Breast Cancer Stem Cells and Tumor Heterogeneity: Characteristics and Therapeutic Strategies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Breast Cancer Characteristics and Classification
3. Breast Cancer Stem Cell Subpopulation
3.1. Methods for BCSC Detection
3.2. Characteristics of BCSCs in Different BC Molecular Subtypes
4. Therapeutic Strategies for the Treatment of Specific Types of Breast Cancer
4.1. BCSC Treatment Strategy
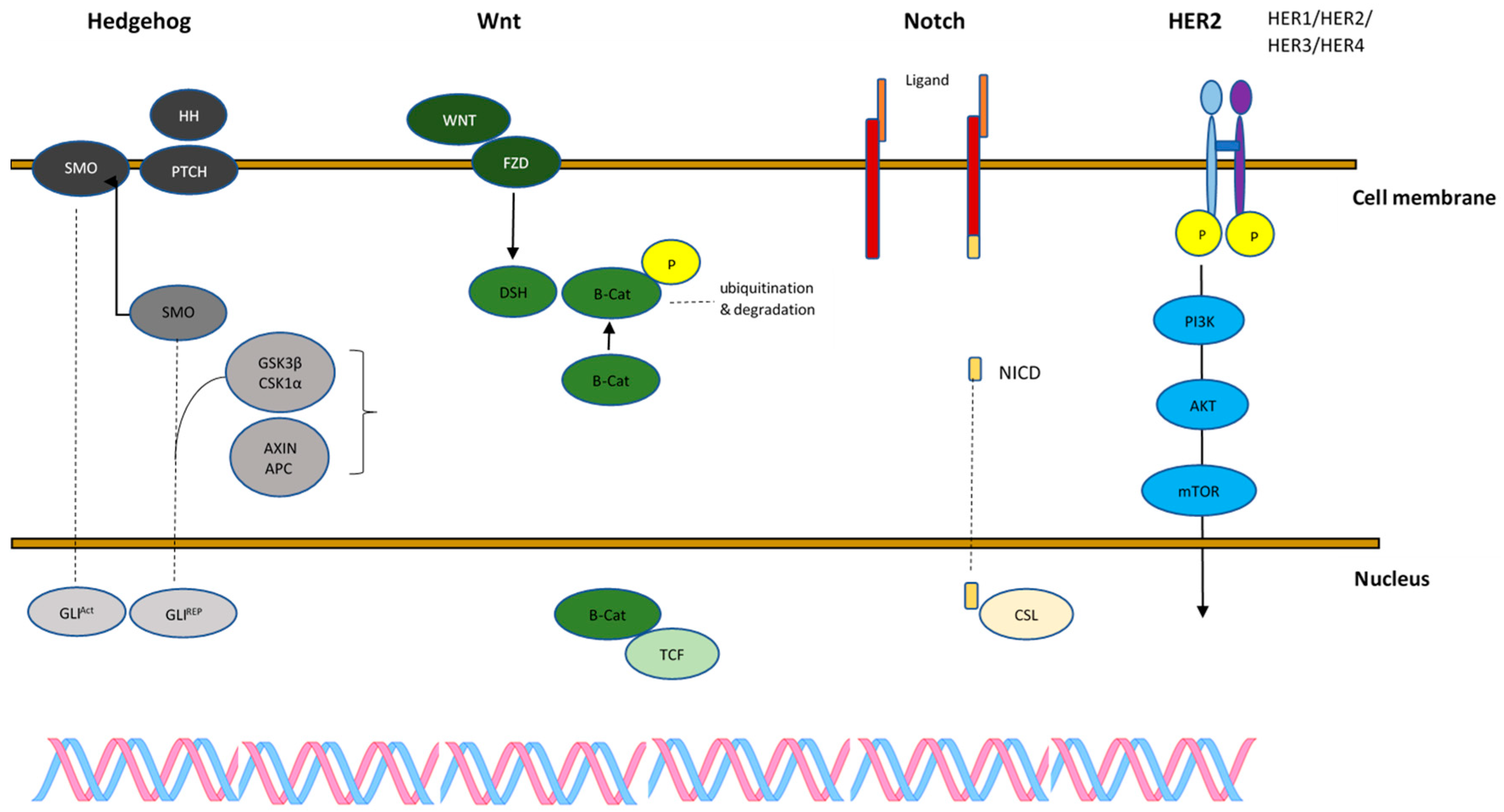
4.1.1. Wnt
4.1.2. Notch
4.1.3. Hedgehog
4.1.4. PI3K/Akt/mTOR
4.1.5. HER2
| Signaling Pathways Targeting BCSCs | |||||
|---|---|---|---|---|---|
| BC Subtype | Wnt | Notch | Hedgehog | PI3K/AKT/mTOR | HER2 |
| Luminal | MK-0752 | Vismodegib | Alpelisib | ||
| RO-4929097 | GANT61 | Everolimus | |||
| XL147 | |||||
| NVP-BKM120 | |||||
| LY-294002 | |||||
| Perifosine | |||||
| Flubendazole | |||||
| HER2+ | Trastuzumab | ||||
| Pertuzumab | |||||
| Lapatinib | |||||
| TDM-1 | |||||
| TNBC | LGK-974 | MK-0752 | Vismodegib | ||
| PF-03084014 | Sonidegib | ||||
| RO-4929097 | GANT61 | ||||
| nirogacestat | |||||
| Basal/ ER- HER2− | Foxy-5 | Sonidegib | GDC-0941 | ||
| Cirmtuzumab | |||||
| Vantictumab | |||||
4.1.6. Active Clinical Trials
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABCG2 | ATP-binding cassette subfamily G member 2 |
| ADAM | a disintegrin and metalloproteinase protein |
| Ais | aromatase inhibitors |
| AKT | a serine/threonine protein kinase |
| ALDH1 | aldehyde dehydrogenase 1 |
| APC | adenomatous polyposis coli |
| BC | breast cancer |
| BCSCs | breast cancer stem cells |
| BL1 | basal-like 1 subtype of TNBC |
| BL2 | basal-like 2 subtype of TNBC |
| BLBC | basal-like breast cancer |
| BLIA | basal-like immune-activated subtype of TNBC |
| BLIS | basal-like immune-suppressed subtype of TNBC |
| BRCA1 | BReast CAncer gene 1 |
| BRCA2 | BReast CAncer gene 2 |
| CB-103/LIMANTRAFIN | an orally active inhibitor of the Notch transcription activation complex |
| CD24 | a cluster of differentiation 24 |
| CD44 | a cluster of differentiation 44 |
| CD70 | a cluster of differentiation 70 |
| CD133 | a cluster of differentiation 133 |
| CK | cytokeratin |
| CK1 | casein kinase 1 |
| CSC | cancer stem cells |
| CSL | CBF1, Suppressor of Hairless, Lag-1 |
| CTFs | carboxy-terminal truncations |
| Dsh | Disheveled |
| EGFR | epidermal growth factor receptor |
| EMT | epithelial–mesenchymal transition |
| EPCAM | epithelial cell adhesion molecule |
| ER | estrogen receptor |
| ESMO | European Society for Medical Oncology |
| FUSCC | Fudan University Shanghai Cancer Center |
| FZD | Frizzled proteins |
| GANT61 | an inhibitor for GLI1 as well as GLI2-induced transcription |
| GDC-0941 (Pictilisib) | a potent inhibitor of PI3Kα/δ |
| GLI | glioma-associated oncogene |
| GLI2/3R | GLI2/3 repressors |
| GLIACT | GLI activators |
| GSK3β | glycogen synthase kinase 3β |
| HER2 | human epidermal growth factor 2 |
| Hh | Hedgehog |
| IM | an immunomodulatory subtype of TNBC |
| LAR | luminal androgen receptor subtype of TNBC |
| LEF | lymphatic enhancer factor |
| LGK-974 | specific PORCN inhibitor |
| lncRNA | long noncoding |
| Lrg5 | leucine-rich repeat-containing G protein-coupled receptor 5 |
| LY-294002 | a PI3K inhibitor |
| M | mesenchymal subtype of TNBC |
| MaSCs | mammary gland stem cells |
| MBC | metastatic breast cancer |
| MES | mesenchymal subtype of TNBC |
| MET | mesenchymal–epithelial transition |
| MK-0752 | γ-secretase inhibitor |
| MS | mammosphere |
| MSL | mesenchymal stem-like subtype of TNBC |
| mTOR | mammalian target of rapamycin |
| MUC1 | mucin 1 |
| NICD | Notch intracellular domain |
| NVP-BKM120 (Buparlisib) | a pan-class I PI3K inhibitor |
| PD-L1 | programmed death-ligand 1 |
| PF-03084014 | γ secretase inhibitor |
| PI3K | phosphatidylinositol 3-kinase |
| PIK3CA | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| PKA | protein kinase A |
| PR | progesterone receptor |
| PTCH1 | Patched 1 |
| RBP-J | recombining binding protein J |
| RO-4929097 (RG-4733) | γ secretase inhibitor |
| SMO | Smoothened SOX9 SRY-box transcription factor 9 |
| SSEA-3 | stage-specific embryonic antigen-3 |
| STAT | signal transducer and activator of transcription |
| SUFU | suppressor of fused homolog |
| TCF | T-cell factor |
| T-DM1 | Trastuzumab emtansine |
| TICs | tumor-initiating cells |
| TNBC | triple-negative breast cancer |
| TWIST | Twist-related protein 1 |
| WNT5A | Wnt family member 5A |
| XL147 (Pilaralisib) | a potent and highly selective class I PI3K inhibitor |
References
- Available online: https://www.who.int/news/item/01-02-2024-global-cancer-burden-growing--amidst-mounting-need-for-services (accessed on 10 May 2024).
- Sun, Y.-S.; Zhao, Z.; Yang, Z.-N.; Xu, F.; Lu, H.-J.; Zhu, Z.-Y.; Shi, W.; Jiang, J.; Yao, P.-P.; Zhu, H.-P. Risk Factors and Preventions of Breast Cancer. Int. J. Biol. Sci. 2017, 13, 1387–1397. [Google Scholar] [CrossRef]
- Jia, T.; Liu, Y.; Fan, Y.; Wang, L.; Jiang, E. Association of Healthy Diet and Physical Activity with Breast Cancer: Lifestyle Interventions and Oncology Education. Front. Public Health. 2022, 23, 797794. [Google Scholar] [CrossRef]
- Momenimovahed, Z.; Salehiniya, H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer 2019, 11, 151–164. [Google Scholar] [CrossRef]
- Oshiro, C.; Yamasaki, M.; Noda, Y.; Nishimae, A.; Takahashi, H.; Inaji, H. Comparative evaluation of nuclear and histological grades as prognostic factors for invasive breast cancer. Breast Cancer 2020, 27, 947–953. [Google Scholar] [CrossRef]
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast cancer heterogeneity and its implication in personalized precision therapy. Exp. Hematol. Oncol. 2023, 12, 3. [Google Scholar] [CrossRef]
- Crabtree, J.S.; Miele, L. Breast Cancer Stem Cells. Biomedicines 2018, 6, 77. [Google Scholar] [CrossRef]
- Woei, C.S.; Chooi, L. Breast cancer stem cells—From origins to targeted therapy. Stem Cell Investig. 2017, 4, 96. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 5416923. [Google Scholar] [CrossRef]
- Available online: https://www.esmo.org/guidelines/guidelines-by-topic/esmo-clinical-practice-guidelines-breast-cancer (accessed on 28 June 2024).
- Zhang, X. Molecular Classification of Breast Cancer: Relevance and Challenges. Arch. Pathol. Lab. Med. 2023, 147, 46–51. [Google Scholar] [CrossRef]
- Oh, H.A.; Eliassen, H.; Wang, M.; Smith-Warner, S.A.; Beck, A.H.; Schnitt, S.J.; Collins, L.C.; Connolly, J.L.; Montaser-Kouhsari, L.; Polyak, K.; et al. Expression of estrogen receptor, progesterone receptor, and Ki67 in normal breast tissue in relation to subsequent risk of breast cancer. Breast Cancer 2016, 2, 16032. [Google Scholar] [CrossRef]
- Sareyeldin, R.M.; Gupta, I.; Al-Hashimi, I.; Al-Thawadi, H.A.; Al Farsi, H.F.; Vranic, S.; Al Moustafa, A.-E. Gene Expression and miRNAs Profiling: Function and Regulation in Human Epidermal Growth Factor Receptor 2 (HER2)-Positive Breast Cancer. Cancers 2019, 11, 646. [Google Scholar] [CrossRef]
- Mohammed, A.A. The clinical behavior of different molecular subtypes of breast cancer. Cancer Treat. Res. Commun. 2021, 29, 100469. [Google Scholar] [CrossRef]
- Gil, R.S.; Vagnarelli, P. Ki-67: More Hidden behind a ‘Classic Proliferation Marker’. Trends Biochem. Sci. 2018, 43, 747–748. [Google Scholar]
- Fragomeni, S.M.; Sciallis, A.; Jeruss, J.S. Molecular Subtypes and Local-Regional Control of Breast Cancer. Surg. Oncol. Clin. N. Am. 2018, 27, 95–120. [Google Scholar] [CrossRef]
- Vallejos, C.; Gómez, H.; Cruz, W.; Pinto, J.A.; Dyer, R.R.; Velarde, R.; Suazo, J.F.; Neciosup, S.P.; León, M.; de la Cruz, M.A.; et al. Breast cancer classification according to immunohistochemistry markers: Subtypes and association with clinicopathologic variables in a Peruvian hospital database. Clin. Breast Cancer 2010, 10, 294–300. [Google Scholar] [CrossRef]
- Xu, M.; Tang, Q.; Li, M.; Liu, Y.; Li, F. An analysis of Ki-67 expression in stage 1 invasive ductal breast carcinoma using apparent diffusion coefficient histograms. Quant. Imaging Med. Surg. 2021, 11, 1518–1531. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Hu, P.H.; Tu, J.H.; Yu, N.S. Luminal B breast cancer: Patterns of recurrence and clinical outcome. Oncotarget 2016, 7, 65024–65033. [Google Scholar] [CrossRef]
- Ontario Health (Quality). Gene Expression Profiling Tests for Early-Stage Invasive Breast Cancer: A Health Technology Assessment. Ont. Health Technol. Assess. Ser. 2020, 20, 1–234. [Google Scholar]
- Miricescu, D.; Totan, A.; Stanescu-Spinu, I.I.; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2021, 22, 173. [Google Scholar] [CrossRef]
- Alexandrou, S.; George, S.M.; Ormandy, C.J.; Lim, E.; Oakes, S.R.; Caldon, E. The Proliferative and Apoptotic Landscape of Basal-like Breast Cancer. Int. J. Mol. Sci. 2019, 20, 667. [Google Scholar] [CrossRef]
- Rakha, E.A.; Elsheikh, S.E.; Aleskandarany, M.A.; Habashi, H.O.; Green, A.R.; Powe, D.G.; El-Sayed, M.E.; Benhasouna, A.; Brunet, J.S.; Akslen, L.A.; et al. Triple-Negative Breast Cancer: Distinguishing between Basal and Nonbasal Subtypes. Clin. Cancer Res. 2009, 15, 2302–2310. [Google Scholar] [CrossRef] [PubMed]
- McGinn, O.; Riley, D.; Finlay-Schultz, J.; Paul, K.V.; Kabos, P.; Sartorius, C.A. Cytokeratins 5 and 17 Maintain an Aggressive Epithelial State in Basal-Like Breast Cancer. Mol. Cancer Res. 2022, 20, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Dogra, A.; Mehta, A.; Doval, D.C. Are Basal-Like and Non-Basal-Like Triple-Negative Breast Cancers Really Different? J. Oncol. 2020, 4061063. [Google Scholar] [CrossRef] [PubMed]
- Lachapelle, J.; Foulkes, W.D. Triple-negative and basal-like breast cancer: Implications for oncologists. Curr. Oncol. 2011, 18, 161–164. [Google Scholar] [CrossRef]
- Dey, N.; Aske, J.; De, P. Therapeutic Strategies for Metastatic Triple-Negative Breast Cancers: From Negative to Positive. Pharmaceuticals 2021, 14, 455. [Google Scholar] [CrossRef] [PubMed]
- Dass, S.A.; Tan, K.L.; Rajan, R.S.; Mokhtar, N.F.; Mohd Adzmi, E.R.; Rahman, W.F.W.A.; Tengku Din, T.A.D.A.T.; Balakrishnan, V. Triple Negative Breast Cancer: A Review of Present and Future Diagnostic Modalities. Medicina 2021, 57, 62. [Google Scholar] [CrossRef] [PubMed]
- Ensenyat-Mendez, M.; Llinàs-Arias, P.; Orozco, J.I.J.; Íñiguez-Muñoz, S.; Salomon, M.P.; Sesé, B.; DiNome, M.L.; Marzese, D.M. Current Triple-Negative Breast Cancer Subtypes: Dissecting the Most Aggressive Form of Breast Cancer. Front. Oncol. 2021, 11, 681476. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.R.; Jiang, Y.Z.; Xu, X.E.; Yu, K.D.; Jin, X.; Hu, X.; Zuo, W.J.; Hao, S.; Wu, S.; Liu, G.Y.; et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res. 2016, 18, 33. [Google Scholar] [CrossRef]
- Bissanum, R.; Chaichulee, S.; Kamolphiwong, R.; Navakanitworakul, R.; Kanokwiroon, K. Molecular Classification Models for Triple Negative Breast Cancer Subtype Using Machine Learning. J. Pers. Med. 2021, 11, 881. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Chiodi, I.; Belgiovine, C.; Donà, F.; Scovassi, A.I.; Mondello, C. Drug Treatment of Cancer Cell Lines: A Way to Select for Cancer Stem Cells? Cancers 2011, 3, 1111–1128. [Google Scholar] [CrossRef] [PubMed]
- Palomeras, S.; Ruiz-Martínez, S.; Puig, T.; Puig, M. Targeting Breast Cancer Stem Cells to Overcome Treatment Resistance. Molecules 2018, 23, 2193. [Google Scholar] [CrossRef] [PubMed]
- Dick, J.E. Looking ahead in cancer stem cell research. Nat. Biotechnol. 2009, 27, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Qureshi-Baig, K.; Ullmann, P.; Haan, S.; Letellier, E. Tumor-Initiating Cells: A criTICal review of isolation approaches and new challenges in targeting strategies. Mol. Cancer 2017, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ahmad, A.; Azmi, A.S.; Ali, S.; Sarkar, F.H. Overview of cancer stem cells (CSCs) and mechanisms of their regulation: Implications for cancer therapy. Curr. Protoc. Pharmacol. 2013, 61, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.S.; Huang, Y.-H.; Luo, M.-H. Cancer stem cell markers are associated with adverse biomarker profiles and molecular subtypes of breast cancer. Breast Cancer Res. Treat. 2012, 136, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Escudero Mendez, L.; Srinivasan, M.; Hamouda, R.K.; Ambedkar, B.; Arzoun, H.; Sahib, I.; Fondeur, J.; Mohammed, L. Evaluation of CD44+/CD24− and Aldehyde Dehydrogenase Enzyme Markers in Cancer Stem Cells as Prognostic Indicators for Triple-Negative Breast Cancer. Cureus 2022, 14, e28056. [Google Scholar] [CrossRef]
- Zhang, X.; Powell, K.; Li, L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers 2020, 12, 3765. [Google Scholar] [CrossRef] [PubMed]
- Pinto, C.A.; Widodo, E.; Waltham, M.; Thompson, E.W. Breast Cancer Stem Cells and Epithelial Mesenchymal Plasticity—Implications for Chemoresistance. Cancer Lett. 2013, 341, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Cianciosi, D.; Ansary, J.; Forbes-Hernandez, T.Y.; Regolo, L.; Quinzi, D.; Gracia Villar, S.; Garcia Villena, E.; Tutusaus Pifarre, K.; Alvarez-Suarez, J.M.; Battino, M.; et al. The Molecular Basis of Different Approaches for the Study of Cancer Stem Cells and the Advantages and Disadvantages of a Three-Dimensional Culture. Molecules 2021, 26, 2615. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2013, 2, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Jiang, J.; Liang, X.H.; Tang, Y.L. Links between cancer stem cells and epithelial-mesenchymal transition. OncoTargets Ther. 2015, 8, 2973–2980. [Google Scholar] [PubMed]
- Chen, J.; Liu, S.; Su, Y.; Zhang, X. ALDH1+ stem cells demonstrate more stem cell-like characteristics than CD44+/CD24−/low stem cells in different molecular subtypes of breast cancer. Transl. Cancer Res. 2020, 9, 1652–1659. [Google Scholar] [CrossRef]
- Zhong, Y.; Shen, S.; Zhou, Y.; Mao, F.; Guan, J.; Lin, Y.; Xu, Y.; Sun, Q. ALDH1 is a better clinical indicator for relapse of invasive ductal breast cancer than the CD44+/CD24− phenotype. Med. Oncol. 2014, 31, 864. [Google Scholar] [CrossRef] [PubMed]
- Chute, J.P.; Muramoto, G.G.; Whitesides, J.; McDonnell, D.P. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 11707–11712. [Google Scholar] [CrossRef]
- Dionísio, M.R.; Vieira, A.F.; Carvalho, R.; Conde, I.; Oliveira, M.; Gomes, M.; Pinto, M.T.; Pereira, P.; Pimentel, J.; Souza, C.; et al. BR-BCSC Signature: The Cancer Stem Cell Profile Enriched in Brain Metastases that Predicts a Worse Prognosis in Lymph Node-Positive Breast Cancer. Cells 2020, 9, 2442. [Google Scholar] [CrossRef]
- Olsson, M.; Larsson, P.; Johansson, J.; Sah, V.R.; Parris, T.Z. Cancer stem cells are prevalent in the basal-like 2 and mesenchymal triple-negative breast cancer subtypes in vitro. Front. Cell Dev. Biol. 2023, 11, 1237673. [Google Scholar] [CrossRef] [PubMed]
- Althobiti, M.; El Ansari, R.; Aleskandarany, M.; Joseph, C.; Toss, M.S.; Green, A.R.; Rakha, E.A. The prognostic significance of ALDH1A1 expression in early invasive breast cancer. Histopathology. 2020, 77, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.G.; Liao, R.X.; Qiu, J.; Jin, J.U.; Wang, X.X.; Duan, Y.Z.; Chen, F.L.; Hao, P.; Xie, X.C.; Wang, Z.X.; et al. Microarray-based analysis of microRNA expression in breast cancer stem cells. J. Exp. Clin. Cancer Res. 2010, 29, 174. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Morita, M.; Kuba, S.; Hayashi, H.; Otsubo, R.; Matsumoto, M.; Yamanouchi, K.; Kobayashi, K.; Soyama, A.; Hidaka, M.; et al. Association of quantitative analysis of intratumoral reduced E-cadherin expression with lymph node metastasis and prognosis in patients with breast cancer. Sci. Rep. 2023, 13, 10434. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.W.; Yu, J.C.; Hsieh, Y.H.; Liao, W.L.; Shieh, J.C.; Yao, C.C.; Lee, H.J.; Chen, P.M.; Wu, P.E.; Shen, C.Y. Increased Cellular Levels of MicroRNA-9 and MicroRNA-221 Correlate with Cancer Stemness and Predict Poor Outcome in Human Breast Cancer. Cell Physiol. Biochem. 2018, 48, 2205–2218. [Google Scholar] [CrossRef] [PubMed]
- Gwak, J.M.; Kim, H.J.; Kim, E.J.; Chung, Y.R.; Yun, S.; Seo, A.N.; Lee, H.J.; Park, S.Y. MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res. Treat. 2014, 147, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Wu, J.; Lin, J.; Liu, W.; Chen, P.; Yu, M.; Zhou, D.; Yao, G. Graphene-based nanomaterials for breast cancer treatment: Promising therapeutics strategies. J. Nanobiotechnol. 2021, 19, 211. [Google Scholar] [CrossRef] [PubMed]
- Kreutzfeldt, J.; Rozeboom, B.; Dey, N.; De, P. The trastuzumab era: Current and upcoming targeted HER2+ breast cancer therapies. Am. J. Cancer Res. 2020, 10, 1045–1067. [Google Scholar] [PubMed]
- Yao, H.; He, G.; Yan, S.; Chen, C.; Song, L.; Rosol, T.J.; Deng, X. Triple-negative breast cancer: Is there a treatment on the horizon? Oncotarget 2017, 8, 1913–1924. [Google Scholar] [CrossRef]
- Haddad, S.; Zemni, I.; Bettaieb, I.; Adouni, O.; Saadallah, F.; Slimane, M.; Chargui, R.; Rahal, K. Basal-Like Breast Cancer: Main Features of a Very Particular Entity of Breast Cancer. Clin. Med. Rev. Case Rep. 2019, 6, 272. [Google Scholar] [CrossRef]
- Cho, N. Molecular subtypes and imaging phenotypes of breast cancer. Ultrasonography 2016, 35, 281–288. [Google Scholar] [CrossRef]
- Vidra, R.; Nemes, A.; Vidrean, A.; Pintea, S.; Tintari, S.; Deac, A.; Ciuleanu, T. Pathological complete response following cisplatin or carboplatin-based neoadjuvant chemotherapy for triple-negative breast cancer: A systematic review and meta-analysis. Exp. Ther. Med. 2022, 23, 91. [Google Scholar] [CrossRef]
- Shi, F.; Zhang, J.; Liu, H.; Wu, L.; Jiang, H.; Wu, Q.; Liu, T.; Lou, M.; Wu, H. The dual PI3K/mTOR inhibitor dactolisib elicits anti-tumor activity in vitro and in vivo. Oncotarget 2018, 9, 706–717. [Google Scholar] [CrossRef]
- Lu, Q.; Xia, W.; Lee, K.; Zhang, J.; Yuan, H.; Yuan, H.; Shi, J.; Wang, S.; Xu, F. Bicalutamide plus Aromatase Inhibitor in Patients with Estrogen Receptor-Positive/Androgen Receptor-Positive Advanced Breast Cancer. Oncologist 2020, 25, 21-e15. [Google Scholar] [CrossRef]
- Cortesi, L.; Rugo, H.S.; Jackisch, C. An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Target. Oncol. 2021, 16, 255–282. [Google Scholar] [CrossRef]
- Prasad, C.P.; Chaurasiya, S.K.; Axelsson, L.; Andersson, T. WNT-5A triggers Cdc42 activation leading to an ERK1/2 dependent decrease in MMP9 activity and invasive migration of breast cancer cells. Mol. Oncol. 2013, 7, 870–883. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Shipitsin, M.; Campbell, L.L.; Argani, P.; Weremowicz, S.; Bloushtain-Qimron, N.; Yao, J.; Nikolskaya, T.; Serebryiskaya, T.; Beroukhim, R.; Hu, M.; et al. Molecular definition of breast tumor heterogeneity. Cancer Cell 2007, 11, 259–273. [Google Scholar] [CrossRef]
- Diamond, J.R.; Becerra, C.; Richards, D.; Mita, A.; Osborne, C.; O’Shaughnessy, J.; Zhang, C.; Henner, R.; Kapoun, A.M.; Xu, L.; et al. Phase Ib clinical trial of the anti-frizzled antibody vantictumab (OMP-18R5) plus paclitaxel in patients with locally advanced or metastatic HER2-negative breast cancer. Breast Cancer Res. Treat. 2020, 184, 53–62. [Google Scholar] [CrossRef]
- Abreu de Oliveira, W.A.; Moens, S.; El Laithy, Y.; van der Veer, B.K.; Athanasouli, P.; Cortesi, E.E.; Baietti, M.F.; Koh, K.P.; Ventura, J.J.; Amant, F.; et al. Wnt/β-Catenin Inhibition Disrupts Carboplatin Resistance in Isogenic Models of Triple-Negative Breast Cancer. Front. Oncol. 2021, 11, 705384. [Google Scholar] [CrossRef]
- Rodon, J.; Argilés, G.; Connolly, R.M.; Vaishampayan, U.; Jonge, M.; Garralda, E.; Giannakis, M.; Smith, D.C.; Dobson, J.R.; McLaughlin, M.E.; et al. Phase 1 study of single-agent WNT974, a first-in-class Porcupine inhibitor, in patients with advanced solid tumours. Br. J. Cancer 2021, 125, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pan, S.; Hsieh, M.H.; Harris, J.H. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. USA 2013, 110, 20224–20229. [Google Scholar] [CrossRef] [PubMed]
- Säfholm, A.; Tuomela, J.; Rosenkvist, J.; Dejmek, J.; Harkonen, P.; Andersson, T. The Wnt-5a–Derived Hexapeptide Foxy-5 Inhibits Breast Cancer Metastasis In vivo by Targeting Cell Motility. Clin. Cancer Res. 2008, 14, 6556–6563. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.E.; Ekström, E.J.; Howlin, J.; Andersson, T. The WNT-5a derived peptide, Foxy-5, possesses dual properties that impair progression of ERα negative breast cancer. Cell Cycle 2009, 8, 1838–1842. [Google Scholar] [CrossRef] [PubMed]
- Shatsky, R.A.; Batra-Sharma, H.; Helsten, T.; Schwab, R.B.; Pittman, E.I.; Pu, M.; Weihe, E.; Ghia, E.M.; Rassenti, L.Z.; Molinolo, A.; et al. A phase 1b study of zilovertamab in combination with paclitaxel for locally advanced/unresectable or metastatic HER2-negative breast cancer. Breast Cancer Res. 2024, 26, 32. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.A.; Shatsky, R.A.; Schwab, R.B.; Wallace, A.M.; Wolf, D.M.; Hirst, G.L.; Brown-Swigart, L.; Esserman, L.J.; van ’t Veer, L.J.; Ghia, E.M.; et al. Association of baseline ROR1 and ROR2 gene expression with clinical outcomes in the I-SPY2 neoadjuvant breast cancer trial. Breast Cancer Res. Treat. 2023, 199, 281–291. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- Zhang, J.; Shao, X.; Sun, H.; Liu, K.E.; Ding, Z.; Chen, J.; Fang, L.; Su, W.; Hong, Y.; Li, H.; et al. NUMB negatively regulates the epithelial-mesenchymal transition of triple-negative breast cancer by antagonizing Notch signaling. Oncotarget 2016, 7, 61036–61053. [Google Scholar] [CrossRef] [PubMed]
- Kiaris, H.; Politi, K.; Grimm, L.M.; Szabolcs, M.; Fisher, P.; Efstratiadis, A.; Szabolcs, M.; Fisher, P.; Efstratiadis, A.; Artavanis-Tsakonas, P. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am. J. Pathol. 2004, 165, 695–705. [Google Scholar] [CrossRef]
- Strizzi, L.; Hardy, K.M.; Seftor, E.A.; Costa, F.F.; Kirschmann, D.A.; Seftor, R.E.; Postovit, L.M.; Hendrix, M.J. Development and cancer: At the crossroads of Nodal and Notch signaling. Cancer Res. 2009, 69, 7131–7134. [Google Scholar] [CrossRef]
- Debeb, B.G.; Cohen, E.N.; Boley, K.; Freiter, E.M.; Li, L.; Robertson, F.M.; Reuben, J.M.; Cristofanilli, M.; Buchholz, T.A.; Woodward, W.A. Pre-clinical studies of Notch signaling inhibitor RO4929097 in inflammatory breast cancer cells. Breast Cancer Res. Treat. 2012, 134, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Giuli, M.V.; Giuliani, E.; Screpanti, I.; Bellavia, D.; Checquolo, S. Notch Signaling Activation as a Hallmark for Triple-Negative Breast Cancer Subtype. J. Oncol. 2019, 8707053. [Google Scholar] [CrossRef] [PubMed]
- Means-Powell, J.A.; Mayer, I.A.; Ismail-Khan, R.; Del Valle, L.; Tonetti, D.; Abramson, V.G.; Sanders, M.S.; Lush, R.M.; Sorrentino, C.; Majumder, S.; et al. A Phase Ib Dose Escalation Trial of RO4929097 (a γ-secretase inhibitor) in Combination with Exemestane in Patients with ER + Metastatic Breast Cancer (MBC). Clin. Breast Cancer 2022, 22, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Schott, A.F.; Landis, M.D.; Dontu, G.; Griffith, K.A.; Layman, R.M.; Krop, I.; Paskett, L.A.; Wong, H.; Dobrolecki, L.E.; Lewis, M.T.; et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin. Cancer Res. 2013, 19, 1512–1524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Yan, Z.; Zong, Q.; Fang, D.D.; Painter, C.; Zhang, Q.; Chen, E.; Lira, M.E.; John-Baptiste, A.; Christensen, J.G. Synergistic effect of the γ-secretase inhibitor PF-03084014 and docetaxel in breast cancer models. Stem Cells Transl. Med. 2013, 2, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Marzia, A.; Locatelli, P.A.; Dees, E.C.; LoRusso, P.M.; Pegram, M.D.; Awada, A.; Huang, B.; Cesari, R.; Jiang, Y.; Naveed Shaik, M.; et al. Phase I study of the gamma secretase inhibitor PF-03084014 in combination with docetaxel in patients with advanced triple-negative breast cancer. Oncotarget 2017, 8, 2320–2328. [Google Scholar] [CrossRef]
- Sardesai, S.; Badawi, M.; Mrozek, E.; Morgan, E.; Phelps, M.; Stephens, J.; Wei, L.; Kassem, M.; Ling, Y.; Lustberg, M.; et al. A phase I study of an oral selective gamma secretase (GS) inhibitor RO4929097 in combination with neoadjuvant paclitaxel and carboplatin in triple negative breast cancer. Investig. New Drugs 2020, 38, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, P.; Mou, Y.; Yang, W.; Zhang, J.; Li, Q.; Dou, X. Silencing Notch4 promotes tumorigenesis and inhibits metastasis of triple-negative breast cancer via Nanog and Cdc42. Cell Death Discov. 2023, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Castro, N.P.; Fedorova-Abrams, N.D.; Merchant, A.S.; Rangel, M.C.; Nagaoka, T.; Karasawa, H.; Klauzinska, M.; Hewitt, S.M.; Biswas, K.; Sharan, S.K.; et al. Cripto-1 as a novel therapeutic target for triple negative breast cancer. Oncotarget 2015, 6, 11910–11929. [Google Scholar] [CrossRef]
- Wang, D.; Xu, J.; Liu, B.; He, X.; Zhou, L.; Hu, L.; Qiao, F.; Zhang, A.; Xu, X.; Zhang, H.; et al. IL6 blockade potentiates the anti-tumor effects of γ-secretase inhibitors in Notch3-expressing breast cancer. Cell Death Differ. 2018, 25, 330–339. [Google Scholar] [CrossRef]
- Doi, T.; Tajimi, M.; Mori, J.; Asou, H.; Inoue, K.; Benhadji, K.A.; Naito, Y. A phase 1 study of crenigacestat (LY3039478), the Notch inhibitor, in Japanese patients with advanced solid tumors. Investig. New Drugs 2021, 39, 469–476. [Google Scholar] [CrossRef]
- Azaro, A.; Massard, C.; Tap, W.D.; Cassier, P.A.; Merchan, J.; Italiano, A.; Anderson, B.; Yuen, E.; Yu, D.; Oakley, G.; et al. A phase 1b study of the Notch inhibitor crenigacestat (LY3039478) in combination with other anticancer target agents (taladegib, LY3023414, or abemaciclib) in patients with advanced or metastatic solid tumors. Investig. New Drugs. 2021, 39, 1089–1098. [Google Scholar] [CrossRef]
- Sorrentino, C.; Cuneo, A.; Roti, G. Therapeutic Targeting of Notch Signaling Pathway in Hematological Malignancies. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019037. [Google Scholar] [CrossRef]
- Even, C.; Lassen, U.; Merchan, J.; Tourneau, C.L.; Soria, J.C.; Ferte, C.; Ricci, F.; Diener, J.T.; Yuen, E.; Smith, C.; et al. Safety and clinical activity of the Notch inhibitor, crenigacestat (LY3039478), in an open-label phase I trial expansion cohort of advanced or metastatic adenoid cystic carcinoma. Investig. New Drugs 2020, 38, 402–409. [Google Scholar] [CrossRef]
- Jing, J.; Wu, Z.; Wang, J.; Luo, G.; Lin, H.; Fan, Y.; Zhou, C. Hedgehog signaling in tissue homeostasis, cancers, and targeted therapies. Signal Transduct. Target. Ther. 2023, 8, 315. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Cui, B.; Li, Q.; Zhao, X.; Huang, T.; Ding, X. The emerging roles of Hedgehog signaling in tumor immune microenvironment. Front. Oncol. Sec. Cancer Mol. Targets Ther. 2023, 13, 1171418. [Google Scholar] [CrossRef]
- Koike, Y.; Ohta, Y.; Saitoh, W.; Yamashita, T.; Kanomata, N.; Moriya, T.; Kurebayashi, J. Anti-cell growth and anti-cancer stem cell activities of the non-canonical hedgehog inhibitor GANT61 in triple-negative breast cancer cells. Breast Cancer 2017, 24, 683–693. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Fan, C.; Gao, P.; Wang, X.; Wei, G.; Wei, J. Estrogen promotes stemness and invasiveness of ER-positive breast cancer cells through Gli1 activation. Mol. Cancer 2014, 13, 137. [Google Scholar] [CrossRef]
- Solzak, J.P.; Atale, R.V.; Hancock, B.A.; Sinn, A.L.; Pollok, K.E.; Jones, D.R.; Radovich, M. Dual PI3K and Wnt pathway inhibition is a synergistic combination against triple negative breast cancer. NPJ Breast Cancer 2017, 17. [Google Scholar] [CrossRef]
- Ruiz-Borrego, M.; Jimenez, B.; Antolín, S.; García-Saenz, J.A.; Corral1, J.; Jerez, Y.; Trigo, J.; Urruticoechea, A.; Colom, H.; Gonzalo, N.; et al. A phase Ib study of sonidegib (LDE225), an oral small molecule inhibitor of smoothened or Hedgehog pathway, in combination with docetaxel in triple negative advanced breast cancer patients: GEICAM/2012-12 (EDALINE) study. Investig. New Drugs 2019, 37, 98–108. [Google Scholar] [CrossRef]
- Sekulic, A.; Migden, M.R.; Oro, A.E.; Dirix, L.; Lewis, K.D.; Hainsworth, J.D.; Solomon, J.A.; Yoo, S.; Arron, S.T.; Friedlander, P.A.; et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 2012, 366, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Vagiannis, D.; Budagaga, Y.; Sabet, Z.; Hanke, I.; Rozkoš, T.; Hofman, J. Sonidegib potentiates the cancer cells’ sensitivity to cytostatic agents by functional inhibition of ABCB1 and ABCG2 in vitro and ex vivo. Biochem. Pharmacol. 2022, 199, 115009. [Google Scholar] [CrossRef] [PubMed]
- Kurebayashi, J.; Koike, Y.; Ohta, Y.; Saitoh, W.; Yamashita, T.; Kanomata, N.; Moriya, T. Anti-cancer stem cell activity of a hedgehog inhibitor GANT61 in estrogen receptor-positive breast cancer cells. Cancer Sci. 2017, 108, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.K.; Khan, W.; Wang, F.; Khaliq, T.; Malik, A.; Razia, E.T.; Khan, J.S.; Haque, S.; Hashem, A.M.; Alkhayyat, S.S.; et al. Targeted Inhibition of Fibroblast Growth Factor Receptor 1-GLI Through AZD4547 and GANT61 Modulates Breast Cancer Progression. Front. Cell Dev. Biol. 2021, 9, 758400. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.K.; Ke, Y.; Wang, F.; Kayani, M.A.; Malik, M.F.A. Influence of SHH/GLI1 axis on EMT mediated migration and invasion of breast cancer cells. Sci. Rep. 2019, 9, 6620. [Google Scholar] [CrossRef] [PubMed]
- Mani, C.; Tripathi, K.; Omy, T.R.; Reedy, M.; Manne, U.; Palle, K. GLI1-targeting drugs induce replication stress and homologous recombination deficiency and synergize with PARP-targeted therapies in triple negative breast cancer cells. Biochim. Biophys. Acta Mol. Basis. Dis. 2022, 1868, 166300. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.K.; Khan, J.S.; Shah, S.T.A.; Wang, F.; Ye, L.; Jiang, W.G.; Malik, M.F.A. Involvement of hedgehog pathway in early onset, aggressive molecular subtypes and metastatic potential of breast cancer. Cell Commun. Signal. 2018, 16, 3. [Google Scholar] [CrossRef]
- Benvenuto, M.; Masuelli, L.; De Smaele, E.; Fantini, M.; Mattera, R.; Cucchi, D.; Bonanno, E.; Di Stefano, E.; Frajese, G.V.; Orlandi, A.; et al. In vitro and in vivo inhibition of breast cancer cell growth by targeting the Hedgehog/GLI pathway with SMO (GDC-0449) or GLI (GANT-61) inhibitors. Oncotarget 2016, 7, 9250–9270. [Google Scholar] [CrossRef]
- Bhateja, P.; Cherian, M.; Majumder, S.; Ramaswamy, B. The Hedgehog Signaling Pathway: A Viable Target in Breast Cancer? Cancers 2019, 11, 1126. [Google Scholar] [CrossRef]
- Fath, M.K.; Ebrahimi, M.; Nourbakhsh, E.; Hazara, A.Z.; Mirzaei, A.; Shafieyari, S.; Salehi, S.; Hoseinzadeh, M.; Payandeh, Z.; Barati, G. PI3K/Akt/mTOR signaling pathway in cancer stem cells. Pathol. Res. Pract. 2022, 237, 154010. [Google Scholar] [CrossRef]
- Cerma, K.; Piacentini, F.; Moscetti, L.; Barbolini, M.; Canino, F.; Tornincasa, A.; Caggia, F.; Cerri, S.; Molinaro, A.; Dominici, M.; et al. Targeting PI3K/AKT/mTOR Pathway in Breast Cancer: From Biology to Clinical Challenges. Biomedicines 2023, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Gennari1, A.; André, F.; Barrios, C.H.; Cortés, J.; de Azambuja, E.; DeMichele, A.; Dent, R.; Fenlon, D.; Gligorov, J.; Hurvitz, S.A.; et al. ESMO 1 ESMO Clinical Practice Guideline for the diagnosis, staging and treatment of patients with metastatic breast cancer. Ann. Oncol. 2021, 32, 1475–1495. [Google Scholar] [CrossRef]
- Wylaź, M.; Kaczmarska, A.; Pajor, D.; Hryniewicki, M.; Gil, D.; Dulińska-Litewka, J. Exploring the role of PI3K/AKT/mTOR inhibitors in hormone-related cancers: A focus on breast and prostate cancer. Biomed. Pharmacother. 2023, 168, 115676. [Google Scholar] [CrossRef] [PubMed]
- Moreau-Bachelard, C.; Robert, M.; Gourmelon, C.; Bourbouloux, E.; Patsouris, A.; Frenel, J.S.; Campone, M. Evaluating everolimus for the treatment of breast cancer. Expert Opin Pharmacother. 2023, 24, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Rozenblit, M.; Mun, S.; Soulos, P.; Adelson, K.; Pusztai, L.; Mougalian, S. Patterns of treatment with everolimus exemestane in hormone receptor-positive HER2-negative metastatic breast cancer in the era of targeted therapy. Breast Cancer Res. 2021, 23, 14. [Google Scholar] [CrossRef]
- Bachelot, T.; Cottu, P.; Chabaud, S.; Dalenc, F.; Allouache, D.; Delaloge, S.; Jacquin, J.P.; Grenier, J.; Venat Bouvet, L.; Jegannathen, A.; et al. Everolimus Added to Adjuvant Endocrine Therapy in Patients With High-Risk Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Primary Breast Cancer. J. Clin. Oncol. 2022, 40, 3699–3708. [Google Scholar] [CrossRef] [PubMed]
- François-Martin, H.; Lardy-Cléaud, A.; Pistilli, B.; Levy, C.; Diéras, V.; Frenel, J.S.; Guiu, S.; Mouret-Reynier, M.A.; Mailliez, A.; Eymard, J.C.; et al. Long-Term Results with Everolimus in Advanced Hormone Receptor Positive Breast Cancer in a Multicenter National Real-World Observational Study. Cancers 2023, 15, 1191. [Google Scholar] [CrossRef] [PubMed]
- Royce, M.E.; Osman, D. Everolimus in the Treatment of Metastatic Breast Cancer. Breast Cancer 2015, 9, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Yao, E.; Zhou, W.; Lee-Hoeflich, S.T.; Truong, T.; Haverty, P.M.; Eastham-Anderson, J.; Lewin-Koh, N.; Gunter, B.; Belvin, M.; Murray, L.J.; et al. Suppression of HER2/HER3-mediated growth of breast cancer cells with combinations of GDC-0941 PI3K inhibitor, trastuzumab, and pertuzumab. Clin. Cancer Res. 2009, 15, 4147–4156. [Google Scholar] [CrossRef]
- Schöffski, P.; Cresta, S.; Mayer, I.A.; Wildiers, H.; Damian, S.; Gendreau, S.; Rooney, I.; Morrissey, K.M.; Spoerke, J.M.; Ng, V.M.; et al. A phase Ib study of pictilisib (GDC-0941) in combination with paclitaxel, with and without bevacizumab or trastuzumab, and with letrozole in advanced breast cancer. Breast Cancer Res. 2018, 20, 109. [Google Scholar] [CrossRef]
- Wallin, J.J.; Guan, J.; Prior, W.W.; Lee, L.B.; Berry, L.; Belmont, L.D.; Koeppen, H.; Belvin, M.; Friedman, L.S.; Sampath, D. GDC-0941, a Novel Class I Selective PI3K Inhibitor, Enhances the Efficacy of Docetaxel in Human Breast Cancer Models by Increasing Cell Death In Vitro and In Vivo. Clin. Cancer Res. 2012, 18, 3901–3911. [Google Scholar] [CrossRef]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Armaghani, A.J.; Han, H.S. Alpelisib in the Treatment of Breast Cancer: A Short Review on the Emerging Clinical Data. Breast Cancer 2020, 12, 251–258. [Google Scholar] [CrossRef]
- The Editors of The Lancet Oncology. Expression of concern—Alpelisib plus fulvestrant in PIK3CA-mutated, hormone receptor-positive advanced breast cancer after a CDK4/6 inhibitor (BYLieve): One cohort of a phase 2, multicentre, open-label, non-comparative study. Lancet Oncol. 2023, 22, 489–498. [Google Scholar] [CrossRef]
- Eskiler, G.G.; Ozturk, M. Therapeutic potential of the PI3K inhibitor LY294002 and PARP inhibitor Talazoparib combination in BRCA-deficient triple negative breast cancer cells. Cell. Signal. 2022, 91, 110229. [Google Scholar] [CrossRef] [PubMed]
- Wheler, J.; Mutch, D.; Lager, J.; Castell, C.; Liu, L.; Jiang, J.; Traynor, A.M. Phase I Dose-Escalation Study of Pilaralisib (SAR245408, XL147) in Combination with Paclitaxel and Carboplatin in Patients with Solid Tumors. Oncologist 2017, 22, 377-e37. [Google Scholar] [CrossRef]
- Bendell, J.C.; Rodon, J.; Burris, H.A.; de Jonge, M.; Verweij, J.; Birle, D.; Demanse, D.; De Buck, S.; Ru, Q.C.; Peters, M.; et al. Dose-Escalation Study of BKM120, an Oral Pan-Class I PI3K Inhibitor, in Patients With Advanced Solid Tumors. JCO 2012, 30, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Foster, P.; Yamaguchi, K.; Hsu, P.P.; Qian, F.; Du, X.; Wu, J.; Won, K.-A.; Yu, P.; Jaeger, C.T.; Zhang, W.; et al. The Selective PI3K Inhibitor XL147 (SAR245408) Inhibits Tumor Growth and Survival and Potentiates the Activity of Chemotherapeutic Agents in Preclinical Tumor Models. Mol. Cancer Ther. 2015, 14, 931–940. [Google Scholar] [CrossRef]
- Song, Z.; Tu, X.; Zhou, Q.; Huang, J.; Chen, Y.; Liu, J.; Lee, S.B.; Kim, W.; Nowsheen, S.; Luo, K.; et al. A novel UCHL3 inhibitor, perifosine, enhances PARP inhibitor cytotoxicity through inhibition of homologous recombination-mediated DNA double strand break repair. Cell Death Dis. 2019, 10, 398. [Google Scholar] [CrossRef]
- Zhen, Y.; Yuan, Z.; Zhang, J.; Chen, Y.; Fu, Y.; Liu, Y.; Fu, L.; Zhang, L.; Zhou, X.L. Flubendazole induces mitochondrial dysfunction and DRP1-mediated mitophagy by targeting EVA1A in breast cancer. Cell Death Dis. 2022, 13, 375. [Google Scholar] [CrossRef]
- Hou, Z.-J.; Luo, X.; Zhang, W.; Peng, F.; Cui, B.; Wu, S.J.; Zheng, F.-M.; Xu, J.; Xu, L.-Z.; Long, Z.J.; et al. Flubendazole, FDA-approved anthelmintic, targets breast cancer stem-like cells. Oncotarget 2015, 6, 6326–6340. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, Y.J.; An, H.; Sun, G.D.; Cho, T.M.; Farrand, L.; Jang, S.; Seo, J.H.; Kim, J.Y. Flubendazole elicits anti-metastatic effects in triple-negative breast cancer via STAT3 inhibition. Int. J. Cancer 2018, 143, 1978–1993. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Osipo, C. Cancer stem cells and HER2 positive breast cancer: The story so far. Genes Dis. 2016, 3, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Shin, I. HER2 Signaling in Breast Cancer. Adv. Exp. Med. Biol. 2021, 1187, 53–79. [Google Scholar] [PubMed]
- Kim, Y.J.; Sung, D.; Oh, E.; Cho, Y.; Cho, T.M.; Farrand, L.; Seo, J.H.; Kim, J.Y. Flubendazole overcomes trastuzumab resistance by targeting cancer stem-like properties and HER2 signaling in HER2-positive breast cancer. Cancer Lett. 2018, 412, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative group (EBCTCG). Trastuzumab for early-stage, HER2-positive breast cancer: A meta-analysis of 13864 women in seven randomised trials. Lancet Oncol. 2021, 22, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Piccart-Gebhart, M.J.; Procter, M.; Leyland-Jones, B.; Goldhirsch, A.; Untch, M.; Smith, I.; Gianni, L.; Baselga, J.; Bell, R.; Jackisch, C.; et al. Herceptin Adjuvant (HERA) Trial Study Team. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1659–1672. [Google Scholar] [CrossRef]
- Amiri-Kordestani, L.; Wedam, S.; Zhang, L.; Tang, S.; Tilley, A.; Ibrahim, A.; Justice, R.; Pazdur, R.; Cortazar, P. First FDA approval of neoadjuvant therapy for breast cancer: Pertuzumab for the treatment of patients with HER2-positive breast cancer. Clin. Cancer Res. 2014, 20, 5359–5364. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Morii, N.; Yamashiro, H. Pertuzumab in the treatment of HER2-positive breast cancer: An evidence-based review of its safety, efficacy, and place in therapy. Core Evid. 2019, 14, 51–70. [Google Scholar] [CrossRef]
- Goldlust, S.A.; Chang, J.H.; Narita, Y.; Welch, Y.R.; Green, R.M.; Drappatz, J.; Piccioni, D.E.; Kim, Y.J.; Melear, J.M.; Tanaka, S.; et al. Ombipepimut dosing emulsion (ODE) + bevacizumab (bev) vs bev alone in patients (pts) with recurrent or progressive glioblastoma (rGBM). JCO 2023, 41. [Google Scholar] [CrossRef]
- Opdam, F.L.; Guchelaar, H.J.; Beijnen, J.H.; Schellens, J.H. Lapatinib for advanced or metastatic breast cancer. Oncologist 2012, 17, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Gui, X.; Li, H.; Yan, Y.; Zhang, R. Efficacy of lapatinib combined with capecitabine in patients with HER2-positive metastatic breast cancer in a real-world study. Oncol. Lett. 2020, 20, 378. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huang, X.; Sun, C.; Li, J.; Wang, B.; Yan, M.; Jin, F.; Wang, H.; Zhang, J.; Fu, P.; et al. Lapatinib in combination with capecitabine versus continued use of trastuzumab in breast cancer patients with trastuzumab-resistance: A retrospective study of a Chinese population. BMC Cancer 2020, 20, 255. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Liu, X.; Cai, Y.; Li, W. Lapatinib and lapatinib plus trastuzumab therapy versus trastuzumab therapy for HER2 positive breast cancer patients: An updated systematic review and meta-analysis. Syst. Rev. 2022, 11, 264. [Google Scholar] [CrossRef] [PubMed]
- von Minckwitz, G.; Huang, C.-S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Loibl, S.; Mamounas, E.; Untch, M.; et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; André, F.; Bachelot, T.; Barrios, C.H.; Bergh, J.; Burstein, H.J.; Cardoso, M.J.; Carey, L.A.; Dawood, S.; Del Mastro, L.; et al. Early breast cancer: ESMO Clinical Practice Guideline for diagnosis treatment and follow-up. Ann. Oncol. 2023, 35, 159–182. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Yu, Q.; Liu, Z.; Li, C.; Wang, F.; Yu, Z. The Effectiveness of Lapatinib in HER2-Positive Metastatic Breast Cancer Patients Pretreated with Multiline Anti-HER2 Treatment: A Retrospective Study in China. Technol. Cancer Res. Treat. 2021, 20, 15330338211037812. [Google Scholar] [CrossRef]
| Molecular Subtype | Frequency | IHC Characteristics | Prognosis |
|---|---|---|---|
| Luminal A | 40% | ER+/PR+/Ki-67-low | Very good |
| Luminal B | 20% | ER+/PR+/Ki-67-high | Good |
| HER2-enriched | 10–15% | ER−/PR−/HER2+ | Poor |
| Basal/TNBC | 15–20% | ER−/PR−/HER2− | Very poor |
| BCSC Subtypes | |||||
|---|---|---|---|---|---|
| Biomarkers | Luminal A | Luminal B | HER-2 | Basal | TNBC |
| CD44+/CD24− | - | - | - | √ | √ BL2, M |
| CD133 | √ | √ | √ | √ | √ |
| ALDH1+ | - | √ | - | √ | √ BL1 |
| miR-200a | √ | - | - | - | - |
| miR-200c | √ | - | - | - | - |
| miR-9 | - | - | √ | - | √ |
| miR-141 | √ | - | - | - | - |
| miR-155 | - | - | - | - | √ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romaniuk-Drapała, A.; Totoń, E.; Taube, M.; Idzik, M.; Rubiś, B.; Lisiak, N. Breast Cancer Stem Cells and Tumor Heterogeneity: Characteristics and Therapeutic Strategies. Cancers 2024, 16, 2481. https://doi.org/10.3390/cancers16132481
Romaniuk-Drapała A, Totoń E, Taube M, Idzik M, Rubiś B, Lisiak N. Breast Cancer Stem Cells and Tumor Heterogeneity: Characteristics and Therapeutic Strategies. Cancers. 2024; 16(13):2481. https://doi.org/10.3390/cancers16132481
Chicago/Turabian StyleRomaniuk-Drapała, Aleksandra, Ewa Totoń, Magdalena Taube, Malgorzata Idzik, Błażej Rubiś, and Natalia Lisiak. 2024. "Breast Cancer Stem Cells and Tumor Heterogeneity: Characteristics and Therapeutic Strategies" Cancers 16, no. 13: 2481. https://doi.org/10.3390/cancers16132481







