A Molecular Hypothesis on Malignant Transformation of Oral Lichen Planus: A Systematic Review and Meta-Analysis of Cancer Hallmarks Expression in This Oral Potentially Malignant Disorder
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol
2.2. Search Strategy
2.3. Eligibility Criteria
2.4. Study Selection Process
2.5. Data Extraction
2.6. Appraisal of Quality and Risk of Bias
2.7. Statistical Analysis
3. Results
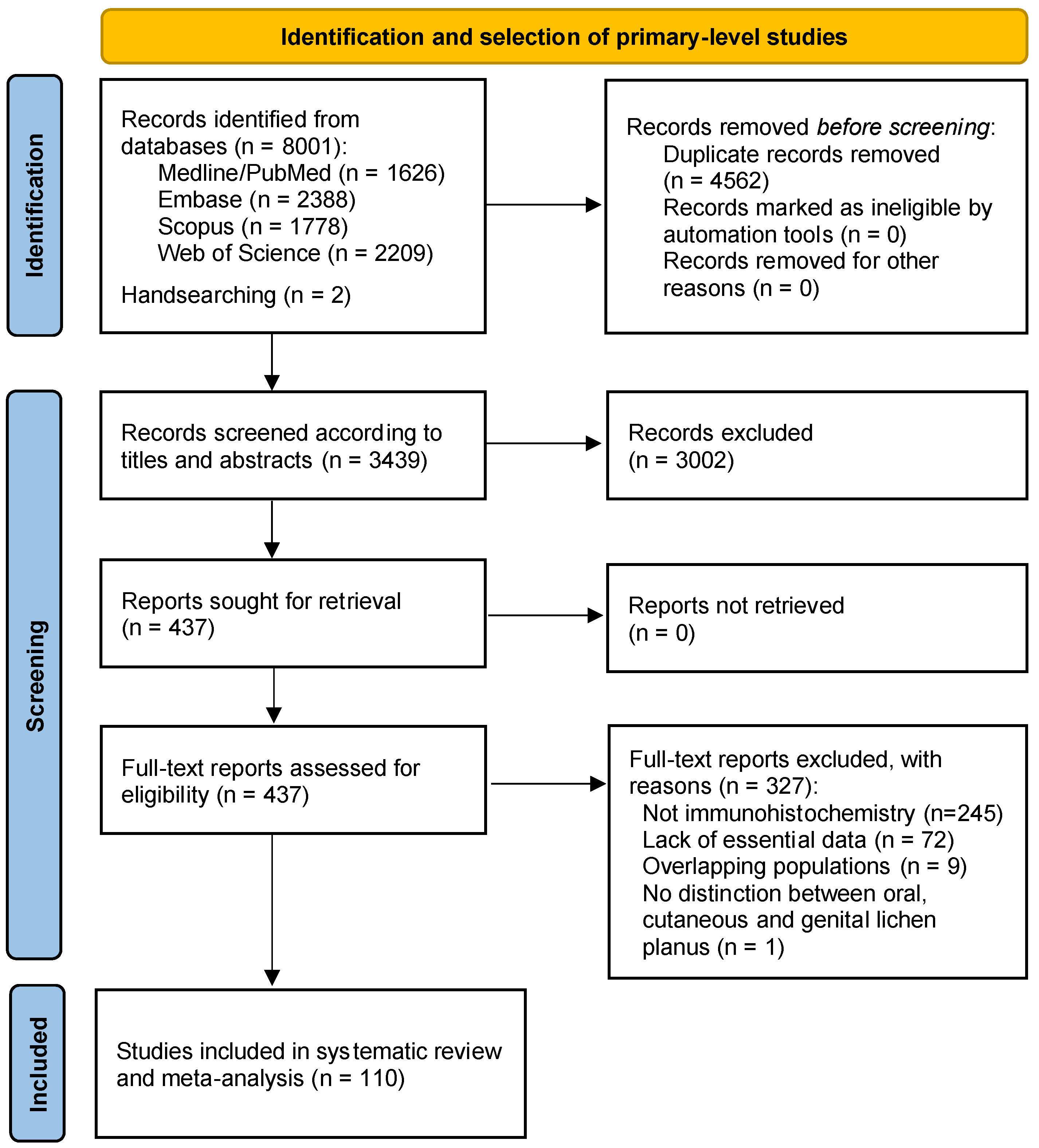
3.1. Results of the Literature Search
3.2. Study Characteristics
3.3. Qualitative Evaluation
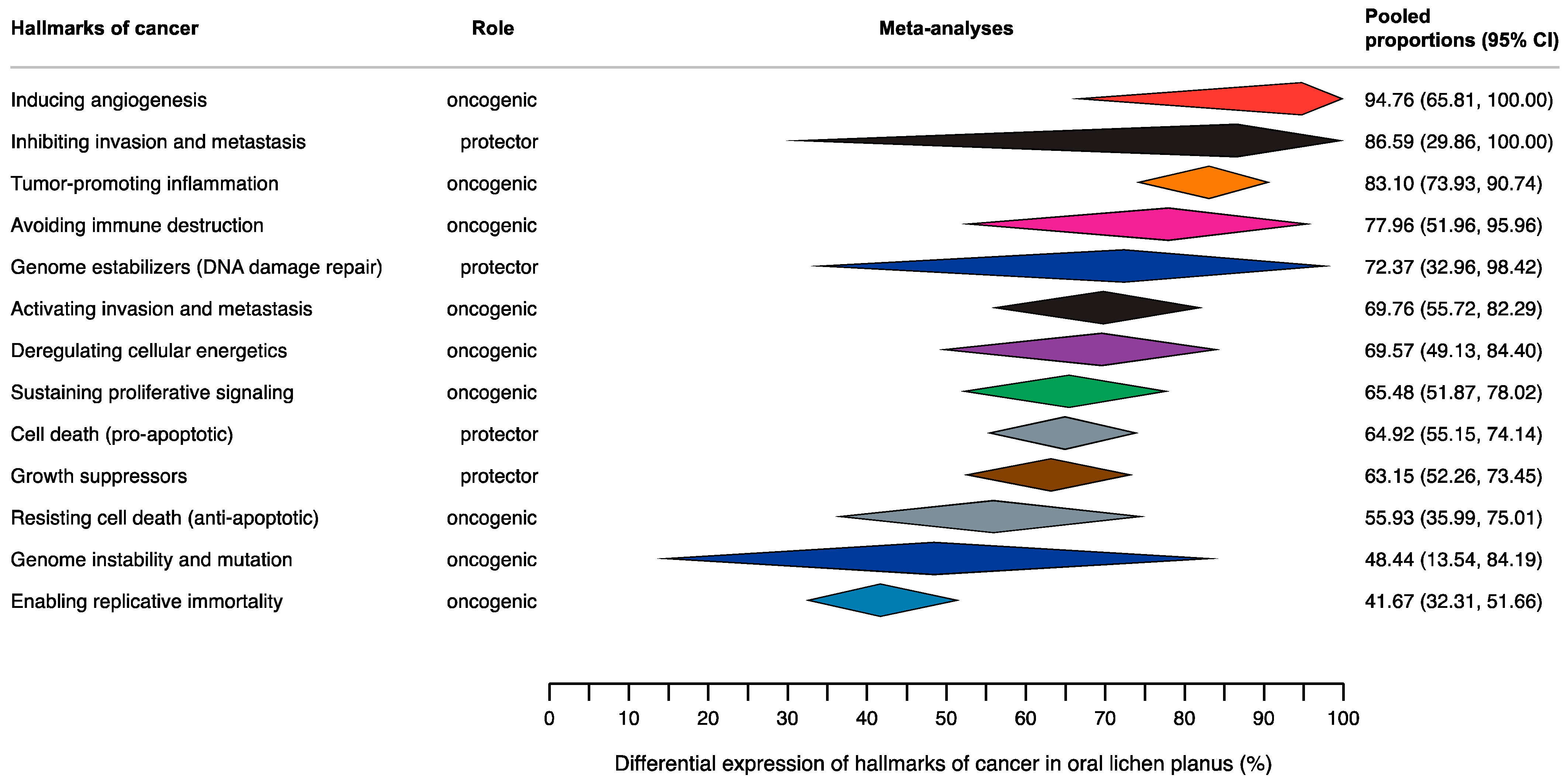
3.4. Quantitative Evaluation (Meta-Analysis)
3.4.1. Hallmark 1: Sustaining Proliferative Signaling
3.4.2. Hallmark 2: Evading Growth Suppressors
3.4.3. Hallmark 3: Resisting Cell Death
3.4.4. Hallmark 4: Enabling Replicative Immortality
3.4.5. Hallmark 5: Inducing Angiogenesis
3.4.6. Hallmark 6: Activating Invasion and Metastasis
3.4.7. Hallmark 7: Avoiding Immune Destruction
3.4.8. Hallmark 8: Deregulating Cellular Energetics
3.4.9. Hallmark 9: Genome Instability and Mutation
3.4.10. Hallmark 10: Tumor-Promoting Inflammation
3.4.11. Unspecified
3.5. Analysis of Small-Study Effects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- González-Moles, M.Á.; Ramos-García, P. Oral lichen planus and related lesions. What should we accept based on the available evidence? Oral Dis. 2023, 29, 2624–2637. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Warnakulasuriya, S.; González-Ruiz, I.; González-Ruiz, L.; Ayén, Á.; Lenouvel, D.; Ruiz-Ávila, I.; Ramos-García, P. Worldwide prevalence of oral lichen planus: A systematic review and meta-analysis. Oral Dis. 2021, 27, 813–828. [Google Scholar] [CrossRef]
- Gonzalez-Moles, M.A.; Scully, C.; Gil-Montoya, J.A. Oral lichen planus: Controversies surrounding malignant transformation. Oral Dis. 2008, 14, 229–243. [Google Scholar] [CrossRef]
- Warnakulasuriya, S.; Kujan, O.; Aguirre-Urizar, J.M.; Bagan, J.V.; González-Moles, M.Á.; Kerr, A.R.; Lodi, G.; Mello, F.W.; Monteiro, L.; Ogden, G.R.; et al. Oral potentially malignant disorders: A consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. 2021, 27, 1862–1880. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Ruiz-Ávila, I.; González-Ruiz, L.; Ayén, Á.; Gil-Montoya, J.A.; Ramos-García, P. Malignant transformation risk of oral lichen planus: A systematic review and comprehensive meta-analysis. Oral Oncol. 2019, 96, 121–130. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Ramos-García, P.; Warnakulasuriya, S. An appraisal of highest quality studies reporting malignant transformation of oral lichen planus based on a systematic review. Oral Dis. 2021, 27, 1908–1918. [Google Scholar] [CrossRef] [PubMed]
- Iocca, O.; Sollecito, T.P.; Alawi, F.; Weinstein, G.S.; Newman, J.G.; De Virgilio, A.; Di Maio, P.; Spriano, G.; Pardiñas López, S.; Shanti, R.M. Potentially malignant disorders of the oral cavity and oral dysplasia: A systematic review and meta-analysis of malignant transformation rate by subtype. Head Neck 2019, 42, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, M.; Troiano, G.; Cordaro, M.; Corsalini, M.; Gioco, G.; Lo Muzio, L.; Pignatelli, P.; Lajolo, C. Rate of malignant transformation of oral lichen planus: A systematic review. Oral Dis. 2018, 25, 693–709. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Kujan, O.; Shearston, K.; Farah, C.S. Oral lichen planus has a very low malignant transformation rate: A systematic review and meta-analysis using strict diagnostic and inclusion criteria. J. Oral Pathol. Med. 2020, 50, 287–298. [Google Scholar] [CrossRef]
- Aghbari, S.M.H.; Abushouk, A.I.; Attia, A.; Elmaraezy, A.; Menshawy, A.; Ahmed, M.S.; Elsaadany, B.A.; Ahmed, E.M. Malignant transformation of oral lichen planus and oral lichenoid lesions: A meta-analysis of 20095 patient data. Oral Oncol. 2017, 68, 92–102. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Ramos-García, P. An Evidence-Based Update on the Potential for Malignancy of Oral Lichen Planus and Related Conditions: A Systematic Review and Meta-Analysis. Cancers 2024, 16, 608. [Google Scholar] [CrossRef] [PubMed]
- Ramos-García, P.; Gonzalez-Moles, M.A.; Warnakulasuriya, S. Oral cancer development in lichen planus and related conditions -3.0 evidence level-: A systematic review of systematic reviews. Oral Dis. 2021, 27, 1919–1935. [Google Scholar] [CrossRef] [PubMed]
- De Porras-Carrique, T.; Ramos-García, P.; Aguilar-Diosdado, M.; Warnakulasuriya, S.; González-Moles, M.Á. Autoimmune disorders in oral lichen planus: A systematic review and meta-analysis. Oral Dis. 2023, 29, 1382–1394. [Google Scholar] [CrossRef]
- González-Moles, M.A.; Rodríguez-Archilla, A.; Ruiz Avila, I.; Esteban, F.; González-Moles, S.; Bravo, M. Presence of HPV 16 sequences in oral lichen planus lesions. Bull. Group. Int. Rech. Sci. Stomatol. Odontol. 1998, 40, 92–97. [Google Scholar]
- González-Moles, M.A.; González-Moles, S.; Ruiz-Avila, I.; Esteban, F.; Galindo-Moreno, P.; Rodríguez-Archilla, A. Epithelial response to the immunitary aggression in oral lichen planus. Acta Stomatol. Belg. 1996, 93, 119–123. [Google Scholar] [PubMed]
- Pérez-Sayáns, M.; Lorenzo-Pouso, A.I.; Chamorro-Petronacci, C.M.; Suárez-Peñaranda, J.M.; Padín-Iruegas, E.; González-Moles, M.A.; Marichalar-Mendía, X.; García-García, A.; Blanco-Carrión, A. Immunoexpression of Apoptosis and Cell-cycle Arrest Markers in Oral Lichen Planus. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 374–381. [Google Scholar] [CrossRef] [PubMed]
- González-Moles, M.A.; Bascones-Ilundain, C.; Gil Montoya, J.A.; Ruiz-Avila, I.; Delgado-Rodríguez, M.; Bascones-Martínez, A. Cell cycle regulating mechanisms in oral lichen planus: Molecular bases in epithelium predisposed to malignant transformation. Arch. Oral Biol. 2006, 51, 1093–1103. [Google Scholar] [CrossRef]
- Bascones-Ilundain, C.; Gonzalez-Moles, M.A.; Esparza, G.; Gil-Montoya, J.A.; Bascones-Martinez, A. Significance of liquefaction degeneration in oral lichen planus: A study of its relationship with apoptosis and cell cycle arrest markers. Clin. Exp. Dermatol. 2007, 32, 556–563. [Google Scholar] [CrossRef]
- Bascones-Ilundain, C.; Gonzalez-Moles, M.A.; Esparza-Gómez, G.; Gil-Montoya, J.A.; Bascones-Martínez, A. Importance of apoptotic mechanisms in inflammatory infiltrate of oral lichen planus lesions. Anticancer Res. 2006, 26, 357–362. [Google Scholar]
- González Moles, M.A.; Esteban, F.; Ruiz-Ávila, I.; Gil Montoya, J.A.; Brener, S.; Bascones-Martínez, A.; Muñoz, M.; Ruiz-Ávila, I.; Gil Montoya, J.A.; Brener, S.; et al. A role for the substance P/NK-1 receptor complex in cell proliferation and apoptosis in oral lichen planus. Oral Dis. 2009, 15, 162–169. [Google Scholar] [CrossRef]
- Gonzalez-Moles, M.A.; Gil-Montoya, J.A.; Ruiz-Avila, I.; Esteban, F.; Bascones-Martinez, A. Differences in the expression of p53 protein in oral lichen planus based on the use of monoclonal antibodies DO7 and pAb 240. Oral Oncol. 2008, 44, 496–503. [Google Scholar] [CrossRef]
- Ramos-García, P.; González-Moles, M.Á.; Warnakulasuriya, S. Significance of p53 overexpression in the prediction of the malignant transformation risk of oral potentially malignant disorders: A systematic review and meta-analysis. Oral Oncol. 2022, 126, 105734. [Google Scholar] [CrossRef]
- Fitzpatrick, S.G.; Hirsch, S.A.; Gordon, S.C. The malignant transformation of oral lichen planus and oral lichenoid lesions: A systematic review. J. Am. Dent. Assoc. 2014, 145, 45–56. [Google Scholar] [CrossRef]
- Califano, J.; van der Riet, P.; Westra, W.; Nawroz, H.; Clayman, G.; Piantadosi, S.; Corio, R.; Lee, D.; Greenberg, B.; Koch, W.; et al. Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 1996, 56, 2488–2492. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-analysis of observational studies in epidemiology: A proposal for reporting. J. Am. Med. Assoc. 2000, 283, 2008–2012. [Google Scholar] [CrossRef]
- Higgins, J.P.; Green, S. Cochrane Handbook for Systematic Reviews of Interventions; Cochrane Book Series; Cochrane Collaboration: London, UK, 2023. [Google Scholar]
- Aromataris, E.; Munn, Z. (Eds.) JBI Manual for Evidence Synthesis; JBI: Adelaide, Australia, 2020; ISBN 9780648848806. [Google Scholar]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; Altman, D.G.; Booth, A.; et al. Preferred reporting items for systematic review and meta-analysis protocols (prisma-p) 2015: Elaboration and explanation. BMJ 2015, 349, g7647. [Google Scholar] [CrossRef] [PubMed]
- Munn, Z.; Moola, S.; Riitano, D.; Lisy, K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int. J. Health Policy Manag. 2014, 3, 123–128. [Google Scholar] [CrossRef]
- Agresti, A.; Coull, B.A. Approximate is Better than “Exact” for Interval Estimation of Binomial Proportions. Am. Stat. 1998, 52, 119–126. [Google Scholar] [CrossRef]
- Freeman, M.F.; Tukey, J.W. Transformations Related to the Angular and the Square Root. Ann. Math. Stat. 1950, 21, 607–611. [Google Scholar] [CrossRef]
- Miller, J.J. The Inverse of the Freeman—Tukey Double Arcsine Transformation. Am. Stat. 1978, 32, 138. [Google Scholar] [CrossRef]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Borenstein, M.; Hedges, L.V.; Higgins, J.P.T.; Rothstein, H.R. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res. Synth. Methods 2010, 1, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Borenstein, M.; Higgins, J.P.T. Meta-Analysis and Subgroups. Prev. Sci. 2013, 14, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Redder, C.P.; Pandit, S.; Desai, D.; Kandagal, V.S.; Ingaleshwar, P.S.; Shetty, S.J.; Vibhute, N. Comparative analysis of cell proliferation ratio in plaque and erosive oral lichen planus: An immunohistochemical study. Dent. Res. J. 2014, 11, 316–320. [Google Scholar]
- DU, Y.; LI, H. Expression of E-cadherin in oral lichen planus. Exp. Ther. Med. 2015, 10, 1544–1548. [Google Scholar] [CrossRef]
- Radwan-Oczko, M.; Bar, J.; Hałoń, A.; Lis-Nawara, A. Comparison of biomarker expression in oral lichen planus and oral lichenoid lesions. Adv. Clin. Exp. Med. 2022, 31, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Squarzanti, D.F.; Cena, T.; Sorrentino, R.; Migliario, M.; Chiocchetti, A.; Rimondini, L.; Azzimonti, B.; Valente, G. Implications on pathogenesis and risk of oral lichen planus neoplastic transformation: An ex-vivo retrospective immunohistochemical study. Histol. Histopathol. 2019, 34, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Sheelam, S.; Reddy, S.P.; Kulkarni, P.G.; Nandan, S.; Keerthi, M.; Raj, G.S. Role of cell proliferation and vascularity in malignant transformation of potentially malignant disorders. J. Oral Maxillofac. Pathol. 2018, 22, 281. [Google Scholar] [CrossRef]
- Shen, L.; Ruan, P.; Xie, F.; Zhao, T. Expressions of Fas/FasL and granzyme B in oral lichen planus and their significance. Di Yi Jun Yi Da Xue Xue Bao 2004, 24, 1362–1366. [Google Scholar]
- Siponen, M.; Kullaa, A.; Nieminen, P.; Salo, T.; Pasonen-Seppänen, S. Altered expression of hyaluronan, HAS1-2, and HYAL1-2 in oral lichen planus. J. Oral Pathol. Med. 2015, 44, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Ögmundsdóttir, H.M.; Björnsson, J.; Holbrook, W.P. Role of TP53 in the progression of pre-malignant and malignant oral mucosal lesions. A follow-up study of 144 patients. J. Oral Pathol. Med. 2009, 38, 565–571. [Google Scholar] [CrossRef]
- Ghallab, N.A.; Kasem, R.F.; El-Ghani, S.F.A.; Shaker, O.G. Gene expression of miRNA-138 and cyclin D1 in oral lichen planus. Clin. Oral Investig. 2017, 21, 2481–2491. [Google Scholar] [CrossRef] [PubMed]
- Kouhsoltani, M.; Aghbali, A.; Shokoohi, B.; Ahmadzadeh, R. Molecular Targeting of Her-2/neu Protein Is Not Recommended as an Adjuvant Therapy in Oral Squamous Cell Carcinoma and Oral Lichen Planus. Adv. Pharm. Bull. 2015, 5, 649–652. [Google Scholar] [CrossRef]
- Girod, S.C.; Cesarz, D.; Fischer, U.; Krueger, G.R. Detection of p53 and MDM2 protein expression in head and neck carcinogenesis. Anticancer Res. 1995, 15, 1453–1457. [Google Scholar]
- Babiuch, K.; Kuśnierz-Cabala, B.; Kęsek, B.; Okoń, K.; Darczuk, D.; Chomyszyn-Gajewska, M. Evaluation of proinflammatory, nf-kappab dependent cytokines: Il-1α, Il-6, Il-8, and TNF-α in tissue specimens and saliva of patients with oral squamous cell carcinoma and oral potentially malignant disorders. J. Clin. Med. 2020, 9, 867. [Google Scholar] [CrossRef] [PubMed]
- Zyada, M.M.; Fikry, H.E. Immunohistochemical study of syndecan-1 down-regulation and the expression of P35 protein in oral lichen planus: A clinicopathologic correlation with hepatitis C infection in the Egyptian population. Ann. Diagn. Pathol. 2010, 14, 153–161. [Google Scholar] [CrossRef]
- Nafarzadeh, S.; Jafari, S.; Bijani, A. Assessment of bax and bcl-2 immunoexpression in patients with oral lichen planus and oral squamous cell carcinoma. Int. J. Mol. Cell. Med. 2013, 2, 136–142. [Google Scholar] [PubMed]
- Ali, A.; Langdon, J.; Stern, P.; Partridge, M. The pattern of expression of the 5T4 oncofoetal antigen on normal, dysplastic and malignant oral mucosa. Oral Oncol. 2001, 37, 57–64. [Google Scholar] [CrossRef]
- Kitkhajornkiat, A.; Rungsiyanont, S.; Talungchit, S.; Jirawechwongsakul, P.; Taebunpakul, P. The expression of Cathepsin L in oral lichen planus. J. Oral Biol. Craniofacial Res. 2020, 10, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Pigatti, F.M.; Taveira, L.A.d.A.; Soares, C.T. Immunohistochemical expression of Bcl-2 and Ki-67 in oral lichen planus and leukoplakia with different degrees of dysplasia. Int. J. Dermatol. 2015, 54, 150–155. [Google Scholar] [CrossRef]
- Angelin, D.; Nair, B.J. Comparative evaluation of survivin expression in leukoplakia, lichen planus, and oral squamous cell carcinoma: An immunohistochemical study. J. Cancer Res. Ther. 2020, 16, 569–574. [Google Scholar] [CrossRef]
- Goel, S.; Khurana, N.; Marvah, A.; Gupta, S. Expression of cdk4 and p16 in Oral Lichen Planus. J. Oral Maxillofac. Res. 2015, 6, e4. [Google Scholar] [CrossRef][Green Version]
- Danielsson, K.; Ebrahimi, M.; Wahlin, Y.B.; Nylander, K.; Boldrup, L. Increased levels of COX-2 in oral lichen planus supports an autoimmune cause of the disease. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 1415–1419. [Google Scholar] [CrossRef]
- Bascones, C.; Gonzalez-Moles, M.A.; Esparza, G.; Bravo, M.; Acevedo, A.; Gil-Montoya, J.A.; Bascones, A. Apoptosis and cell cycle arrest in oral lichen planus. Arch. Oral Biol. 2005, 50, 873–881. [Google Scholar] [CrossRef]
- Salehinejad, J.; Sharifi, N.; Amirchaghmaghi, M.; Ghazi, N.; Shakeri, M.T.; Ghazi, A. Immunohistochemical expression of p16 protein in oral squamous cell carcinoma and lichen planus. Ann. Diagn. Pathol. 2014, 18, 210–213. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, L.A.; Manni, J.J.; Ramaekers, F.C.; Kuijpers, W. Expression of intermediate filament proteins in benign lesions of the oral mucosa. Eur. Arch. Otorhinolaryngol. 1999, 256, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, W.; Geng, N.; Tian, K.; Jack Windsor, L. MMPs, TIMP-2, and TGF-β1 in the cancerization of oral lichen planus. Head Neck 2008, 30, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Sridevi, U.; Jain, A.; Nagalaxmi, V.; Kumar, U.V.; Goyal, S. Expression of E-cadherin in normal oral mucosa, in oral precancerous lesions and in oral carcinomas. Eur. J. Dent. 2015, 9, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Costa, N.L.; Gonçalves, J.A.M.; de Lima, S.L.G.; de Arruda, J.A.A.; Miranda, A.C.C.; Mesquita, R.A.; da Silveira, É.J.D.; Batista, A.C. Evaluation of PD-L1, PD-L2, PD-1 and cytotoxic immune response in oral lichen planus. Oral Dis. 2020, 26, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Carnelio, S.; Rodrigues, G. Immunohistochemical and clinical significance of matrix metalloproteinase-2 and its inhibitor in oral lichen planus. J. Oral Maxillofac. Pathol. 2019, 23, 476. [Google Scholar] [CrossRef] [PubMed]
- Aghili, S.S.; Zare, R.; Jahangirnia, A. Evaluation of Paxillin Expression in Epithelial Dysplasia, Oral Squamous Cell Carcinoma, Lichen Planus with and without Dysplasia, and Hyperkeratosis: A Retrospective Cross-Sectional Study. Diagnostics 2023, 13, 2476. [Google Scholar] [CrossRef] [PubMed]
- Martín-Ezquerra, G.; Salgado, R.; Toll, A.; Gilaberte, M.; Baró, T.; Alameda Quitllet, F.; Yébenes, M.; Solé, F.; Garcia-Muret, M.; Espinet, B.; et al. Multiple genetic copy number alterations in oral squamous cell carcinoma: Study of MYC, TP53, CCDN1, EGFR and ERBB2 status in primary and metastatic tumours. Br. J. Dermatol. 2010, 163, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Hadzi-Mihailovic, M.; Petrovic, R.; Raybaud, H.; Stanimirovic, D.; Ozar Koray, M. Expression and role of p53 in oral lichen planus patients. J. BUON 2017, 22, 1278–1286. [Google Scholar]
- Yadav, M.; Arivananthan, M.; Chandrashekran, A.; Tan, B.S.; Hashim, B.Y. Human herpesvirus-6 (HHV-6) DNA and virus-encoded antigen in oral lesions. J. Oral Pathol. Med. 1997, 26, 393–401. [Google Scholar] [CrossRef]
- Hu, X.S.; Huang, Y.H.; Liu, X.S.; Hua, H. Expression and significance of p38 mitogen-activated protein kinase in oral lichen planus and oral squamous cell cacinoma. Beijing Da Xue Xue Bao 2016, 48, 310–315. [Google Scholar]
- Sun, L.; Feng, J.; Ma, L.; Liu, W.; Zhou, Z. CD133 expression in oral lichen planus correlated with the risk for progression to oral squamous cell carcinoma. Ann. Diagn. Pathol. 2013, 17, 486–489. [Google Scholar] [CrossRef]
- Li, H.-B.; Zhang, Y.-H.; Chen, H.-Z.; Chen, Y. Expression of human DNA mismatch-repair protein, hMSH2, in patients with oral lichen planus. Exp. Ther. Med. 2015, 9, 203–206. [Google Scholar] [CrossRef][Green Version]
- Thongprasom, K.; Dhanuthai, K.; Sarideechaigul, W.; Chaiyarit, P.; Chaimusig, M. Expression of TNF-α in oral lichen planus treated with fluocinolone acetonide 0.1%. J. Oral Pathol. Med. 2006, 35, 161–166. [Google Scholar] [CrossRef]
- Miri-Moghaddam, M.; Kadeh, H. Immunohistochemical Expression of Stromelysin-2 (St-2) In Patients with Oral Lichen Planus and Its Clinical Significance. J. Dent. 2016, 17, 250–255. [Google Scholar]
- Neppelberg, E.; Johannessen, A.C. DNA content, Cyclooxygenase-2 expression and loss of E-cadherin expression do not predict risk of malignant transformation in oral lichen planus. Eur. Arch. Oto-Rhino-Laryngol. 2007, 264, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Tanda, N.; Mori, S.; Saito, K.; Ikawa, K.; Sakamoto, S. Expression of apoptotic signaling proteins in leukoplakia and oral lichen planus: Quantitative and topographical studies. J. Oral Pathol. Med. 2000, 29, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Pirkić, A.; Biocina-Lukenda, D.; Cekić-Arambasin, A.; Buković, D.; Pavelić, L.; Sakić, S. Changes in the tissue expression of the C-erbB-2 oncogen in the oral lichen ruber. Coll. Antropol. 2004, 28, 455–461. [Google Scholar]
- Fan, Y.; Zhan, Z.; Peng, T.; Song, X.; Feng, Z. The expression of apoptosis-associated proteins Bcl-2, Bax in oral leukoplakia and lichen planus. Shanghai Kou Qiang Yi Xue 2004, 13, 497–501. [Google Scholar]
- Shiva, A.; Zamanian, A.; Arab, S.; Boloki, M. Immunohistochemical Study of p53 Expression in Patients with Erosive and Non-Erosive Oral Lichen Planus. J. Dent. 2018, 19, 118–123. [Google Scholar]
- Satelur, K.P.; Bopaiah, S.; Bavle, R.M.; Ramachandra, P. Role of Cathepsin B as a Marker of Malignant Transformation in Oral Lichen Planus: An Immunohistochemical Study. J. Clin. Diagn. Res. 2017, 11, ZC29–ZC32. [Google Scholar] [CrossRef]
- Shailaja, G.; Kumar, J.V.; Baghirath, P.V.; Kumar, U.; Ashalata, G.; Krishna, A.B. Estimation of malignant transformation rate in cases of oral epithelial dysplasia and lichen planus using immunohistochemical expression of Ki-67, p53, BCL-2, and BAX markers. Dent. Res. J. 2015, 12, 235–242. [Google Scholar]
- Shi, P.; Liu, W.; Zhou, Z.-T.; He, Q.-B.; Jiang, W.-W. Podoplanin and ABCG2: Malignant Transformation Risk Markers for Oral Lichen Planus. Cancer Epidemiol. Biomark. Prev. 2010, 19, 844–849. [Google Scholar] [CrossRef][Green Version]
- Sudha, V.M.; Hemavathy, S. Role of bcl-2 oncoprotein in oral potentially malignant disorders and squamous cell carcinoma: An immunohistochemical study. Indian J. Dent. Res. 2011, 22, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Alves, M.; Balducci, I.; Rodarte Carvalho, Y.; Cabral, L.; Nunes, F.; Almeida, J. Evaluation of the expression of p53, MDM2, and SUMO-1 in oral lichen planus. Oral Dis. 2013, 19, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Peter, C.D.; Shashidara, R.; Jain, V.; Haragannavar, V.C.; Samuel, P.; Nayak, S.R. Senescence in oral lichen planus as assessed by the immunohistochemical evaluation of senescence marker protein-30 (Regucalcin). Indian J. Pathol. Microbiol. 2023, 66, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-R.; Chen, L.-Y.; Qi, H.-Y.; Sun, S.-H. Expression and clinical significance of periostin in oral lichen planus. Exp. Ther. Med. 2018, 15, 5141–5147. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Fu, X.; Lv, H. The expression of EGFR in oral lichen planus, squamous cell papilloma and squamous cell carcinoma. Shanghai Kou Qiang Yi Xue 2012, 21, 673–676. [Google Scholar] [PubMed]
- Basheer, S.; Shameena, P.M.; Sudha, S.; Varma, S.; Vidyanath, S.; Varekar, A. Expression of survivin and p53 in oral lichen planus, lichenoid reaction and lichenoid dysplasia: An immunohistochemical study. J. Oral Maxillofac. Pathol. 2017, 21, 456–457. [Google Scholar] [CrossRef]
- Montebugnoli, L.; Venturi, M.; Gissi, D.B.; Leonardi, E.; Farnedi, A.; Foschini, M.P. Immunohistochemical expression of p16INK4A protein in oral lichen planus. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2011, 112, 222–227. [Google Scholar] [CrossRef]
- Ögmundsdóttir, H.M.; Hilmarsdóttir, H.; Björnsson, J.; Holbrook, W.P. Longitudinal study of TP53 mutations in eight patients with potentially malignant oral mucosal disorders. J. Oral Pathol. Med. 2009, 38, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Ramírez, D.A.; Rodríguez-Tojo, M.J.; Gainza-Cirauqui, M.L.; Martínez-Conde, R.; Aguirre-Urizar, J.M. Overexpression of cyclooxygenase-2 as a biomarker in different subtypes of the oral lichenoid disease. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2010, 110, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Kłosek, S.K.; Sporny, S.; Stasikowska-Kanicka, O.; Kurnatowska, A.J. Cigarette smoking induces overexpression of c-Met receptor in microvessels of oral lichen planus. Arch. Med. Sci. 2011, 4, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Nagao, T.; Maeda, H.; Kameyama, Y.; Warnakulasuriya, K.A.A.S. Epithelial cell proliferation in oral lichen planus. Cell Prolif. 2002, 35 (Suppl. 1), 103–109. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Shen, L.; Yin, C. Expression of caspase-8, receptor interacting protein and nuclear factor-kappaBp65 in oral lichen planus. Zhonghua Kou Qiang Yi Xue Za Zhi 2010, 45, 11–15. [Google Scholar] [PubMed]
- Bombeccari, G.P.; Giannì, A.B.; Spadari, F. Immunoexpression of cytokeratin-19 in the oral lichen planus and related oral squamous cell carcinoma. Ann. Stomatol. 2017, 8, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Jaafari-Ashkavandi, Z.; Aslani, E. Caveolin-1 expression in oral lichen planus, dysplastic lesions and squamous cell carcinoma. Pathol. Res. Pract. 2017, 213, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-J.; Cui, J. COX-2, MMP-7 expression in oral lichen planus and oral squamous cell carcinoma. Asian Pac. J. Trop. Med. 2013, 6, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-M.; Huang, X.-Y.; Guan, W.-Q. Expressions of Interleukin-27 in Oral Lichen Planus, Oral Leukoplakia, and Oral Squamous Cell Carcinoma. Inflammation 2022, 45, 1023–1038. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hu, Q.; Jiang, C.; Guo, F.; Munnee, K.; Jian, X.; Hu, Y.; Tang, Z. Cys-X-Cys ligand 9 might be an immunological factor in the pathogenesis of oral submucous fibrosis and its concomitant oral lichenoid lesion. Clin. Oral Investig. 2013, 17, 1251–1258. [Google Scholar] [CrossRef]
- Hsieh, P.-C.; Chen, Y.-K.; Tsai, K.-B.; Shieh, T.-Y.; Chang, Y.-Y.; Chang, J.-G.; Wu, H.-L.; Lin, S.-F. Expression of BUBR1 in human oral potentially malignant disorders and squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2010, 109, 257–267. [Google Scholar] [CrossRef]
- Siponen, M.; Bitu, C.C.; Al-Samadi, A.; Nieminen, P.; Salo, T. Cathepsin K expression is increased in oral lichen planus. J. Oral Pathol. Med. 2016, 45, 758–765. [Google Scholar] [CrossRef]
- Zhang, L.; Mao, E.; Priddy, R.; Rosin, M. p53 overexpression in oral lichen planus. Oncol. Rep. 1996, 3, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Poomsawat, S.; Buajeeb, W.; Khovidhunkit, S.P.; Punyasingh, J. Overexpression of cdk4 and p16 in oral lichen planus supports the concept of premalignancy. J. Oral Pathol. Med. 2011, 40, 294–299. [Google Scholar] [CrossRef]
- Younes, F.; Quartey, E.L.; Kiguwa, S.; Partridge, M. Expression of TNF and the 55-kDa TNF receptor in epidermis, oral mucosa, lichen planus and squamous cell carcinoma. Oral Dis. 1996, 2, 25–31. [Google Scholar] [CrossRef]
- Pérez, M.Á.; Gandolfo, M.S.; Masquijo Bisio, P.; Paparella, M.L.; Itoiz, M.E. Different expression patterns of carbonic anhydrase IX in oral lichen planus and leukoplakia. Acta Odontol. Latinoam. 2018, 31, 77–81. [Google Scholar] [PubMed]
- de Sousa, F.A.C.G.; Paradella, T.C.; Carvalho, Y.R.; Rosa, L.E.B. Comparative analysis of the expression of proliferating cell nuclear antigen, p53, bax, and bcl-2 in oral lichen planus and oral squamous cell carcinoma. Ann. Diagn. Pathol. 2009, 13, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, A.; Sato, E.; Yasuda, T.; Isomura, T.; Nagao, T.; Chikazu, D. Intraepithelial CD8+ lymphocytes as a predictive diagnostic biomarker for the remission of oral lichen planus. Hum. Pathol. 2018, 74, 43–53. [Google Scholar] [CrossRef]
- Danielsson, K.; Olah, J.; Zohori-Zangeneh, R.; Nylander, E.; Ebrahimi, M. Increased expression of p16 in both oral and genital lichen planus. Med. Oral Patol. Oral Cir. Bucal 2018, 23, e449–e453. [Google Scholar] [CrossRef] [PubMed]
- Baghaei, F.; Shojaei, S.; Afshar-Moghaddam, N.; Zargaran, M.; Rastin, V.; Nasr, M.; Moghimbeigi, A. Study of P21 Expression in Oral Lichen Planus and Oral Squamous Cell Carcinoma by Immunohistochemical Technique. J. Dent. 2015, 16, 156–161. [Google Scholar]
- Girod, S.C.; Pfeiffer, P.; Ries, J.; Pape, H.D. Proliferative activity and loss of function of tumour suppressor genes as “biomarkers” in diagnosis and prognosis of benign and preneoplastic oral lesions and oral squamous cell carcinoma. Br. J. Oral Maxillofac. Surg. 1998, 36, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Arreaza, A.; Rivera, H.; Correnti, M. p53 expression in oral lichenoid lesions and oral lichen planus. Gen. Dent. 2015, 63, 69–72. [Google Scholar] [PubMed]
- Krauss, E.; Rauthe, S.; Gattenlöhner, S.; Reuther, T.; Kochel, M.; Kriegebaum, U.; Kübler, A.C.; Müller-Richter, U.D.A. MAGE-A antigens in lesions of the oral mucosa. Clin. Oral Investig. 2011, 15, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Akama, M.S.K.; Teixeira, L.R.; Innocentini, L.M.A.R.; Gallo, C.d.B.; Pinheiro, T.N.; Ribeiro-Silva, A.; Motta, A.C.F. Laminin-332 expression in oral lichen planus: Preliminary results of a cross-sectional study. Oral Dis. 2021, 27, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Hadzi-Mihailovic, M.; Raybaud, H.; Monteil, R.; Cakic, S.; Djuric, M.; Jankovic, L. Bcl-2 expression and its possible influence on malignant transformation of oral lichen planus. J. BUON 2010, 15, 362–368. [Google Scholar] [PubMed]
- Muraki, Y.; Yoshioka, C.; Fukuda, J.; Haneji, T.; Kobayashi, N. Immunohistochemical detection of Fas antigen in oral epithelia. J. Oral Pathol. Med. 1997, 26, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Oluwadara, O.; Giacomelli, L.; Christensen, R.; Kossan, G.; Avezova, R.; Chiappelli, F. LCK, survivin and PI-3K in the molecular biomarker profiling of oral lichen planus and oral squamous cell carcinoma. Bioinformation 2009, 4, 249–257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zargaran, M.; Baghaei, F.; Moghimbeigi, A. Comparative study of β-catenin and CD44 immunoexpression in oral lichen planus and squamous cell carcinoma. Int. J. Dermatol. 2018, 57, 794–798. [Google Scholar] [CrossRef] [PubMed]
- Jacques, C.M.C.; Pereira, A.L.C.; Maia, V.; Cuzzi, T.; Ramos-e-Silva, M. Expression of cytokeratins 10, 13, 14 and 19 in oral lichen planus. J. Oral Sci. 2009, 51, 355–365. [Google Scholar] [CrossRef]
- Lee, J.-J.; Kuo, M.-Y.; Cheng, S.-J.; Chiang, C.-P.; Jeng, J.-H.; Chang, H.-H.; Kuo, Y.-S.; Lan, W.-H.; Kok, S.-H. Higher expressions of p53 and proliferating cell nuclear antigen (PCNA) in atrophic oral lichen planus and patients with areca quid chewing. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2005, 99, 471–478. [Google Scholar] [CrossRef]
- Pariyawathee, S.; Phattarataratip, E.; Thongprasom, K. CD146 expression in oral lichen planus and oral cancer. Clin. Oral Investig. 2020, 24, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Nagao, Y.; Sata, M.; Kage, M.; Kameyama, T.; Ueno, T. Histopathological and immunohistochemical study of oral lichen planus-associated HCV infection. Eur. J. Intern. Med. 2000, 11, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Du, G.-H.; Qin, X.-P.; Li, Q.; Zhou, Y.-M.; Shen, X.-M.; Tang, G.-Y. The high expression level of programmed death-1 ligand 2 in oral lichen planus and the possible costimulatory effect on human T cells. J. Oral Pathol. Med. 2011, 40, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Acay, R.R.; Felizzola, C.R.; Soares de Araújo, N.; Machado de Sousa, S.O. Evaluation of proliferative potential in oral lichen planus and oral lichenoid lesions using immunohistochemical expression of p53 and Ki67. Oral Oncol. 2006, 42, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, K.; Wahlin, Y.B.; Coates, P.J.; Nylander, K. Increased expression of Smad proteins, and in particular Smad3, in oral lichen planus compared to normal oral mucosa. J. Oral Pathol. Med. 2010, 39, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Arreaza, A.J.; Rivera, H.; Correnti, M. Expression of COX-2 and bcl-2 in oral lichen planus lesions and lichenoid reactions. Ecancermedicalscience 2014, 8, 411. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, G.; Liu, Q.; Tan, J.; Hu, X.; Wang, J.; Wang, Q.; Wang, X. The cellular character of liquefaction degeneration in oral lichen planus and the role of interferon gamma. J. Oral Pathol. Med. 2017, 46, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Bloor, B.K.; Tidman, N.; Leigh, I.M.; Odell, E.; Dogan, B.; Wollina, U.; Ghali, L.; Waseem, A. Expression of keratin K2e in cutaneous and oral lesions: Association with keratinocyte activation, proliferation, and keratinization. Am. J. Pathol. 2003, 162, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Huerta, E.-R.; Ledesma-Montes, C.; Rojo-Botello, R.-E.; Vega-Memije, E. P53 and bcl-2 immunoexpression in patients with oral lichen planus and oral squamous cell carcinoma. Med. Oral Patol. Oral Cir. Bucal 2012, 17, e745–e750. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Lara, I.; González-Moles, M.A.; Ruiz-Avila, I.; Bravo, M.; Ramos, M.C.; Fernández-Martínez, J.A. Proliferating cell nuclear antigen (PCNA) as a marker of dysplasia in oral mucosa. Acta Stomatol. Belg. 1996, 93, 29–32. [Google Scholar]
- Girod, S.C.; Pape, H.D.; Krueger, G.R. p53 and PCNA expression in carcinogenesis of the oropharyngeal mucosa. Eur. J. Cancer B Oral Oncol. 1994, 30B, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Brunotto, M.; Zárate, A.M.; Cismondi, A.; Fernández, M.d.C.; Noher de Halac, R.I. Valuation of exfoliative cytology as prediction factor in oral mucosa lesions. Med. Oral Patol. Oral Cir. Bucal 2005, 10 (Suppl. 2), E92–E102. [Google Scholar] [PubMed]
- Ma, L.; Wang, H.; Yao, H.; Zhu, L.; Liu, W.; Zhou, Z. Bmi1 expression in oral lichen planus and the risk of progression to oral squamous cell carcinoma. Ann. Diagn. Pathol. 2013, 17, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Martín-Ezquerra, G.; Salgado, R.; Toll, A.; Baró, T.; Mojal, S.; Yébenes, M.; Garcia-Muret, M.P.; Solé, F.; Quitllet, F.A.; Espinet, B.; et al. CDC28 protein kinase regulatory subunit 1B (CKS1B) expression and genetic status analysis in oral squamous cell carcinoma. Histol. Histopathol. 2011, 26, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shen, Z.; Shi, L.; Sun, H.; Liu, W.; Zhou, Z. Aldehyde dehydrogenase 1 expression correlated with malignant potential of oral lichen planus. Ann. Diagn. Pathol. 2013, 17, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Prodromidis, G.; Nikitakis, N.G.; Sklavounou, A. Immunohistochemical Analysis of the Activation Status of the Akt/mTOR/pS6 Signaling Pathway in Oral Lichen Planus. Int. J. Dent. 2013, 2013, 743456. [Google Scholar] [CrossRef] [PubMed]
- Rivarola de Gutierrez, E.; Innocenti, A.C.; Cippitelli, M.J.; Salomon, S.; Vargas-Roig, L.M. Determination of cytokeratins 1, 13 and 14 in oral lichen planus. Med. Oral Patol. Oral Cir. Bucal 2014, 19, e359–e365. [Google Scholar] [CrossRef] [PubMed]
- Nafarzadeh, S.; Ejtehadi, S.; Amini Shakib, P.; Fereidooni, M.; Bijani, A. Comparative study of expression of smad3 in oral lichen planus and normal oral mucosa. Int. J. Mol. Cell. Med. 2013, 2, 194–198. [Google Scholar] [PubMed]
- Bascones-Ilundain, C.; González-Moles, M.Á.; Campo-Trapero, J.; Gil-Montoya, J.A.; Esparza-Gómez, G.C.; Cano-Sánchez, J.; Bascones-Martínez, A. No differences in caspase-3 and Bax expression in atrophic-erosive vs. reticular oral lichen planus. J. Eur. Acad. Dermatol. Venereol. 2008, 22, 204–212. [Google Scholar] [CrossRef]
- Kilpi, A.; Rich, A.M.; Reade, P.C.; Konttinen, Y.T. Studies of the inflammatory process and malignant potential of oral mucosal lichen planus. Aust. Dent. J. 1996, 41, 87–90. [Google Scholar] [CrossRef]
- Zolfaghari Saravi, Z.; Seyedmajidi, M.; Sharbatdaran, M.; Bijani, A.; Mozaffari, F.; Aminishakib, P. VEGFR-3 Expression in Oral Lichen Planus. Asian Pac. J. Cancer Prev. 2017, 18, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Favia, G.; Corsalini, M.; Iacobellis, M.; Maiorano, E. Squamous cell carcinoma in oral lichen ruber planus. A clinico-pathological and immunohistochemical study of 11 cases. Minerva Stomatol. 1994, 43, 479–491. [Google Scholar] [PubMed]
- Baddevithana, A.K.; Jayasinghe, R.D.; Tilakaratne, W.M.; Illeperuma, R.P.; Siriwardena, B.S.M.S. Expression of Human Papillomavirus and the p16 Gene in Oral Potentially Malignant Disorders (OPMD): A Comparative Study With Oral Squamous Cell Carcinoma. Appl. Immunohistochem. Mol. Morphol. AIMM 2023, 31, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Ramirez, D.A.; Rodriguez-Tojo, M.J.; Coca-Meneses, J.C.; Marichalar-Mendia, X.; Aguirre-Urizar, J.M. Epidermal growth factor receptor expression in different subtypes of oral lichenoid disease. Med. Oral Patol. Oral Cir. Bucal 2014, 19, e451–e458. [Google Scholar] [CrossRef] [PubMed]
- Cívico-Ortega, J.L.; González-Ruiz, I.; Ramos-García, P.; Cruz-Granados, D.; Samayoa-Descamps, V.; González-Moles, M.Á. Prognostic and Clinicopathological Significance of Epidermal Growth Factor Receptor (EGFR) Expression in Oral Squamous Cell Carcinoma: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 11888. [Google Scholar] [CrossRef] [PubMed]
- Ramos-García, P.; González-Moles, M.Á.; González-Ruiz, L.; Ruiz-Ávila, I.; Ayén, Á.; Gil-Montoya, J.A. Prognostic and clinicopathological significance of cyclin D1 expression in oral squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2018, 83, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Ramos-García, P.; Bravo, M.; González-Ruiz, L.; González-Moles, M. Significance of cytoplasmic cyclin D1 expression in oral oncogenesis. Oral Dis. 2018, 24, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Ramos-García, P.; González-Moles, M.Á.; González-Ruiz, L.; Ayén, Á.; Ruiz-Ávila, I.; Bravo, M.; Gil-Montoya, J.A. Clinicopathological significance of tumor cyclin D1 expression in oral cancer. Arch. Oral Biol. 2019, 99, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Ramos-García, P.; González-Moles, M.Á.; Ayén, Á.; González-Ruiz, L.; Ruiz-Ávila, I.; Lenouvel, D.; Gil-Montoya, J.A.; Bravo, M. Asymmetrical proliferative pattern loss linked to cyclin D1 overexpression in adjacent non-tumour epithelium in oral squamous cell carcinoma. Arch. Oral Biol. 2019, 97, 12–17. [Google Scholar] [CrossRef]
- Ramos-García, P.; Ruiz-Ávila, I.; Gil-Montoya, J.A.; Ayén, Á.; González-Ruiz, L.; Navarro-Triviño, F.J.; González-Moles, M.Á. Relevance of chromosomal band 11q13 in oral carcinogenesis: An update of current knowledge. Oral Oncol. 2017, 72, 7–16. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Warnakulasuriya, S.; López-Ansio, M.; Ramos-García, P. Hallmarks of Cancer Applied to Oral and Oropharyngeal Carcinogenesis: A Scoping Review of the Evidence Gaps Found in Published Systematic Reviews. Cancers 2022, 14, 3834. [Google Scholar] [CrossRef] [PubMed]
- Schifter, M.; Jones, A.M.; Walker, D.M. Epithelial p53 gene expression and mutational analysis, combined with growth fraction assessment, in oral lichen planus. J. Oral Pathol. Med. 1998, 27, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Bates, R.C.; Mercurio, A.M. Tumor necrosis factor-α stimulates the epithelial-to-mesenchymal transition of human colonic organoids. Mol. Biol. Cell 2003, 14, 1790–1800. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Sharma, K. COX-2 signaling and cancer: New players in old arena. Curr. Drug Targets 2014, 15, 347–359. [Google Scholar] [CrossRef]
- Lenouvel, D.; González-Moles, M.Á.; Ruiz-Ávila, I.; Gonzalez-Ruiz, L.; Gonzalez-Ruiz, I.; Ramos-García, P. Prognostic and clinicopathological significance of PD-L1 overexpression in oral squamous cell carcinoma: A systematic review and comprehensive meta-analysis. Oral Oncol. 2020, 106, 104722. [Google Scholar] [CrossRef] [PubMed]
- Lenouvel, D.; González-Moles, M.Á.; Ruiz-Ávila, I.; Chamorro-Santos, C.; González-Ruiz, L.; González-Ruiz, I.; Ramos-García, P. Clinicopathological and prognostic significance of PD-L1 in oral cancer: A preliminary retrospective immunohistochemistry study. Oral Dis. 2021, 27, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Lenouvel, D.; González-Moles, M.Á.; Talbaoui, A.; Ramos-García, P.; González-Ruiz, L.; Ruiz-Ávila, I.; Gil-Montoya, J.A. An update of knowledge on PD-L1 in head and neck cancers: Physiologic, prognostic and therapeutic perspectives. Oral Dis. 2020, 26, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Coghlin, C.; Murray, G.I. Current and emerging concepts in tumour metastasis. J. Pathol. 2010, 222, 1–15. [Google Scholar] [CrossRef]
- Manchanda, Y.; Rathi, S.K.; Joshi, A.; Das, S. Oral Lichen Planus: An Updated Review of Etiopathogenesis, Clinical Presentation, and Management. Indian Dermatol. Online J. 2023, 15, 8–23. [Google Scholar] [CrossRef]
- Arnold, D.L.; Krishnamurthy, K. Lichen Planus; StatPearls Publishing: St. Petersburg, FL, USA, 2023. [Google Scholar]
| Total | 110 studies |
| Year of publication | 1994–2023 |
| Total cases (range) | 7065 * (3–123) |
| Study design | |
| Retrospective cohort | 108 |
| Prospective cohort | 2 |
| Experimental methods | |
| Immunohistochemistry | 110 studies |
| Geographical region | |
| Europe | 37 studies (13 countries) |
| Asia | 54 studies (8 countries) |
| North America | 3 studies (3 countries) |
| South America | 13 studies (3 countries) |
| Africa | 2 studies (1 country) |
| Oceania | 1 study (1 country) |
| Total | 5 continents, 29 countries |
| Pooled Data | Heterogeneity | |||||||
|---|---|---|---|---|---|---|---|---|
| Meta-Analyses | No. of Studies * | No. of Cases * | Stat. Model | Wt | ES (95%CI) | p-Value | Phet | I2 (%) |
| Hallmark 1: Sustaining proliferative signaling | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-proliferative) | 35 | 1011 | REM | D-L | PP = 65.48% (51.87–78.02) | — | <0.001 | 94.5 |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-proliferative) | 18 | 934 | REM | D-L | OR = 4.39 (2.22–8.71) | <0.001 | 0.001 | 58.5 |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-proliferative) | 20 | 932 | REM | D-L | OR = 2.90 (1.27–6.65) | 0.01 | <0.001 | 71.9 |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-proliferative) | 9 | 378 | REM | D-L | OR = 7.50 (2.58–21.73) | <0.001 | 0.05 | 48.4 |
| Hallmark 2: Evading growth suppressors | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | ||||||||
| Protector (growth suppressor) | 36 | 1096 | REM | D-L | PP = 63.15% (52.26–73.45) | — | <0.001 | 91.9 |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Protector (growth suppressor) | 18 | 1278 | REM | D-L | OR = 2.16 (1.26–3.69) | 0.005 | 0.009 | 50.8 |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Protector (growth suppressor) | 25 | 1034 | REM | D-L | OR = 11.43 (6.89–18.95) | <0.001 | 0.30 | 11.4 |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Protector (growth suppressor) | 11 | 577 | REM | D-L | OR = 19.18 (8.25–44.61) | <0.001 | 0.74 | 0.0 |
| Hallmark 3: Resisting cell death | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | 0.41 ** | |||||||
| Oncogenic (anti-apoptotic) | 22 | 537 | REM | D-L | PP = 55.93% (35.99–75.01) | <0.001 | 95.0 | |
| Protector (pro-apoptotic) | 18 | 636 | REM | D-L | PP = 64.92% (55.15–74.14) | <0.001 | 83.8 | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | 0.18 ** | |||||||
| Oncogenic (anti-apoptotic) | 12 | 657 | REM | D-L | OR = 2.34 (1.16–4.70) | 0.02 | 0.09 | 39.1 |
| Protector (pro-apoptotic) | 5 | 281 | REM | D-L | OR = 0.90 (0.27–3.03) | 0.87 | 0.05 | 57.0 |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | 0.73 ** | |||||||
| Oncogenic (anti-apoptotic) | 14 | 444 | REM | D-L | OR = 3.95 (1.07–14.63) | 0.04 | <0.001 | 72.2 |
| Protector (pro-apoptotic) | 12 | 600 | REM | D-L | OR = 5.25 (2.07–13.31) | <0.001 | 0.001 | 66.8 |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | 0.08 ** | |||||||
| Oncogenic (anti-apoptotic) | 8 | 331 | REM | D-L | OR = 8.16 (2.19–30.35) | 0.002 | 0.04 | 52.6 |
| Protector (pro-apoptotic) | 3 | 138 | REM | D-L | OR = 48.53 (10.52–223.82) | <0.001 | 0.63 | 0.0 |
| Hallmark 4: Enabling replicative immortality | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-survival/immortalization) | 1 | 96 | — | — | PP = 41.67% (32.31–51.66) | — | — | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-survival/immortalization) | 1 | 102 | — | — | OR = 18.14 (0.99–331.13) | 0.051 | — | — |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-survival/immortalization) | 1 | 106 | — | — | OR = 15.05 (0.86–264.32) | 0.06 | — | — |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic | 1 | 16 | — | — | OR = 273.00 (4.80–15,515) | 0.007 | — | — |
| Hallmark 5: Inducing angiogenesis | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-angiogenic) | 3 | 96 | REM | D-L | PP = 94.76% (65.81–100) | <0.001 | 91.0 | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-angiogenic) | 0 | 0 | — | — | — | — | — | — |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-angiogenic) | 1 | 46 | — | — | OR = 2.40 (0.62–9.27) | 0.20 | — | — |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-angiogenic) | 0 | 0 | — | — | — | — | — | — |
| Hallmark 6: Activating invasion and metastasis | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | 0.52 ** | |||||||
| Oncogenic (pro-invasive) | 21 | 914 | REM | D-L | PP = 69.76% (55.72–82.29) | <0.001 | 94.2 | |
| Protector (anti-invasive) | 2 | 57 | REM | D-L | PP = 86.59% (29.86–100) | <0.001 | 95.3 | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | 0.04 ** | |||||||
| Oncogenic (pro-invasive) | 14 | 911 | REM | D-L | OR = 6.95 (3.20–15.10) | <0.001 | 0.08 | 37.5 |
| Protector (anti-invasive) | 1 | 42 | — | — | OR = 1.38 (0.37–5.15) | 0.64 | — | — |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | 0.89 ** | |||||||
| Oncogenic (pro-invasive) | 17 | 954 | REM | D-L | OR = 13.50 (5.12–35.59) | <0.001 | <0.001 | 66.0 |
| Protector (anti-invasive) | 2 | 78 | REM | D-L | OR = 15.59 (2.58–93.99) | 0.003 | 0.91 | 0.0 |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | 0.80 ** | |||||||
| Oncogenic (pro-invasive) | 11 | 328 | REM | D-L | OR = 28.04 (8.71–90.28) | <0.001 | 0.02 | 51.6 |
| Protector (anti-invasive) | 1 | 26 | — | — | OR = 20.00 (1.97–203.32) | 0.01 | — | — |
| Hallmark 7: Avoiding immune destruction | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (anti-tumor arrest) | 4 | 186 | REM | D-L | PP = 77.96% (51.96–95.96) | <0.001 | 92.8 | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (anti-tumor arrest) | — | — | — | — | — | — | — | |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (anti-tumor arrest) | 2 | 140 | REM | D-L | OR = 107.92 (13.63–843.45) | <0.001 | 0.96 | 0.0 |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (anti-tumor arrest) | 0 | 0 | — | — | — | — | — | — |
| Hallmark 8: Deregulating cellular energetics | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (enhancing tumor acidosis) | 1 | 23 | — | — | PP = 69.57% (49.13–84.40) | — | — | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (enhancing tumor acidosis) | 0 | 0 | — | — | — | — | — | — |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (enhancing tumor acidosis) | 1 | 30 | — | — | OR = 33.00 (1.66–656.23) | 0.02 | — | — |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (enhancing tumor acidosis) | 0 | 0 | — | — | — | — | — | — |
| Hallmark 9: Genome instability and mutation | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | 0.39 ** | |||||||
| Oncogenic (DNA instability) | 5 | 157 | REM | D-L | PP = 48.44% (13.54–84.19) | <0.001 | 95.3 | |
| Protector (DNA damage repair) | 2 | 79 | REM | D-L | PP = 72.37% (32.96–98.42) | 0.001 | 91.6 | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (DNA instability) | — | — | — | — | — | — | — | — |
| Protector (DNA damage repair) | 1 | 38 | — | — | OR = 2.88 (0.14–60.81) | 0.50 | — | — |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (DNA instability) | — | — | — | — | — | — | — | — |
| Protector (DNA damage repair) | 1 | 91 | — | — | OR = 0.28 (0.11–0.73) | 0.009 | — | — |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (DNA instability) | 1 | 52 | — | — | OR = 1653.0 (30.82–88,665) | <0.001 | — | — |
| Protector (DNA damage repair) | 0 | 0 | — | — | — | — | — | — |
| Hallmark 10: Tumor-promoting inflammation | ||||||||
| Differential expression in OLP | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-inflammatory) | 29 | 1386 | REM | D-L | PP = 83.10% (73.93–90.74) | <0.001 | 93.7 | |
| Magnitude of association (OSCC vs. OLP) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-inflammatory) | 8 | 301 | REM | D-L | OR = 2.40 (0.88–6.51) | 0.09 | 0.06 | 49.2 |
| Magnitude of association (OLP vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-inflammatory) | 14 | 691 | REM | D-L | OR = 7.50 (1.97–28.56) | 0.003 | <0.001 | 74.5 |
| Magnitude of association (OSCC vs. healthy controls) | ||||||||
| Subgroup analysis by role | ||||||||
| Oncogenic (pro-inflammatory) | 6 | 193 | REM | D-L | OR = 15.24 (2.54–91.34) | 0.003 | 0.02 | 62.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keim-del Pino, C.; Ramos-García, P.; González-Moles, M.Á. A Molecular Hypothesis on Malignant Transformation of Oral Lichen Planus: A Systematic Review and Meta-Analysis of Cancer Hallmarks Expression in This Oral Potentially Malignant Disorder. Cancers 2024, 16, 2614. https://doi.org/10.3390/cancers16152614
Keim-del Pino C, Ramos-García P, González-Moles MÁ. A Molecular Hypothesis on Malignant Transformation of Oral Lichen Planus: A Systematic Review and Meta-Analysis of Cancer Hallmarks Expression in This Oral Potentially Malignant Disorder. Cancers. 2024; 16(15):2614. https://doi.org/10.3390/cancers16152614
Chicago/Turabian StyleKeim-del Pino, Carmen, Pablo Ramos-García, and Miguel Ángel González-Moles. 2024. "A Molecular Hypothesis on Malignant Transformation of Oral Lichen Planus: A Systematic Review and Meta-Analysis of Cancer Hallmarks Expression in This Oral Potentially Malignant Disorder" Cancers 16, no. 15: 2614. https://doi.org/10.3390/cancers16152614
APA StyleKeim-del Pino, C., Ramos-García, P., & González-Moles, M. Á. (2024). A Molecular Hypothesis on Malignant Transformation of Oral Lichen Planus: A Systematic Review and Meta-Analysis of Cancer Hallmarks Expression in This Oral Potentially Malignant Disorder. Cancers, 16(15), 2614. https://doi.org/10.3390/cancers16152614








