, 38–41. [Google Scholar] [CrossRef]
- Fraser, R.D.B.; MacRae, T.P.; Miller, A. The coiled-coil model of α-keratin structure. J. Mol. Biol. 1964, 10, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, W.; McKittrick, J.; Meyers, M.A. Keratin: Structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog. Mat. Sci. 2016, 76, 229–318. [Google Scholar] [CrossRef]
- Kidane, G.; Speller, R.D.; Royle, G.J.; Hanby, A.M. X-ray scatter signatures for normal and neoplastic breast tissues. Phys. Med. Biol. 1999, 44, 1791. [Google Scholar] [CrossRef]
- Poletti, M.E.; Gonçalves, O.D.; Mazzaro, I. Coherent and incoherent scattering of 17.44 and 6.93 keV X-ray photons scattered from biological and biological-equivalent samples: Characterization of tissues. X-ray Spectrom. 2002, 31, 57–61. [Google Scholar] [CrossRef]
- Cunha, D.M.; Oliveira, O.R.; Pérez, C.A.; Poletti, M.E. X-ray scattering profiles of some normal and malignant human breast tissues. X-ray Spectrom. 2006, 35, 370–374. [Google Scholar] [CrossRef]
- Moss, R.M.; Amin, A.S.; Crews, C.; Purdie, C.A.; Jordan, L.B.; Iacoviello, F.; Evans, A.; Speller, R.D.; Vinnicombe, S.J. Correlation of X-ray diffraction signatures of breast tissue and their histopathological classification. Sci. Rep. 2017, 7, 12998. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Blinchevsky, B.; Slight, M.; Tanaka, A.; Lazarev, A.; Zhang, W.; Aram, B.; Ghadimi, M.; Lomis, T.; Murokh, L.; et al. Structural Biomarkers for Breast Cancer Determined by X-ray Diffraction. In Quantum Effects and Measurement Techniques in Biology and Biophotonics; Aiello, C., Polyakov, S.V., Derr, P., Eds.; SPIE: California, CA, USA, 2024; Volume 12863, p. 1286302. [Google Scholar]
- James, V.; Kearsley, J.; Irving, T.; Amemiya, Y.; Cookson, D. Using hair to screen for breast cancer. Nature 1999, 398, 33–34. [Google Scholar] [CrossRef]
- James, V.J. Synchrotron fibre diffraction identifies and locates foetal collagenous breast tissue associated with breast carcinoma. J. Synchrotron Rad. 2002, 9, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Briki, F.; Busson, B.; Salicru, B.; Estève, F.; Doucet, J. Breast-cancer diagnosis using hair. Nature 1999, 400, 226. [Google Scholar] [CrossRef] [PubMed]
- Suortti, P.; Fernandez, M.; Urban, V. Comments on Synchrotron fibre diffraction identifies and locates foetal collagenous breast tissue associated with breast carcinoma by V. J. James (2002). J. Synchrotron Rad. 9, 71–76. J. Synchrotron Rad. 2003, 10, 198. [Google Scholar] [CrossRef]
- Iniewski, K. (Ed.) Advanced X-ray Detector Technologies, Design and Applications; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Swanson, K.; Wu, E.; Zhang, A.; Alizadeh, A.A.; Zou, J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 2023, 186, 1772–1791. [Google Scholar] [CrossRef] [PubMed]
- Pacurari, A.C.; Bhattarai, S.; Muhammad, A.; Avram, C.; Mederle, A.O.; Rosca, O.; Bratosin, F.; Bogdan, I.; Fericean, R.M.; Biris, M.; et al. Diagnostic Accuracy of Machine Learning AI Architectures in Detection and Classification of Lung Cancer: A Systematic Review. Diagnostics 2023, 13, 2145. [Google Scholar] [CrossRef] [PubMed]
- Rai, H.M.; Yoo, J. A comprehensive analysis of recent advancements in cancer detection using machine learning and deep learning models for improved diagnostics. J. Cancer Res. Clin. Oncol. 2023, 149, 14365–14408. [Google Scholar] [CrossRef] [PubMed]
- Warren, B.E.; Averbach, B.L. The Effect of Cold-Work Distortion on X-ray Patterns. J. Appl. Phys. 1950, 21, 595–599. [Google Scholar] [CrossRef]
- Benedetti, A.; Fagherazzi, G.; Enzo, S.; Battagliarin, M. A profile-fitting procedure for analysis of broadened X-ray diffraction peaks. II. Application and discussion of the methodology. J. Appl. Cryst. 1988, 21, 543–549. [Google Scholar] [CrossRef]
- Yao, W.; Weng, Y.; Catchmark, J.M. Improved cellulose X-ray diffraction analysis using Fourier series modeling. Cellulose 2020, 27, 5563–5579. [Google Scholar] [CrossRef]
- Montoya-Escobar, N.; Ospina-Acero, D.; Velásquez-Cock, J.A.; Gómez-Hoyos, C.; Serpa Guerra, A.; Gañan Rojo, P.F.; Vélez Acosta, L.M.; Escobar, J.P.; Correa-Hincapié, N.; Triana-Chávez, O.; et al. Use of Fourier Series in X-ray Diffraction (XRD) Analysis and Fourier-Transform Infrared Spectroscopy (FTIR) for Estimation of Crystallinity in Cellulose from Different Sources. Polymers 2022, 14, 5199. [Google Scholar] [CrossRef] [PubMed]
- Flory, A.; Kruglyak, K.M.; Tynan, J.A.; McLennan, L.M.; Rafalko, J.M.; Fiaux, P.C.; Hernandez, G.E.; Marass, F.; Nakashe, P.; Ruiz-Perez, C.A.; et al. Clinical validation of a next-generation sequencing-based multi-cancer early detection “liquid biopsy” blood test in over 1,000 dogs using an independent testing set: The CANcer Detection in Dogs (CANDiD) study. PLoS ONE 2022, 17, e0266623. [Google Scholar] [CrossRef]
- Flory, A.; McLennan, L.; Peet, B.; Kroll, M.; Stuart, D.; Brown, D.; Stuebner, K.; Phillips, B.; Coomber, B.L.; Woods, J.P.; et al. Cancer detection in clinical practice and using blood-based liquid biopsy: A retrospective audit of over 350 dogs. J. Vet. Int. Med. 2023, 37, 258–267. [Google Scholar] [CrossRef] [PubMed]



| 40 Radial Features | 40 Circular Features | 40 Radial and 40 Circular Features | |
|---|---|---|---|
| Threshold | 0.48 | 0.46 | 0.46 |
| Specificity, % | 97.4 | 92.3 | 94.9 |
| Sensitivity, % | 72.4 | 44.8 | 58.6 |
| Balanced accuracy, % | 84.9 | 68.6 | 76.7 |
| ROC-AUC (samples/patients) | 0.83/0.91 | 0.69/0.77 | 0.80/0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alekseev, A.; Yuk, D.; Lazarev, A.; Labelle, D.; Mourokh, L.; Lazarev, P. Canine Cancer Diagnostics by X-ray Diffraction of Claws. Cancers 2024, 16, 2422. https://doi.org/10.3390/cancers16132422
Alekseev A, Yuk D, Lazarev A, Labelle D, Mourokh L, Lazarev P. Canine Cancer Diagnostics by X-ray Diffraction of Claws. Cancers. 2024; 16(13):2422. https://doi.org/10.3390/cancers16132422
Chicago/Turabian StyleAlekseev, Alexander, Delvin Yuk, Alexander Lazarev, Daizie Labelle, Lev Mourokh, and Pavel Lazarev. 2024. "Canine Cancer Diagnostics by X-ray Diffraction of Claws" Cancers 16, no. 13: 2422. https://doi.org/10.3390/cancers16132422
APA StyleAlekseev, A., Yuk, D., Lazarev, A., Labelle, D., Mourokh, L., & Lazarev, P. (2024). Canine Cancer Diagnostics by X-ray Diffraction of Claws. Cancers, 16(13), 2422. https://doi.org/10.3390/cancers16132422





