The Role of AKR1B10 in Lung Cancer Malignancy Induced by Sublethal Doses of Chemotherapeutic Drugs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Transfection and Chemical Reagents
2.2. Cell Viability Assay and Clonogenic Survival Assay
2.3. Cell Migration Assay
2.4. Matrigel Invasion Assay
2.5. Microarray Analysis
2.6. AKR1B10 Plasmid Construction
2.7. AKR1B10 Activity Assay
2.8. Data Mining of Clinical Information on AKR1B10
2.9. Western Blot Analysis
2.10. In Vivo Metastases Assay
2.11. Statistical Analysis
3. Results
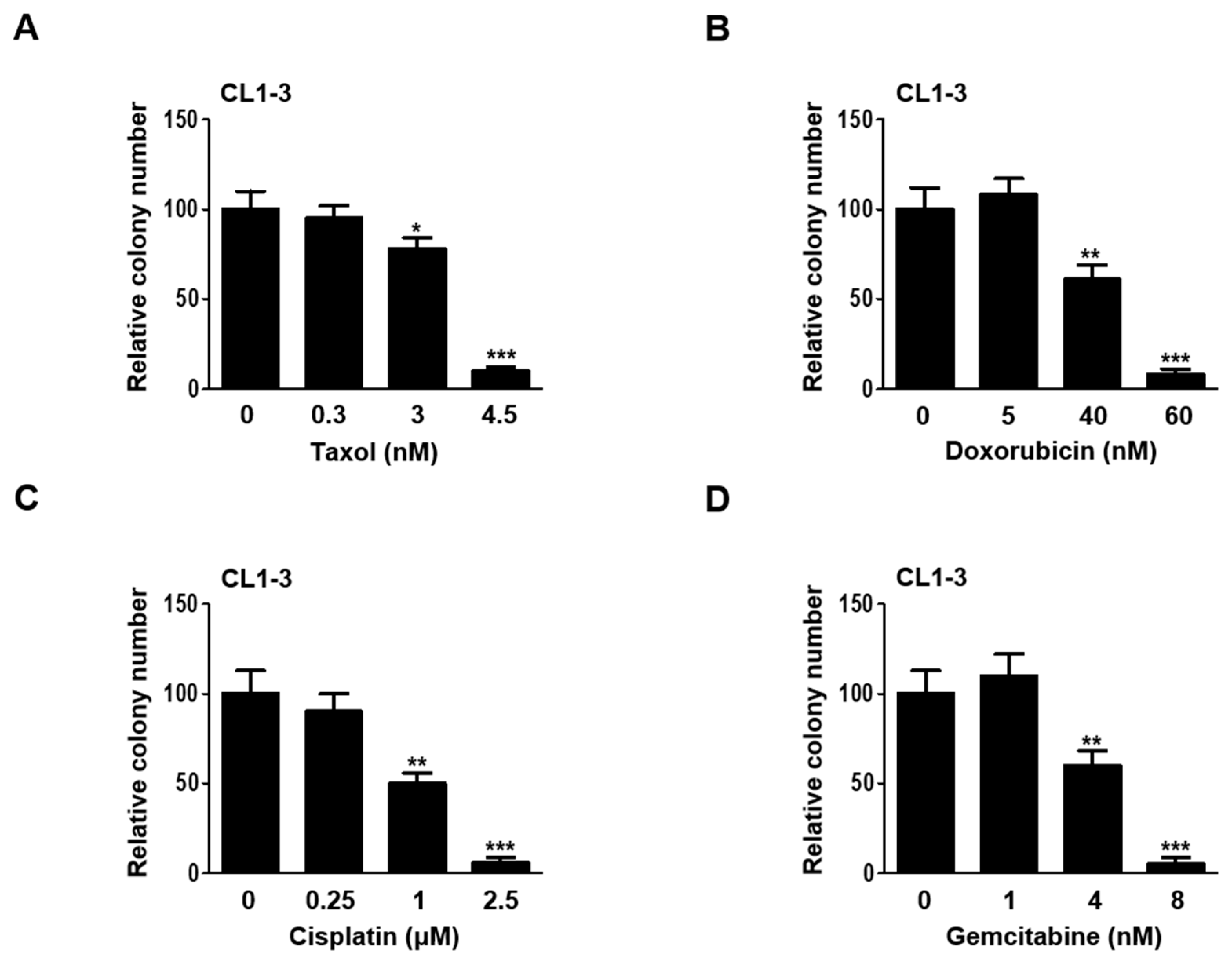
3.1. Establishing Sublethal Doses of Chemotherapy Drugs for Lung Adenocarcinoma Cells
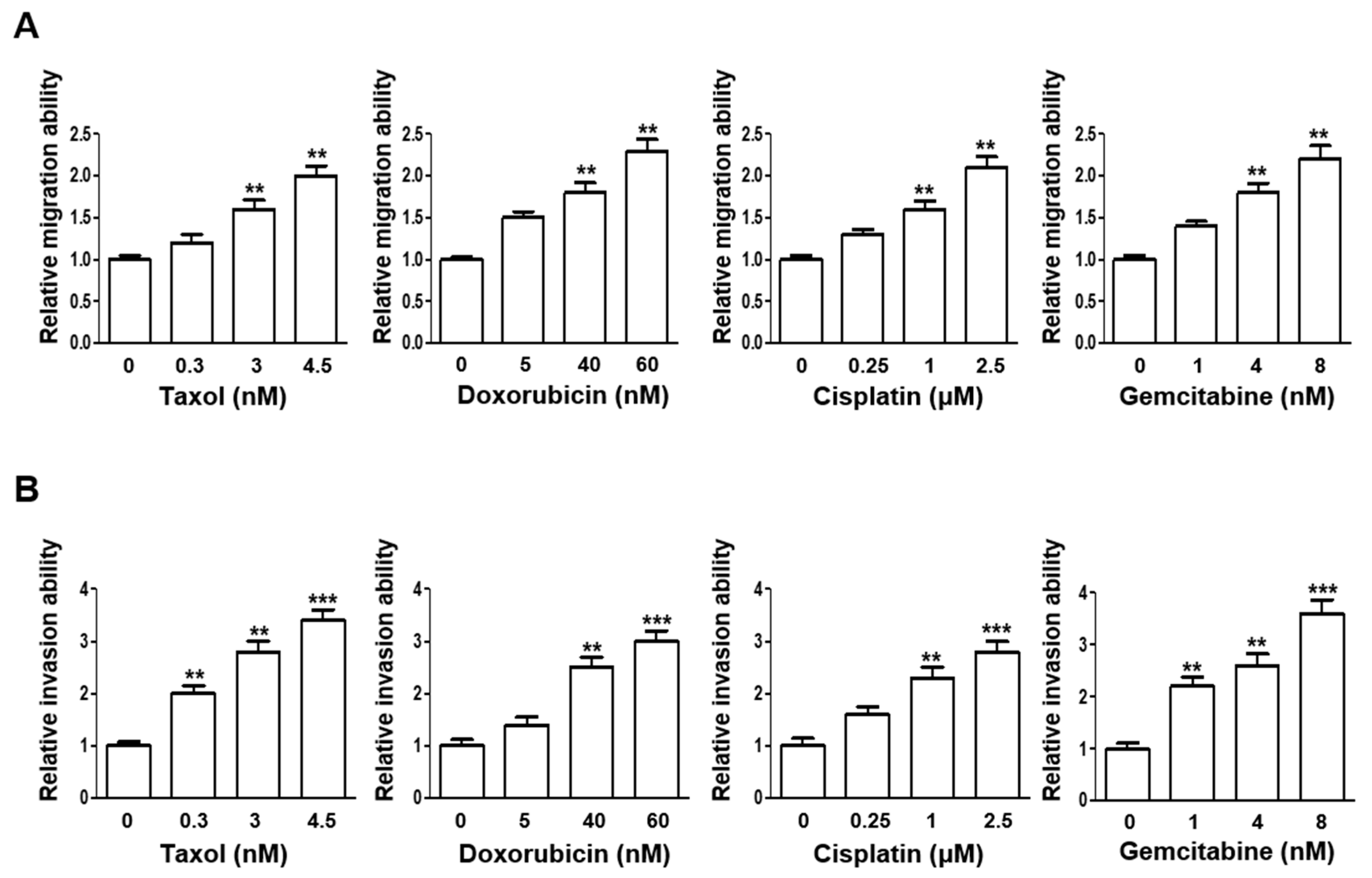
3.2. Escalating Sublethal Doses of Chemotherapeutics Elevates Migration and Invasion Ability of Lung Adenocarcinoma Cells
3.3. NSCLC In Vivo Lung Metastasis Is Augmented upon Sublethal Dose Treatments
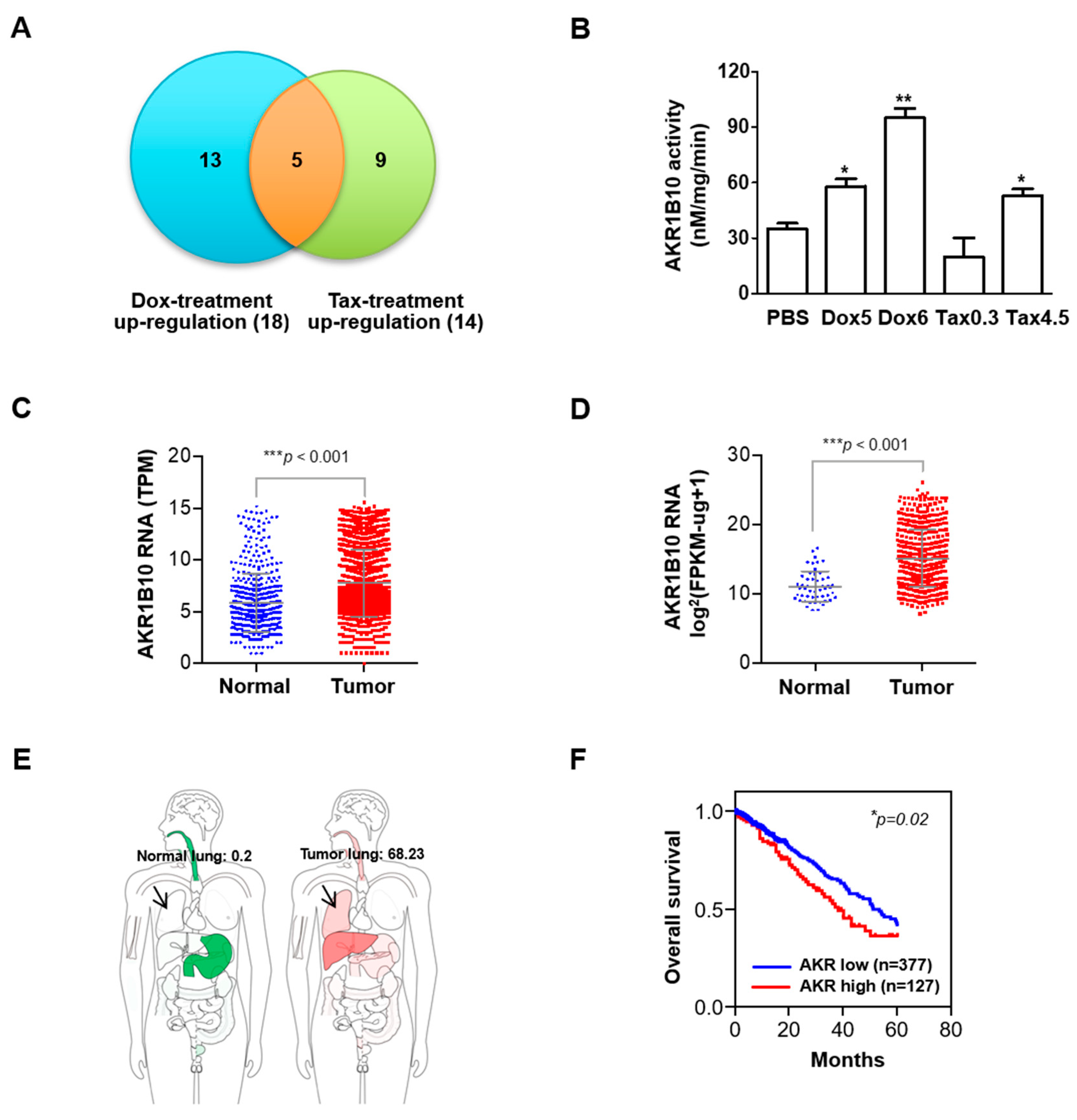
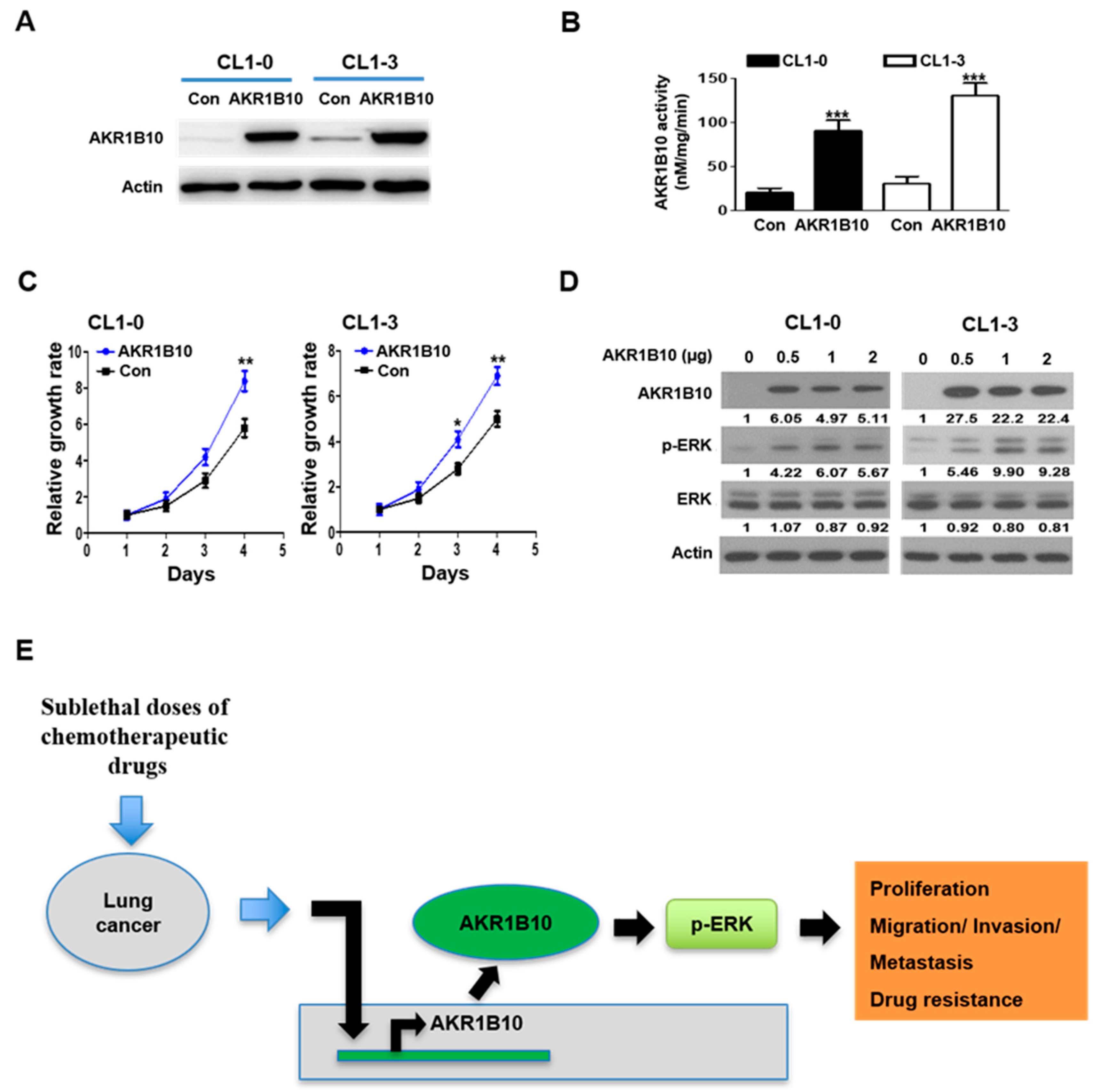
3.4. Expression and Activity of AKR1B10 Is Upregulated in Response to Sublethal Chemotherapeutic Regimens
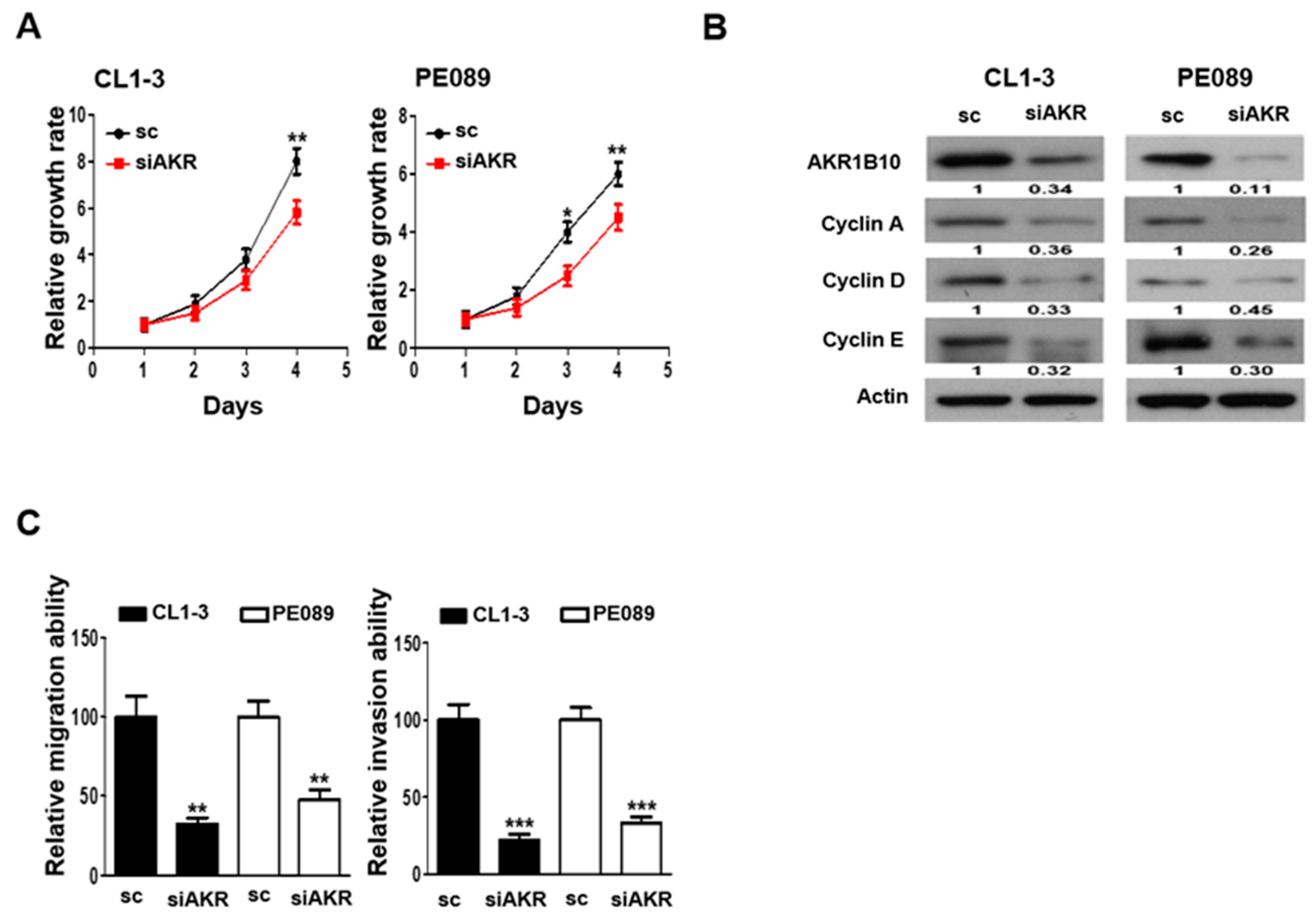
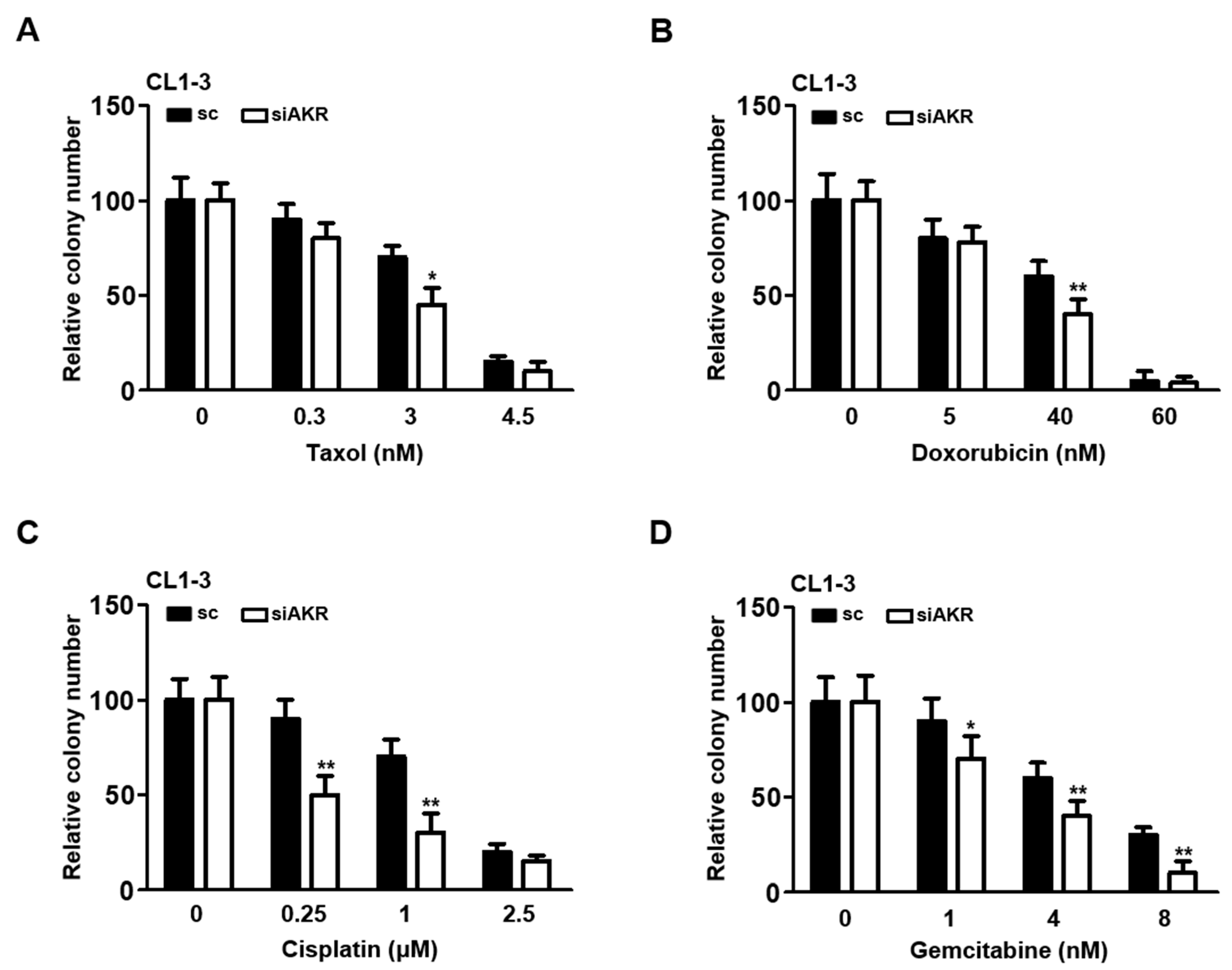
3.5. AKR1B10 Silencing Reduces Cell Proliferation, Migration, and Invasion in NSCLC
3.6. Silencing AKR1B10 Greatly Ameliorates Cellular Sensitivity to Chemotherapeutics
3.7. AKR1B10 Expression Modulates ERK Activation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef]
- Lungcancer-Non-Smallcell: Statistics. Available online: https://www.cancer.net/cancer-types/lung-cancer-non-small-cell/statistics (accessed on 12 December 2023).
- Yang, C.-Y.; Yang, J.C.-H.; Yang, P.-C. Precision Management of Advanced Non–Small Cell Lung Cancer. Annu. Rev. Med. 2020, 71, 117–136. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef] [PubMed]
- Min, H.Y.; Lee, H.Y. Mechanisms of resistance to chemotherapy in non-small cell lung cancer. Arch. Pharm. Res. 2021, 44, 146–164. [Google Scholar] [CrossRef]
- Chatterjee, N.; Bivona, T.G. Polytherapy and Targeted Cancer Drug Resistance. Trends Cancer 2019, 5, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Galván, S.; Carnero, A. Leveraging Genomics, Transcriptomics, and Epigenomics to Understand the Biology and Chemoresistance of Ovarian Cancer. Cancers 2021, 13, 4029. [Google Scholar] [CrossRef]
- Keating, P.; Cambrosio, A.; Nelson, N.; Mogoutov, A.; Cointet, J.-P. Therapy’s Shadow: A Short History of the Study of Resistance to Cancer Chemotherapy. Front. Pharmacol. 2013, 4, 58. [Google Scholar] [CrossRef] [PubMed]
- Assaraf, Y.G.; Brozovic, A.; Gonçalves, A.C.; Jurkovicova, D.; Linē, A.; Machuqueiro, M.; Saponara, S.; Sarmento-Ribeiro, A.B.; Xavier, C.P.R.; Vasconcelos, M.H. The multi-factorial nature of clinical multidrug resistance in cancer. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2019, 46, 100645. [Google Scholar] [CrossRef]
- Riganti, C.; Contino, M. New Strategies to Overcome Resistance to Chemotherapy and Immune System in Cancer. Int. J. Mol. Sci. 2019, 20, 4783. [Google Scholar] [CrossRef]
- Lane, R.J.; Khin, N.Y.; Pavlakis, N.; Hugh, T.J.; Clarke, S.J.; Magnussen, J.; Rogan, C.; Flekser, R.L. Challenges in chemotherapy delivery: Comparison of standard chemotherapy delivery to locoregional vascular mass fluid transfer. Future Oncol. 2018, 14, 647–663. [Google Scholar] [CrossRef]
- Primeau, A.J.; Rendon, A.; Hedley, D.; Lilge, L.; Tannock, I.F. The Distribution of the Anticancer Drug Doxorubicin in Relation to Blood Vessels in Solid Tumors. Clin. Cancer Res. 2005, 11, 8782–8788. [Google Scholar] [CrossRef]
- Audet-Delage, Y.; St-Louis, C.; Minarrieta, L.; McGuirk, S.; Kurreal, I.; Annis, M.G.; Mer, A.S.; Siegel, P.M.; St-Pierre, J. Spatiotemporal modeling of chemoresistance evolution in breast tumors uncovers dependencies on SLC38A7 and SLC46A1. Cell Rep. 2023, 42, 113191. [Google Scholar] [CrossRef]
- Dalvi, M.P.; Wang, L.; Zhong, R.; Kollipara, R.K.; Park, H.; Bayo, J.; Yenerall, P.; Zhou, Y.; Timmons, B.C.; Rodriguez-Canales, J.; et al. Taxane-Platin-Resistant Lung Cancers Co-develop Hypersensitivity to JumonjiC Demethylase Inhibitors. Cell Rep. 2017, 19, 1669–1684. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Shamseddine, A.A.; Newcomb, B.; Chavez, R.S.; Panzner, T.D.; Lee, A.H.; Canals, D.; Okeoma, C.M.; Clarke, C.J.; Hannun, Y.A. Sublethal doxorubicin promotes migration and invasion of breast cancer cells: Role of Src Family non-receptor tyrosine kinases. Breast Cancer Res. 2021, 23, 76. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Dong, C.; Ruan, X.; Yan, W.; Cao, M.; Pizzo, D.; Wu, X.; Yang, L.; Liu, L.; Ren, X.; et al. Chemotherapy-Induced Extracellular Vesicle miRNAs Promote Breast Cancer Stemness by Targeting ONECUT2. Cancer Res. 2019, 79, 3608–3621. [Google Scholar] [CrossRef]
- Luo, D.-x.; Huang, M.C.; Ma, J.; Gao, Z.; Liao, D.-f.; Cao, D. Aldo-keto reductase family 1, member B10 is secreted through a lysosome-mediated non-classical pathway. Biochem. J. 2011, 438, 71–80. [Google Scholar] [CrossRef]
- Cao, D.; Fan, S.T.; Chung, S.S. Identification and characterization of a novel human aldose reductase-like gene. J. Biol. Chem. 1998, 273, 11429–11435. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Wu, J.; Yu, S.; Zhou, Y.; Zhou, C. AKR1B10 inhibits the proliferation and migration of gastric cancer via regulating epithelial-mesenchymal transition. Aging 2021, 13, 22298–22314. [Google Scholar] [CrossRef]
- Endo, S.; Matsunaga, T.; Nishinaka, T. The Role of AKR1B10 in Physiology and Pathophysiology. Metabolites 2021, 11, 332. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.J.; Yeh, Y.C.; Hsu, W.H. Prognostic significance of AKR1B10 in patients with resected lung adenocarcinoma. Thorac. Cancer 2018, 9, 1492–1499. [Google Scholar] [CrossRef]
- Soares, C.T.; Fachin, L.R.V.; Trombone, A.P.F.; Rosa, P.S.; Ghidella, C.C.; Belone, A.F.F. Potential of AKR1B10 as a Biomarker and Therapeutic Target in Type 2 Leprosy Reaction. Front. Med. 2018, 5, 263. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ban, H.; Liu, Y.; Ni, J. The expression and significance of AKR1B10 in laryngeal squamous cell carcinoma. Sci. Rep. 2021, 11, 18228. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Song, J.; Du, X.; Zhou, Y.; Li, Y.; Li, R.; Lyu, L.; He, Y.; Hao, J.; Ben, J.; et al. AKR1B10 (Aldo-keto reductase family 1 B10) promotes brain metastasis of lung cancer cells in a multi-organ microfluidic chip model. Acta Biomater. 2019, 91, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Endo, S.; Xia, S.; Suyama, M.; Morikawa, Y.; Oguri, H.; Hu, D.; Ao, Y.; Takahara, S.; Horino, Y.; Hayakawa, Y. Synthesis of potent and selective inhibitors of aldo-keto reductase 1B10 and their efficacy against proliferation, metastasis, and cisplatin resistance of lung cancer cells. J. Med. Chem. 2017, 60, 8441–8455. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.H.; Liu, K.J.; Tsai, M.F.; Wu, C.R.; Feng, A.C.; Chu, N.M.; Kao, S.H. Estrogen adversely affects the prognosis of patients with lung adenocarcinoma. Cancer Sci. 2015, 106, 51–59. [Google Scholar] [CrossRef]
- Yang, P.C.; Luh, K.T.; Wu, R.; Wu, C.W. Characterization of the mucin differentiation in human lung adenocarcinoma cell lines. Am. J. Respir. Cell Mol. Biol. 1992, 7, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.W.; Yang, P.C.; Yang, S.C.; Shyu, Y.C.; Hendrix, M.J.; Wu, R.; Wu, C.W. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am. J. Respir. Cell Mol. Biol. 1997, 17, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Geng, N.; Jin, Y.; Li, Y.; Zhu, S.; Bai, H. AKR1B10 Inhibitor Epalrestat Facilitates Sorafenib-Induced Apoptosis and Autophagy Via Targeting the mTOR Pathway in Hepatocellular Carcinoma. Int. J. Med. Sci. 2020, 17, 1246–1256. [Google Scholar] [CrossRef]
- Kosacka, M.; Piesiak, P.; Porebska, I.; Korzeniewska, A.; Dyla, T.; Jankowska, R. Cyclin A and Cyclin E expression in resected non-small cell lung cancer stage I-IIIA. In Vivo 2009, 23, 519–525. [Google Scholar]
- Pang, W.; Li, Y.; Guo, W.; Shen, H. Cyclin E: A potential treatment target to reverse cancer chemoresistance by regulating the cell cycle. Am. J. Transl. Res. 2020, 12, 5170–5187. [Google Scholar]
- Goh, K.Y.; Lim, W.-T. Cyclin D1 expression in KRAS mutant non-small cell lung cancer—Old wine into new skins. Transl. Lung Cancer Res. 2020, 9, 2302–2304. [Google Scholar] [CrossRef] [PubMed]
- Volm, M.; Koomägi, R.; Mattern, J.; Stammler, G. Cyclin A is associated with an unfavourable outcome in patients with non-small-cell lung carcinomas. Br. J. Cancer 1997, 75, 1774–1778. [Google Scholar] [CrossRef]
- Ramos, A.; Sadeghi, S.; Tabatabaeian, H. Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them. Int. J. Mol. Sci. 2021, 22, 9451. [Google Scholar] [CrossRef] [PubMed]
- Finn, K.J.; Martin, S.E.; Settleman, J. A Single-Step, High-Dose Selection Scheme Reveals Distinct Mechanisms of Acquired Resistance to Oncogenic Kinase Inhibition in Cancer Cells. Cancer Res. 2020, 80, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Luo, D.X.; Huang, C.; Shen, Y.; Bu, Y.; Markwell, S.; Gao, J.; Liu, J.; Zu, X.; Cao, Z.; et al. AKR1B10 overexpression in breast cancer: Association with tumor size, lymph node metastasis and patient survival and its potential as a novel serum marker. Int. J. Cancer 2012, 131, E862–E871. [Google Scholar] [CrossRef]
- Chung, Y.T.; Matkowskyj, K.A.; Li, H.; Bai, H.; Zhang, W.; Tsao, M.S.; Liao, J.; Yang, G.Y. Overexpression and oncogenic function of aldo-keto reductase family 1B10 (AKR1B10) in pancreatic carcinoma. Mod. Pathol. 2012, 25, 758–766. [Google Scholar] [CrossRef]
- Matsunaga, T.; Yamaji, Y.; Tomokuni, T.; Morita, H.; Morikawa, Y.; Suzuki, A.; Yonezawa, A.; Endo, S.; Ikari, A.; Iguchi, K.; et al. Nitric oxide confers cisplatin resistance in human lung cancer cells through upregulation of aldo-keto reductase 1B10 and proteasome. Free Radic. Res. 2014, 48, 1371–1385. [Google Scholar] [CrossRef]
- Chewchuk, S.; Guo, B.; Parissenti, A.M. Alterations in estrogen signalling pathways upon acquisition of anthracycline resistance in breast tumor cells. PLoS ONE 2017, 12, e0172244. [Google Scholar] [CrossRef]
- Morikawa, Y.; Kezuka, C.; Endo, S.; Ikari, A.; Soda, M.; Yamamura, K.; Toyooka, N.; El-Kabbani, O.; Hara, A.; Matsunaga, T. Acquisition of doxorubicin resistance facilitates migrating and invasive potentials of gastric cancer MKN45 cells through up-regulating aldo-keto reductase 1B10. Chem. Biol. Interact. 2015, 230, 30–39. [Google Scholar] [CrossRef]
- Qu, J.; Liu, X.; Li, J.; Gong, K.; Duan, L.; Luo, W.; Luo, D. AKR1B10 promotes proliferation of breast cancer cells by activating Wnt/β-catenin pathway. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2019, 35, 1094–1100. [Google Scholar]
- Huang, Z.; Yan, Y.; Zhu, Z.; Liu, J.; He, X.; Dalangood, S.; Li, M.; Tan, M.; Cai, J.; Tang, P.; et al. CBX7 suppresses urinary bladder cancer progression via modulating AKR1B10–ERK signaling. Cell Death Dis. 2021, 12, 537. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yan, R.; Al-Salman, A.; Shen, Y.; Bu, Y.; Ma, J.; Luo, D.X.; Huang, C.; Jiang, Y.; Wilber, A.; et al. Epidermal growth factor induces tumour marker AKR1B10 expression through activator protein-1 signalling in hepatocellular carcinoma cells. Biochem. J. 2012, 442, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Qi, Y.; Bai, Y.; Zhang, H.; Zhu, W.; Zhou, S.; Zhang, Y. LncRNA 1700020I14Rik promotes AKR1B10 expression and activates Erk pathway to induce hepatocyte damage in alcoholic hepatitis. Cell Death Discov. 2022, 8, 374. [Google Scholar] [CrossRef] [PubMed]
- Gkountakos, A.; Centonze, G.; Vita, E.; Belluomini, L.; Milella, M.; Bria, E.; Milione, M.; Scarpa, A.; Simbolo, M. Identification of Targetable Liabilities in the Dynamic Metabolic Profile of EGFR-Mutant Lung Adenocarcinoma: Thinking beyond Genomics for Overcoming EGFR TKI Resistance. Biomedicines 2022, 10, 277. [Google Scholar] [CrossRef]
- Naveena, H.A.; Bhatia, D. Hypoxia Modulates Cellular Endocytic Pathways and Organelles with Enhanced Cell Migration and 3D Cell Invasion. ChemBioChem 2023, 24, e202300506. [Google Scholar] [CrossRef]
| Number of Lung Nodules (Mean ± SD) | p Value | n | |
|---|---|---|---|
| PBS | 2.9 ± 1.5 | 14 | |
| Tax 0.3 | 3.9 ± 3.4 | 0.309 | 11 |
| Tax 4.5 | 6.8 ± 2.9 | 0.000 ** | 11 |
| Dox 5 | 4.2 ± 1.2 | 0.021 * | 12 |
| Dox 60 | 6.0 ± 1.7 | 0.000 ** | 13 |
| Cis 0.25 | 5.0 ± 2.2 | 0.006 ** | 13 |
| Cis 2.5 | 5.8 ± 2.6 | 0.001 ** | 11 |
| Gem 1 | 5.6 ± 1.9 | 0.000 ** | 12 |
| Gem 8 | 6.5 ± 3.8 | 0.003 ** | 13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, T.-H.; Lin, S.-C.; Yang, Y.-Y.; Lay, J.-D.; Chang, C.-L.; Yao, C.-J.; Huang, J.-S.; Chuang, S.-E. The Role of AKR1B10 in Lung Cancer Malignancy Induced by Sublethal Doses of Chemotherapeutic Drugs. Cancers 2024, 16, 2428. https://doi.org/10.3390/cancers16132428
Jang T-H, Lin S-C, Yang Y-Y, Lay J-D, Chang C-L, Yao C-J, Huang J-S, Chuang S-E. The Role of AKR1B10 in Lung Cancer Malignancy Induced by Sublethal Doses of Chemotherapeutic Drugs. Cancers. 2024; 16(13):2428. https://doi.org/10.3390/cancers16132428
Chicago/Turabian StyleJang, Te-Hsuan, Sheng-Chieh Lin, Ya-Yu Yang, Jong-Ding Lay, Chih-Ling Chang, Chih-Jung Yao, Jhy-Shrian Huang, and Shuang-En Chuang. 2024. "The Role of AKR1B10 in Lung Cancer Malignancy Induced by Sublethal Doses of Chemotherapeutic Drugs" Cancers 16, no. 13: 2428. https://doi.org/10.3390/cancers16132428







