Enhanced Lung Cancer Detection Using a Combined Ratio of Antigen–Autoantibody Immune Complexes against CYFRA 21-1 and p53
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Methods
- Preparation—A plasma sample (20 µL) is mixed with the detection mixture (containing CYFRA 21-1-cAb-DNA (p53-cAb-DNA) and anti-human-IgG-FB).
- Incubation—The mixture is incubated to allow for the formation of the CIC or PIC complexes.
- Loading—The mixture is loaded onto a 9G DNA membrane.
- Hybridization and washing—The membrane is hybridized with a probe specific for CIC or PIC, followed by washing to remove unbound probes.
- Scanning—The membrane is scanned using a fluorescence scanner to detect the presence of the CIC complex.
- Preparation—A plasma sample (20 µL) is mixed with the detection mixture (containing CYFRA 21-1-dAb-FB or p53-dAb-FB).
- Incubation—The mixture is incubated to allow for the formation of the CYFRA 21-1-dAb-FB or p53-dAB-FB complexes.
- Loading—The mixture is loaded onto a 9G DNA membrane.
- Hybridization and washing—The membrane is hybridized with a probe specific for CYFRA 21-1 or p53, followed by washing to remove unbound probes.
- Scanning—The membrane is scanned using a fluorescence scanner to detect the presence of CYFRA 21-1 or p53.
2.3. Statistical Analysis
3. Results
3.1. Diagnostic Performance
3.2. Utility as a Complementary Test to Radiologic Exams
3.3. LC Index Level According to the Basic Characteristics in Lung Cancer Patients and Healthy Controls
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Jeon, D.S.; Kim, H.C.; Kim, S.H.; Kim, T.J.; Kim, H.K.; Moon, M.H.; Beck, K.S.; Suh, Y.G.; Song, C.; Ahn, J.S.; et al. Five-Year Overall Survival and Prognostic Factors in Patients with Lung Cancer: Results from the Korean Association of Lung Cancer Registry (KALC-R) 2015. Cancer Res. Treat. 2023, 55, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Jonas, D.E.; Reuland, D.S.; Reddy, S.M.; Nagle, M.; Clark, S.D.; Weber, R.P.; Enyioha, C.; Malo, T.L.; Brenner, A.T.; Armstrong, C.; et al. Screening for Lung Cancer with Low-Dose Computed Tomography: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2021, 325, 971–987. [Google Scholar] [CrossRef]
- Bonney, A.; Malouf, R.; Marchal, C.; Manners, D.; Fong, K.M.; Marshall, H.M.; Irving, L.B.; Manser, R. Impact of low-dose computed tomography (LDCT) screening on lung cancer-related mortality. Cochrane Database Syst. Rev. 2022, 8, CD013829. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.W.; Li, W.; Han, F.J.; Liu, Y.D. Screening for lung cancer using low-dose computed tomography: Concerns about the application in low-risk individuals. Transl. Lung Cancer Res. 2015, 4, 275–286. [Google Scholar] [CrossRef] [PubMed]
- The American Cancer Society Medical and Editorial Content Team. Lung Cancer Risk Factors. Available online: https://www.cancer.org/cancer/types/lung-cancer/causes-risks-prevention/risk-factors.html (accessed on 29 June 2024).
- Lee, J.Y.; Bhandare, R.R.; Boddu, S.H.S.; Shaik, A.B.; Saktivel, L.P.; Gupta, G.; Negi, P.; Barakat, M.; Singh, S.K.; Dua, K.; et al. Molecular mechanisms underlying the regulation of tumour suppressor genes in lung cancer. Biomed. Pharmacother. 2024, 173, 116275. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, A.; D’Ascanio, M.; Ricci, A.; Pagliuca, A.; Carico, E. Expression and role of p16 and GLUT1 in malignant diseases and lung cancer: A review. Thorac. Cancer 2020, 11, 3060–3070. [Google Scholar] [CrossRef] [PubMed]
- Zaenker, P.; Gray, E.S.; Ziman, M.R. Autoantibody Production in Cancer—The Humoral Immune Response toward Autologous Antigens in Cancer Patients. Autoimmun. Rev. 2016, 15, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.W.; Wandall, H.H. Autoantibodies as Biomarkers in Cancer. Lab. Med. 2011, 42, 623–628. [Google Scholar] [CrossRef]
- Heo, C.K.; Bahk, Y.Y.; Cho, E.W. Tumor-associated autoantibodies as diagnostic and prognostic biomarkers. BMB Rep. 2012, 45, 677–685. [Google Scholar] [CrossRef]
- Mohamed, E.; Garcia Martinez, D.J.; Hosseini, M.S.; Yoong, S.Q.; Fletcher, D.; Hart, S.; Guinn, B.A. Identification of biomarkers for the early detection of non-small cell lung cancer: A systematic review and meta-analysis. Carcinogenesis 2024, 45, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Pujol, J.L.; Molinier, O.; Ebert, W.; Daures, J.P.; Barlesi, F.; Buccheri, G.; Paesmans, M.; Quoix, E.; Moro-Sibilot, D.; Szturmowicz, M.; et al. CYFRA 21-1 is a prognostic determinant in non-small-cell lung cancer: Results of a meta-analysis in 2063 patients. Br. J. Cancer 2004, 90, 2097–2105. [Google Scholar] [CrossRef] [PubMed]
- Trulson, I.; Holdenrieder, S. Prognostic value of blood-based protein biomarkers in non-small cell lung cancer: A critical review and 2008–2022 update. Tumour. Biol. 2024, 46, S111–S161. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.S.; Hsu, H.K.; Lu, J.Y.; Ger, L.P.; Lai, N.S. CYFRA 21-1 enzyme-linked immunosorbent assay. Evaluation as a tumor marker in non-small cell lung cancer. Chest 1996, 109, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, Y.; Sone, K.; Oguri, T.; Horiuchi, M.; Fukuda, S.; Uemura, T.; Takakuwa, O.; Maeno, K.; Fukumitsu, K.; Kanemitsu, Y.; et al. Predictive role of CYFRA 21-1 for S-1 monotherapy in non-small cell lung cancer patients. Respir. Investig. 2022, 60, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, D.C.; Davidson, A.G.; Summers, C.L.; Warden, H.E.; Doshi, H.M. Accumulation of p53 protein correlates with a poor prognosis in human lung cancer. Cancer Res. 1992, 52, 4828–4831. [Google Scholar] [PubMed]
- Hou, Y.; Tan, S.; Wang, G. Significance of TP53 Mutation in Cellular Process and Disease Progression in Lung Adenocarcinoma. Genet. Test. Mol. Biomarkers 2021, 25, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Mogi, A.; Kuwano, H. TP53 mutations in nonsmall cell lung cancer. J. Biomed. Biotechnol. 2011, 2011, 583929. [Google Scholar] [CrossRef]
- Li, Y.; Karjalainen, A.; Koskinen, H.; Hemminki, K.; Vainio, H.; Shnaidman, M.; Ying, Z.; Pukkala, E.; Brandt-Rauf, P.W. p53 autoantibodies predict subsequent development of cancer. Int. J. Cancer 2005, 114, 157–160. [Google Scholar] [CrossRef]
- Zalcman, G.; Schlichtholz, B.; Tredaniel, J.; Urban, T.; Lubin, R.; Dubois, I.; Milleron, B.; Hirsch, A.; Soussi, T. Monitoring of p53 autoantibodies in lung cancer during therapy: Relationship to response to treatment. Clin. Cancer Res. 1998, 4, 1359–1366. [Google Scholar] [PubMed]
- Mack, U.; Ukena, D.; Montenarh, M.; Sybrecht, G.W. Serum anti-p53 antibodies in patients with lung cancer. Oncol. Rep. 2000, 7, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Mao, C.; Chen, Q.; Cu, X.W.; Zhang, W.S. Serum p53 protein and anti-p53 antibodies are associated with increased cancer risk: A case-control study of 569 patients and 879 healthy controls. Mol. Biol. Rep. 2010, 37, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.M.; Ling, Z.G.; Wang, C.M.; Wu, Y.B.; Kong, J.L. Serum tumor-associated autoantibodies as diagnostic biomarkers for lung cancer: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0182117. [Google Scholar] [CrossRef] [PubMed]
- Nimse, S.B.; Song, K.S.; Kim, J.; Sayyed, D.R.; Kim, T. 9G DNAChip Technology: Self-Assembled Monolayer (SAM) of ssDNA for Ultra-Sensitive Detection of Biomarkers. Int. J. Mol. Sci. 2013, 14, 5723–5733. [Google Scholar] [CrossRef] [PubMed]
- Choe, W.; Chae, J.D.; Lee, B.H.; Kim, S.H.; Park, S.Y.; Nimse, S.B.; Kim, J.; Warkad, S.D.; Song, K.S.; Oh, A.C.; et al. 9G Test(TM) Cancer/Lung: A Desirable Companion to LDCT for Lung Cancer Screening. Cancers 2020, 12, 3192. [Google Scholar] [CrossRef]
- Kim, H.; Lee, J.K.; Oh, A.C.; Kim, H.R.; Hong, Y.J. The Usefulness of the Ratio of Antigen-Autoantibody Immune Complexes to Their Free Antigens in the Diagnosis of Non-Small Cell Lung Cancer. Diagnostics 2023, 13, 2999. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Nimse, S.B.; Warkad, S.D.; Kim, J.H.; Kim, H.J.; Kim, T. Detection and Quantification of Tp53 and p53-Anti-p53 Autoantibody Immune Complex: Promising Biomarkers in Early Stage Lung Cancer Diagnosis. Biosensors 2022, 12, 127. [Google Scholar] [CrossRef]
- Song, K.S.; Nimse, S.B.; Warkad, S.D.; Oh, A.C.; Kim, T.; Hong, Y.J. Quantification of CYFRA 21-1 and a CYFRA 21-1-anti-CYFRA 21-1 autoantibody immune complex for detection of early stage lung cancer. Chem. Commun. 2019, 55, 10060–10063. [Google Scholar] [CrossRef]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef]
- Micke, P.; Faldum, A.; Metz, T.; Beeh, K.M.; Bittinger, F.; Hengstler, J.G.; Buhl, R. Staging small cell lung cancer: Veterans Administration Lung Study Group versus International Association for the Study of Lung Cancer—What limits limited disease? Lung Cancer 2002, 37, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.I.; Barderas, R. Identification of cancer autoantigens in serum: Toward diagnostic/prognostic testing? Mol. Diagn. Ther. 2010, 14, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Nimse, S.B.; Kim, J.; Kim, J.; Nguyen, V.T.; Ta, V.T.; Kim, T. 9G DNAChip: Microarray based on the multiple interactions of 9 consecutive guanines. Chem. Commun. 2011, 47, 7101–7103. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.; Li, Q.; Freeburg, N.F.; Murali, N.; Indeglia, A.; Grothusen, G.P.; Cicchini, M.; Mai, H.; Gladstein, A.C.; Adler, K.M.; et al. p53 restoration in small cell lung cancer identifies a latent cyclophilin-dependent necrosis mechanism. Nat. Commun. 2023, 14, 4403. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, T.; Su, W.; Dou, Z.; Zhao, D.; Jin, X.; Lei, H.; Wang, J.; Xie, X.; Cheng, B.; et al. Mutant p53 in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 2022, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Synnott, N.C.; O’Grady, S.; Crown, J. Targeting p53 for the treatment of cancer. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2022; Volume 79, pp. 58–67. [Google Scholar] [CrossRef]
- Petitjean, A.; Mathe, E.; Kato, S.; Ishioka, C.; Tavtigian, S.V.; Hainaut, P.; Olivier, M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007, 28, 622–629. [Google Scholar] [CrossRef]
- Molina, R.; Holdenrieder, S.; Auge, J.M.; Schalhorn, A.; Hatz, R.; Stieber, P. Diagnostic relevance of circulating biomarkers in patients with lung cancer. Cancer Biomark. 2010, 6, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Pujol, J.L.; Boher, J.M.; Grenier, J.; Quantin, X. Cyfra 21-1, neuron specific enolase and prognosis of non-small cell lung cancer: Prospective study in 621 patients. Lung Cancer 2001, 31, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Stampfli, M.R.; Anderson, G.P. How cigarette smoke skews immune responses to promote infection, lung disease and cancer. Nat. Rev. Immunol. 2009, 9, 377–384. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretic, L.; Kong, G.; Leenders, F.; Lu, X.; Fernandez-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef]
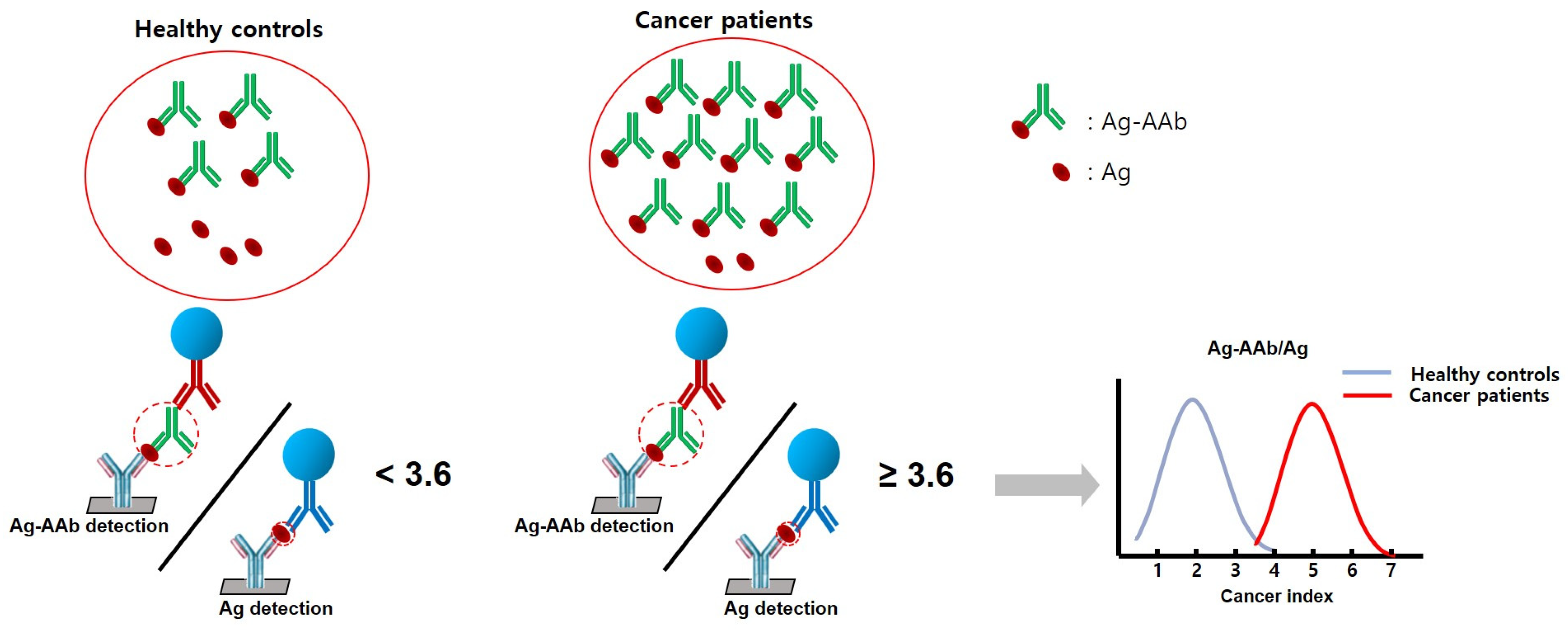
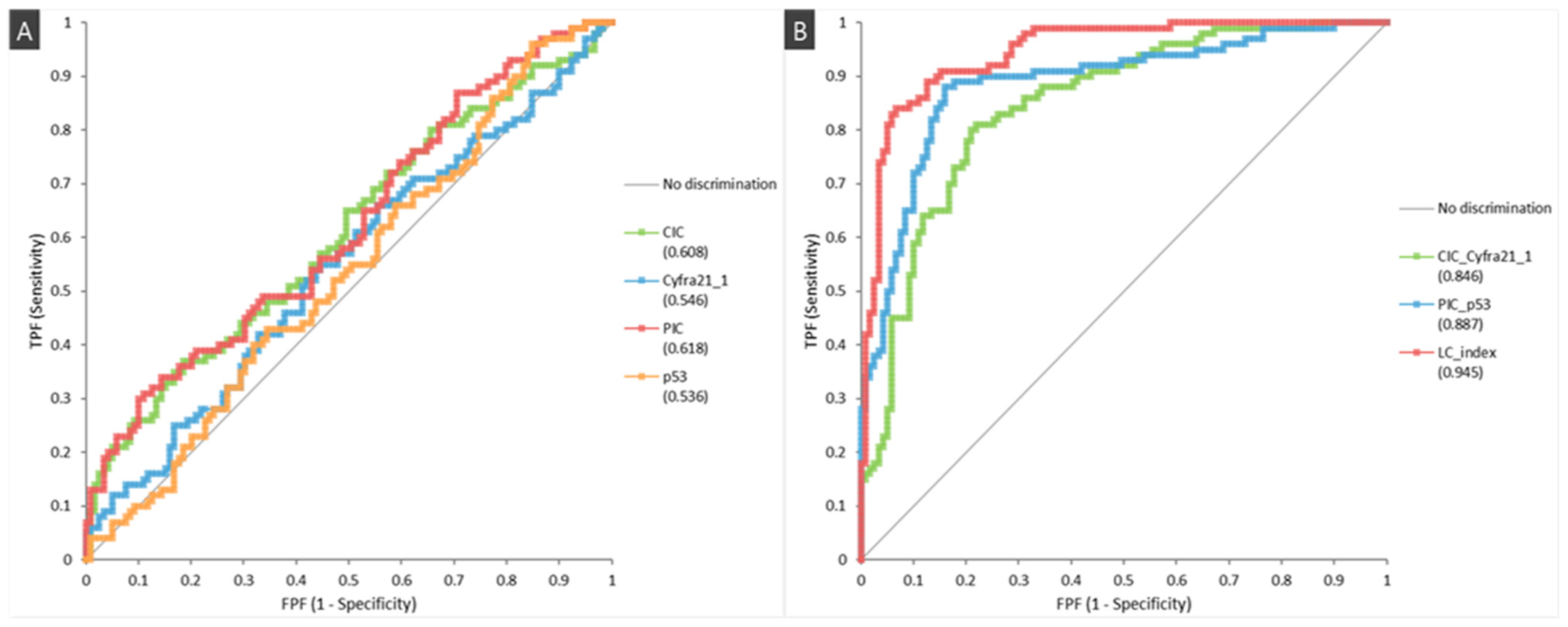
| Characteristics | Lung Cancer (n = 100) | Healthy Controls (n = 119) | p-Value |
|---|---|---|---|
| Age, years (median, IQR) | 68 (62–75) | 57 (55–60) | <0.001 |
| Male gender (%) | 66 (66%) | 79 (66.4%) | 1.000 |
| Smoking status * | |||
| - Former smoker (%) | 65 (65.7%) | 36 (30.5%) | <0.001 |
| - Current smoker (%) | 5 (5.1%) | 14 (11.9%) | 0.1266 |
| - Never smoker (%) | 29 (29.3%) | 69 (58.5%) | <0.001 |
| - Pack year (median, IQR) | 19 (0, 46) | 0 (0, 1) | <0.001 |
| <20 pack year | 20 (29.9%) | 31 (64.6%) | <0.001 |
| ≥20 pack year | 47 (70.2%) | 17 (35.4%) | |
| Stage | |||
| Non-small cell lung cancer | 90 | ||
| - Stage I | 39 | ||
| - Stage II | 12 | ||
| - Stage III | 20 | ||
| - Stage IV | 19 | ||
| Small cell lung cancer | 10 | ||
| - LD | 5 | ||
| - ED | 5 | ||
| Pathologic diagnosis | |||
| Non-small cell lung cancer | |||
| - Squamous cell carcinoma | 22 | ||
| - Adenocarcinoma | 61 | ||
| - Other types | |||
| Large cell carcinoma | 3 | ||
| Pleomorphic carcinoma | 3 | ||
| Adenosquamous carcinoma | 1 | ||
| Small cell lung cancer | 10 | ||
| Size of lung mass or nodule, cm (median, IQR) | 3.05 (1.9, 4.32) |
| Characteristics | Lung Cancer (n = 100) | Healthy Controls (n = 119) | p-Value |
|---|---|---|---|
| CIC (median (IQR), pg/mL) | 2.23 (1.33–4.29) | 1.78 (0.92–2.97) | 0.0062 |
| CYFRA 21-1 (median (IQR), pg/mL) | 0.97 (0.46–2.00) | 1.23 (0.52–2.16) | 0.2369 |
| PIC (median (IQR), pg/mL) | 1232.87 (650.04–3286.74) | 990.65 (424.94–2046.01) | 0.0026 |
| p53 (median (IQR), pg/mL) | 720.50 (353.06–1588.69) | 817.93 (374.63–1690.86) | 0.3601 |
| CIC/CYFRA 21-1 ratio (median (IQR)) | 2.28 (1.92–2.78) | 1.46 (1.16–1.78) | <0.0001 |
| PIC/p53 ratio (median (IQR)) | 2.08 (1.76–2.47) | 1.19 (0.94–1.38) | <0.0001 |
| LC Index (median (IQR)) | 4.72 (3.75–5.99) | 1.70 (1.30–2.37) | <0.0001 |
| Variables | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|
| CIC/CYFRA 21-1 | 82.0 (72.8–88.7) | 78.2 (69.5–85.0) | 75.9 (66.6–83.4) | 83.8 (75.3–89.9) |
| PIC/p53 | 83.0 (73.9–89.5) | 90.8 (83.7–95.1) | 88.3 (79.6–93.7) | 89.8 (78.8–91.7) |
| LC Index | 81.0 (71.7–87.9) | 95.0 (88.9–97.9) | 93.1 (85.0–97.2) | 85.6 (78.2–90.9) |
| Type | Variables | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|---|
| NSCLC | Stage I (n = 39) | 87.2 (71.8–95.2) | 95.0 (88.9–97.9) | 85.0 (69.5–93.8) | 95.8 (89.9–98.4) |
| Stage II (n = 12) | 91.7 (59.8–99.6) | 64.7 (38.6–84.7) | 99.1 (94.5–100.0) | ||
| Stage III (n = 20) | 75.0 (50.6–90.4) | 71.4 (47.7–87.8) | 95.8 (89.9–98.4) | ||
| Stage IV (n = 19) | 73.7 (48.6–89.9) | 70.0 (45.7–87.2) | 95.8 (89.9–98.4) | ||
| Stage I~II (n = 51) | 88.2 (75.4–95.1) | 88.2 (75.4–95.1) | 95.0 (88.9–97.9) | ||
| Stage III~IV (n = 39) | 74.4 (57.6–86.4) | 82.9 (65.7–92.8) | 91.9 (85.2–95.8) | ||
| SCLC | Limited (n = 5) | 80.0 (29.9–99.0) | 95.0 (88.9–97.9) | 36.4 (13.7–72.6) | 99.1 (94.5–100.0) |
| Extensive (n = 5) | 60.0 (17.0–92.7) | 40.0 (9.0–69.1) | 99.1 (93.3–99.7) | ||
| Early LC (I~II and LD) (n = 56) | 87.5 (75.3–94.4) | 95.0 (88.9–97.9) | 94.2 (87.9–97.4) | ||
| Advanced LC (III~IV and ED) (n = 44) | 72.7 (76.0–84.5) | 90.4 (83.5–94.7) | |||
| Pathological Type | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | |
|---|---|---|---|---|---|
| NSCLC (n = 90, 51/39) | 82.2 (72.4–89.2) | 92.5 (83.8–96.9) | 87.6 (80.4–92.5) | ||
| Squamous carcinoma (n = 22, 11/11 *) | 72.7 (49.6–88.4) | 95.0 (88.9–97.9) | 72.7 (49.6–88.4) | 95.0 (88.9–97.9) | |
| Adenocarcinoma (n = 62, 34/28 *) | 85.5 (73.7–92.8) | 89.8 (78.5–95.8) | 92.6 (86.7–96.4) | ||
| Other NSCLCs (n = 6, 6/0 *) | 83.3 (36.5–99.1) | 45.5 (18.1–75.4) | 99.1 (94.5–100.0) | ||
| SCLC (n = 10, 5/5) | 70.0 (35.4–91.9) | 53.9 (26.2–79.6) | 97.4 (92.1–99.3) | ||
| Test | Chest X-ray (n = 100) | LDCT (n = 100) | Chest X-ray & LDCT (n = 84 †) | |||
|---|---|---|---|---|---|---|
| Lung Mass | Small Nodules /Others § | Lung Mass | Small Nodules /Others § | Lung Mass | Small Nodules /Others § | |
| LC index (+) | 66 | 15 | 71 | 10 | 61 | 5 |
| LC index (−) | 17 | 2 | 18 | 1 | 17 | 1 |
| Total | 83 | 17 | 89 | 11 | 78 | 6 |
| Accuracy | 79.5% | 88.2% | 79.8% | 90.9% | 78.2% | 83.3% |
| Characteristics | Lung Cancer (n = 100) | p-Value | Healthy Controls (n = 119) | p-Value | |
|---|---|---|---|---|---|
| LC Index (IQR) | LC Index (IQR) | ||||
| Age | 0.3873 | 0.5548 | |||
| Age < 60 year | 5.03 (4.07, 6.54) | 1.74 (1.23, 2.52) | |||
| Age ≥ 60 year | 4.71 (3.75, 5.97) | 1.63 (1.36, 2) | |||
| Gender | 0.2548 | 0.6507 | |||
| Male | 4.53 (3.74, 5.9) | 1.74 (1.32, 2.26) | |||
| Female | 4.77(4.44, 6.29) | 1.58 (1.15, 2.64) | |||
| Smoking | 0.1082 | 0.5049 | |||
| Former | 4.37 (3.69, 5.52) | 1.74 (1.25, 2.02) | |||
| Current | 4.95 (3.65, 5.06) | 2 (1.44, 2.5) | |||
| Never | 4.9 (4.49, 6.36) | 1.65 (1.31, 2.55) | |||
| Former smoker | 0.8996 | 0.6533 | |||
| P-Y < 20 year | 4.47 (3.59, 5.14) | 1.86 (1.16, 2.15) | |||
| P-Y ≥ 20 year | 4.37 (3.72, 5.79) | 1.7 (1.38, 1.96) | |||
| Current | 0.5 | 0.4136 | |||
| P-Y < 20 year | 3.65 (3.65, 3.65) | 2.23 (1.49, 4.39) | |||
| P-Y ≥ 20 year | 5.06 (5.01, 5.39) | 2 (1.44, 2.41) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Lee, J.K.; Kim, H.-R.; Hong, Y.J. Enhanced Lung Cancer Detection Using a Combined Ratio of Antigen–Autoantibody Immune Complexes against CYFRA 21-1 and p53. Cancers 2024, 16, 2661. https://doi.org/10.3390/cancers16152661
Kim H, Lee JK, Kim H-R, Hong YJ. Enhanced Lung Cancer Detection Using a Combined Ratio of Antigen–Autoantibody Immune Complexes against CYFRA 21-1 and p53. Cancers. 2024; 16(15):2661. https://doi.org/10.3390/cancers16152661
Chicago/Turabian StyleKim, Heyjin, Jin Kyung Lee, Hye-Ryoun Kim, and Young Jun Hong. 2024. "Enhanced Lung Cancer Detection Using a Combined Ratio of Antigen–Autoantibody Immune Complexes against CYFRA 21-1 and p53" Cancers 16, no. 15: 2661. https://doi.org/10.3390/cancers16152661
APA StyleKim, H., Lee, J. K., Kim, H.-R., & Hong, Y. J. (2024). Enhanced Lung Cancer Detection Using a Combined Ratio of Antigen–Autoantibody Immune Complexes against CYFRA 21-1 and p53. Cancers, 16(15), 2661. https://doi.org/10.3390/cancers16152661





