Comparison between Three Radiomics Models and Clinical Nomograms for Prediction of Lymph Node Involvement in PCa Patients Combining Clinical and Radiomic Features
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Magnetic Resonance Imaging
2.2. Segmentation, Feature Extraction
- Shape features: These describe the geometric properties of the region of interest (ROI), such as the surface area, total volume, diameter, elongation, sphericity, and surface-to-volume ratio.
- First-order statistics (histogram-based features): These detail the distribution of voxel intensities within the image ROI, using conventional parameters like energy, entropy, mean, interquartile range, skewness, kurtosis, and uniformity.
- Second-order statistics (textural features): These capture the statistical interrelationships between neighboring voxels. Notable methods include:
- ○
- Gray-level Cooccurrence Matrix (GLCM): Analyzes the spatial distribution of gray-level intensities in a 3D image.
- ○
- Gray-Level Run-length Matrix (GLRLM): Measures contiguous voxels with the same gray-level value, characterizing the gray-level run lengths in various directions.
- ○
- Gray-Level Size-Zone Matrix (GLSZM): Quantifies the zones of connected voxels sharing the same gray-level intensity in a 3D image.
- ○
- Neighboring Gray-Tone Difference Matrix (NGTMD): Calculates the difference between a voxel’s gray value and the average gray value of its neighbors within a specified distance.
- ○
- Gray-level Dependence Matrix (GLDM): Assesses the number of connected voxels within a certain distance that depend on the center voxel.
2.3. Radiomics Analysis and Model Development
2.4. Clinical Nomograms
- Briganti: Preoperative PSA, clinical stage T, Gleason score of the biopsy, percentage of positive cores with a high level of prostate cancer, percentage of positive cores with a low level of prostate cancer.
- MSKCC: Age, thickness, Clark level, localization, ulceration.
- Yale: PSA, Gleason score, clinical T stage.
- Roach: PSA, Gleason score.
3. Results
4. Discussion
- The Briganti model [23] is used for cancer involvement and grading heterogeneity in biopsy samples and improves the accuracy in estimating the risk of lymph node invasion (LNI), suggesting changes in staging approaches.
- Due to the risk of overtreatment with the only Briganti model, the MSKCC (Memorial Sloan Kattering Cancer Center) calculate another nomogram with a minimalist approach. In this case, the evaluation includes only the PSA, age, and biopsy Gleason score.
- The Yale is a linear model based on PSA levels, the T stage, and the Gleason score. This model more successfully classifies high-risk categories patients (>15%). Unlike previous models, Yale does not underestimate the risks associated with lymph node involvement.
- The Roach formula [24] that includes only the PSA and the Gleason score can be used for the evaluation of lymph node involvement and the seminal vesicle and capsular involvement.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miyahira, A.K.; Sharp, A.; Ellis, L.; Jones, J.; Kaochar, S.; Larman, H.B.; Quigley, D.A.; Ye, H.; Simons, J.W.; Soule, H.R.; et al. Prostate cancer research: The next generation, report from the 2019 Coffey-Holden Prostate Cancer Academy Meeting. Prostate 2020, 80, 113–132. [Google Scholar] [CrossRef] [PubMed]
- European Commission’s Joint Research Centre. Available online: https://visitors-centre.jrc.ec.europa.eu/en/media/infographics/prostate-cancer-burden-eu-27 (accessed on 23 June 2024).
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Preisser, F.; van den Bergh, R.C.; Gandaglia, G.; Ost, P.; Surcel, C.I.; Sooriakumaran, P.; Montorsi, F.; Graefen, M.; van der Poel, H.; Tilki, D.; et al. Effect of Extended Pelvic Lymph Node Dissection on Oncologic Outcomes in Patients with D’Amico Intermediate and High Risk Prostate Cancer Treated with Radical Prostatectomy: A Multi-Institutional Study. J. Urol. 2020, 203, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, L.; Gandaglia, G.; Fossati, N.; Suardi, N.; Moschini, M.; Cucchiara, V.; Bianchi, M.; Damiano, R.; Schiavina, R.; Shariat, S.F.; et al. Pelvic Lymph Node Dissection in Prostate Cancer: Indications, Extent and Tailored Approaches. Urol. J. 2017, 84, 9–19. [Google Scholar] [CrossRef]
- Van Huele, A.; Poelaert, F.; Fonteyne, V.; Decaestecker, K.; Ost, P.; Lumen, N. Pelvic lymph node dissection in prostate cancer staging: Evaluation of morbidity and oncological outcomes. Acta Chir. Belg. 2019, 119, 103–109. [Google Scholar] [CrossRef] [PubMed]
- De Hertogh, O.; Le Bihan, G.; Zilli, T.; Palumbo, S.; Jolicoeur, M.; Crehange, G.; Derashodian, T.; Roubaud, G.; Salembier, C.; Supiot, S.; et al. Consensus Delineation Guidelines for Pelvic Lymph Node Radiation Therapy of Prostate Cancer: On Behalf of the Francophone Group of Urological Radiation Therapy (GFRU). Int. J. Radiat. Oncol. Biol. Phys. 2024, 118, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Bao, J.; Song, Y.; Bao, M.-L.; Jiang, K.-W.; Zhang, J.; Yang, G.; Hu, C.-H.; Shi, H.-B.; Wang, X.-M.; et al. Integration of clinicopathologic identification and deep transferrable image feature representation improves predictions of lymph node metastasis in prostate cancer. eBioMedicine 2021, 68, 103395. [Google Scholar] [CrossRef] [PubMed]
- Gandaglia, G.; Fossati, N.; Zaffuto, E.; Bandini, M.; Dell’Oglio, P.; Bravi, C.A.; Fallara, G.; Pellegrino, F.; Nocera, L.; Karakiewicz, P.I.; et al. Development and Internal Validation of a Novel Model to Identify the Candidates for Extended Pelvic Lymph Node Dissection in Prostate Cancer. Eur. Urol. 2017, 72, 632–640. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.J.; Eastham, J.A.; Scardino, P.T.; Lilja, H. The Memorial Sloan Kettering Cancer Center Recommendations for Prostate Cancer Screening. Urology 2016, 91, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Tosoian, J.J.; Chappidi, M.; Feng, Z.; Humphreys, E.B.; Han, M.; Pavlovich, C.P.; Epstein, J.I.; Partin, A.W.; Trock, B.J. Prediction of pathological stage based on clinical stage, serum prostate-specific antigen, and biopsy Gleason score: Partin Tables in the contemporary era. BJU Int. 2017, 119, 676–683. [Google Scholar] [CrossRef]
- Wang, J.; Wu, C.; Bao, M.; Zhang, J.; Shi, H.; Zhang, Y. Using support vector machine analysis to assess PartinMR: A new prediction model for organ-confined prostate cancer. J. Magn. Reson. Imaging 2018, 48, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.B.; Makarov, D.V.; Gross, C. A New Formula for Prostate Cancer Lymph Node Risk. Int. J. Radiat. Oncol. Biol. Phys. 2011, 80, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jian, T.; Chi, C.; Liang, Y.; Liang, X.; Yu, Y.; Jiang, F.; Lu, J. Machine Learning-Based Models Enhance the Prediction of Prostate Cancer. Front. Oncol. 2022, 12, 941349. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, A.; Santucci, D.; Landi, R.; Beomonte Zobel, B.; Faiella, E.; de Felice, C. Radiomics MRI for lymph node status prediction in breast cancer patients: The state of art. J. Cancer Res. Clin. Oncol. 2021, 147, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Faiella, E.; Santucci, D.; D’Amone, G.; Cirimele, V.; Vertulli, D.; Bruno, A.; Zobel, B.B.; Grasso, R.F. Focal Minimally Invasive Treatment in Localized Prostate Cancer: Comprehensive Review of Different Possible Strategies. Cancers 2024, 16, 765. [Google Scholar] [CrossRef] [PubMed]
- Faiella, E.; Santucci, D.; Greco, F.; Frauenfelder, G.; Giacobbe, V.; Muto, G.; Zobel, B.B.; Gralsso, R.F. Analysis of histological findings obtained combining US/mp-MRI fusion-guided biopsies with systematic US biopsies: Mp-MRI role in prostate cancer detection and false negative. Radiol. Med. 2018, 123, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Faiella, E.; Santucci, D.; Calabrese, A.; Russo, F.; Vadalà, G.; Zobel, B.B.; Soda, P.; Iannello, G.; de Felice, C.; Denaro, V. Artificial Intelligence in Bone Metastases: An MRI and CT Imaging Review. Int. J. Environ. Res. Public Health 2022, 19, 1880. [Google Scholar] [CrossRef] [PubMed]
- Faiella, E.; Vaccarino, F.; Ragone, R.; D’amone, G.; Cirimele, V.; Piccolo, C.L.; Vertulli, D.; Grasso, R.F.; Zobel, B.B.; Santucci, D.; et al. Can Machine Learning Models Detect and Predict Lymph Node Involvement in Prostate Cancer? A Comprehensive Systematic Review. J. Clin. Med. 2023, 12, 7032. [Google Scholar] [CrossRef] [PubMed]
- 3D Slicer. Available online: https://www.slicer.org/ (accessed on 23 June 2024).
- Cheung, D.C.; Fleshner, N.; Sengupta, S.; Woon, D. A narrative review of pelvic lymph node dissection in prostate cancer. Transl. Androl. Urol. 2020, 9, 3049–3055. [Google Scholar] [CrossRef]
- Mohler, J.L.; Armstrong, A.J.; Bahnson, R.R.; D’Amico, A.V.; Davis, B.J.; Eastham, J.A.; Enke, C.A.; Farrington, T.A.; Higano, C.S.; Horwitz, E.M.; et al. Prostate Cancer, Version 1.2016. J. Natl. Compr. Cancer Netw. 2016, 14, 19–30. [Google Scholar] [CrossRef]
- Gandaglia, G.; Martini, A.; Ploussard, G.; Fossati, N.; Stabile, A.; De Visschere, P.; Borgmann, H.; Heidegger, I.; Steinkohl, F.; Kretschmer, A.; et al. External Validation of the 2019 Briganti Nomogram for the Identification of Prostate Cancer Patients Who Should Be Considered for an Extended Pelvic Lymph Node Dissection. Eur. Urol. 2020, 78, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Roach, M.; Marquez, C.; Yuo, H.S.; Narayan, P.; Coletman, L.; Nseyo, U.O.; Navvab, Z.; Carroll, P.R. Predicting the risk of lymph node involvement using the pre-treatment prostate specific antigen and gleason score in men with clinically localized prostate cancer. Int. J. Radiat. Oncol. Biol. Phys. 1994, 28, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Faiella, E.; Santucci, D.; Vertulli, D.; Esperto, F.; Messina, L.; Castiello, G.; Papalia, R.; Flammia, G.; Scarpa, R.; Fiore, M.; et al. The role of multiparametric mri in the diagnosis of local recurrence after radical prostatectomy and before salvage radiotherapy. Actas Urol. Esp. 2022, 46, 397–406. [Google Scholar] [CrossRef]
- Arslan, A.; Karaarslan, E.; Güner, A.L.; Sağlıcan, Y.; Tuna, M.B.; Kural, A.R. Comparing the Diagnostic Performance of Multiparametric Prostate MRI Versus 68Ga-PSMA PET-CT in the Evaluation Lymph Node Involvement and Extraprostatic Extension. Acad. Radiol. 2022, 29, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Choyke, P.L.; Eberhardt, S.C.; Tempany, C.M.; Turkbey, B.; Rosenkrantz, A.B. The Current State of MR Imaging–targeted Biopsy Techniques for Detection of Prostate Cancer. Radiology 2017, 285, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Huan, Y.; Wang, H.; Zhao, H.; Get, Y.; Chang, Y.; Liu, Y. Diffusion-weighted imaging in normal prostate and differential diagnosis of prostate diseases. Abdom. Imaging 2008, 33, 724–728. [Google Scholar] [CrossRef]
- Chen, M.; Dang, H.D.; Wang, J.Y.; Zhou, C.; Li, S.-Y.; Wang, W.-C.; Zhao, W.-F.; Yang, Z.-H.; Zhong, C.-Y.; Li, G.-Z. Prostate cancer detection: Comparison of t2-weighted imaging, diffusion-weighted imaging, proton magnetic resonance spectroscopic imaging, and the three techniques combined. Acta Radiol. 2008, 49, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Sanda, M.G.; Cadeddu, J.A.; Kirkby, E.; Chen, R.C.; Crispino, T.; Fontanarosa, J.; Freedland, S.J.; Greene, K.; Klotz, L.H.; Makarov, D.V.; et al. Clinically Localized Prostate Cancer: AUA/ASTRO/SUO Guideline. Part I: Risk Stratification, Shared Decision Making, and Care Options. J. Urol. 2018, 199, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Faiella, E.; Vertulli, D.; Esperto, F.; Cordelli, E.; Soda, P.; Muraca, R.M.; Moramarco, L.P.; Grasso, R.F.; Zobel, B.B.; Santucci, D.; et al. Quantib Prostate Compared to an Expert Radiologist for the Diagnosis of Prostate Cancer on mpMRI: A Single-Center Preliminary Study. Tomography 2022, 8, 2010–2019. [Google Scholar] [CrossRef]
- Ferro, M.; de Cobelli, O.; Musi, G.; del Giudice, F.; Carrieri, G.; Busetto, G.M.; Falagario, U.G.; Sciarra, A.; Maggi, M.; Crocetto, F.; et al. Radiomics in prostate cancer: An up-to-date review. Ther. Adv. Urol. 2022, 14, 175628722211090. [Google Scholar] [CrossRef]
- Sánchez Iglesias, Á.; Morillo Macías, V.; Picó Peris, A.; Fuster-Matanzo, A.; Infante, A.N.; Soria, R.M.; Bataller, F.B.; Pomar, M.D.; Meléndez, C.C.; Huertas, R.Y.; et al. Prostate Region-Wise Imaging Biomarker Profiles for Risk Stratification and Biochemical Recurrence Prediction. Cancers 2023, 15, 4163. [Google Scholar] [CrossRef]
- Chiacchio, G.; Castellani, D.; Nedbal, C.; De Stefano, V.; Brocca, C.; Tramanzoli, P.; Galosi, A.B.; da Silva, R.D.; Teoh, J.Y.-C.; Tiong, H.Y.; et al. Radiomics vs radiologist in prostate cancer. Results from a systematic review. World J. Urol. 2023, 41, 709–724. [Google Scholar] [CrossRef]
- Nketiah, G.; Elschot, M.; Kim, E.; Teruel, J.R.; Scheenen, T.W.; Bathen, T.F.; Selnæs, K.M. T2-weighted MRI-derived textural features reflect prostate cancer aggressiveness: Preliminary results. Eur. Radiol. 2017, 27, 3050–3059. [Google Scholar] [CrossRef]
- Li, Q.; Lu, H.; Choi, J.; Gaget, K.; Feuerlein, S.; Pow-Sang, J.M.; Gillies, R.; Balagurunathan, Y. Radiological semantics discriminate clinically significant grade prostate cancer. Cancer Imaging 2019, 19, 81. [Google Scholar] [CrossRef]
- Duenweg, S.R.; Bobholz, S.A.; Barrett, M.J.; Lowman, A.K.; Winiarz, A.; Nath, B.; Stebbins, M.; Bukowy, J.; Iczkowski, K.A.; Jacobsohn, K.M.; et al. T2-Weighted MRI Radiomic Features Predict Prostate Cancer Presence and Eventual Biochemical Recurrence. Cancers 2023, 15, 4437. [Google Scholar] [CrossRef]
- Rigatti, S.J. Random Forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef]
- Dikaios, N.; Alkalbani, J.; Sidhu, H.S.; Fujiwara, T.; Abd-Alazeez, M.; Kirkham, A.; Allen, C.; Ahmed, H.; Emberton, M.; Freeman, A.; et al. Logistic regression model for diagnosis of transition zone prostate cancer on multi-parametric MRI. Eur. Radiol. 2015, 25, 523–532. [Google Scholar] [CrossRef]
- Ertas, G. Detection of high GS risk group prostate tumors by diffusion tensor imaging and logistic regression modelling. Magn. Reson. Imaging 2018, 50, 125–133. [Google Scholar] [CrossRef]
- Lay, N.; Tsehay, Y.; Greer, M.D.; Turkbey, B.; Kwak, J.T.; Choyke, P.L.; Pinto, P.; Wood, B.J.; Summers, R.M. Detection of prostate cancer in multiparametric MRI using random forest with instance weighting. J. Med. Imaging 2017, 4, 024506. [Google Scholar] [CrossRef]
- Liu, X.; He, W. Adaptive kernel scaling support vector machine with application to a prostate cancer image study. J. Appl. Stat. 2022, 49, 1465–1484. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Zhang, Y.; Sun, Z.; Zhang, X.; Wang, X. Preoperative prediction of pelvic lymph nodes metastasis in prostate cancer using an ADC-based radiomics model: Comparison with clinical nomograms and PI-RADS assessment. Abdom. Radiol. 2022, 47, 3327–3337. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tian, J.; Wu, J.; Zhang, Y.; Walng, X.; Zhang, X.; Wang, X. Utility of diffusion weighted imaging-based radiomics nomogram to predict pelvic lymph nodes metastasis in prostate cancer. BMC Med. Imaging 2022, 22, 190. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Miao, Q.; Liu, Y.; Mirak, S.A.; Hosseiny, M.; Scalzo, F.; Raman, S.S.; Sung, K. Multiparametric MRI-based radiomics model to predict pelvic lymph node invasion for patients with prostate cancer. Eur. Radiol. 2022, 32, 5688–5699. [Google Scholar] [CrossRef]
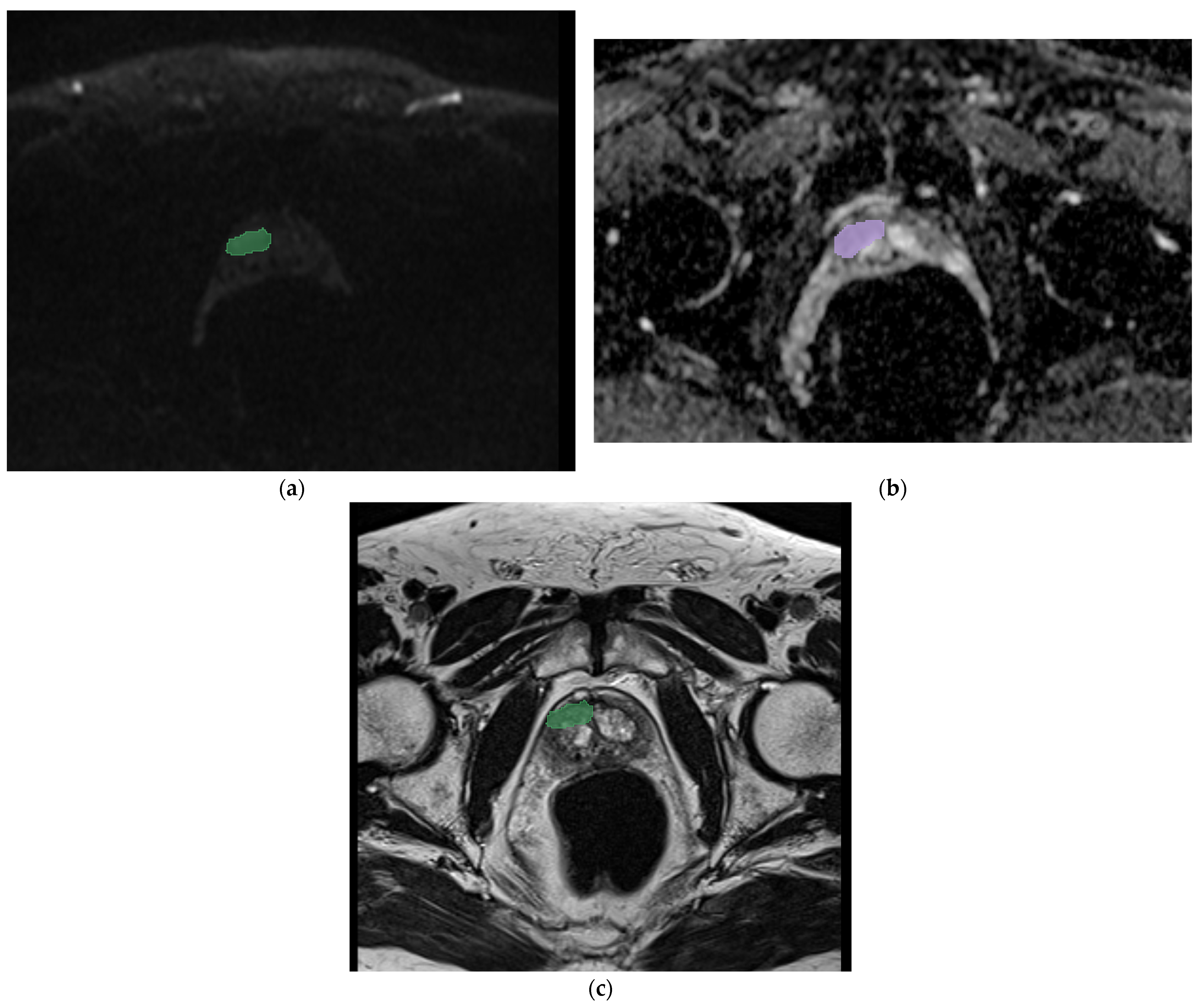
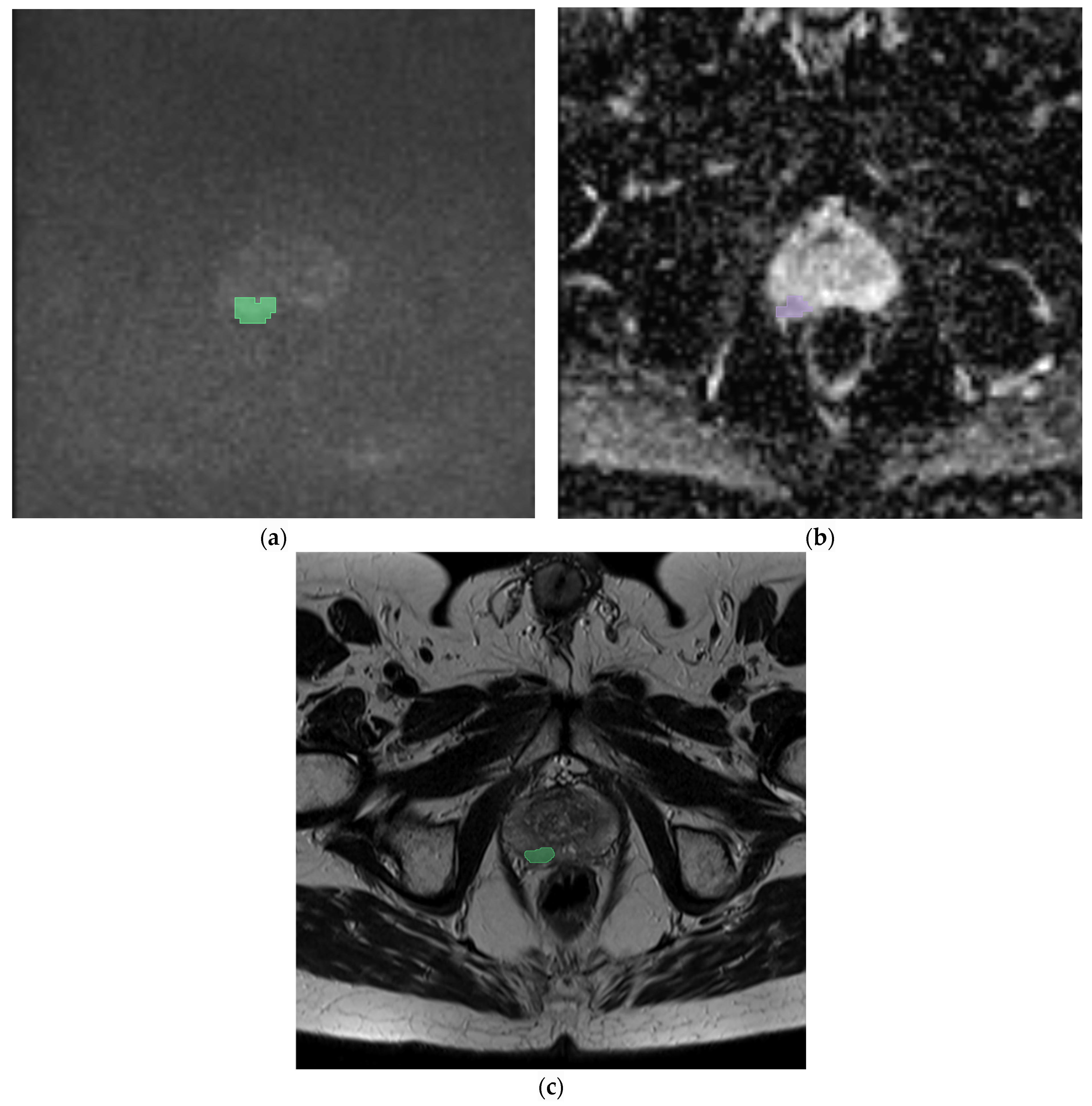
| Category | Number |
|---|---|
| Number of patients | 95 |
| Patients with metastatic nodule at lymphadenectomy | 35 |
| Patients without metastatic nodule at lymphadenectomy | 60 |
| Race | Caucasian |
| Age | 40–80 |
| PSA [ng/mL] (Median, range) | 4.5 (7.1) |
| Period of mp-MRI | 2016–2023 |
| Gleason Grade (Mediana) | 7 |
| Tumor target Zone Peripheral | 33 |
| Tumor target Zone Transition | 12 |
| Disease Grade | T3 (16) T2 (10) T3,5 (19) |
| (a) | ||||
| # | Features | LogRegCoef | SVMCoef | RFImportance |
| 1 | T2_noduloglcmContrast | 0.0084 | 0.0427 | 0.0457 |
| 2 | T2_nodulofirstorderKurtosis | −0.5975 | −0.2378 | 0.0395 |
| 3 | T2_nodulofirstorderMeanAbsoluteDeviation | −0.0009 | 0.0231 | 0.0364 |
| 4 | T2_nodulofirstorderVariance | 0.4459 | 0.1540 | 0.0332 |
| 5 | T2_noduloglcmIdm | 0.1337 | 0.0445 | 0.0320 |
| 6 | T2_noduloshapeSphericity | 0.0420 | 0.0148 | 0.0000 |
| 7 | T2_nodulofirstorder10Percentile | −0.1215 | −0.0385 | 0.0000 |
| 8 | T2_noduloglcmDifferenceVariance | −0.3277 | −0.1183 | 0.0000 |
| 9 | T2_noduloglrlmGrayLevelNonUniformityNormalized | 0.0420 | 0.0148 | 0.0000 |
| 10 | T2_noduloglrlmLowGrayLevelRunEmphasis | 0.0420 | 0.0148 | 0.0000 |
| (b) | ||||
| # | Feature | RFImportance | LogRegCoef | |
| 1 | DWIfirstorderEnergy | 0.0770 | 3.50 × 10−8 | |
| 2 | DWIglcmIdn | 0.0693 | −4.46 × 10−13 | |
| 3 | DWIglrlmRunLengthNonUniformityNormalized | 0.0663 | −1.39 × 10−13 | |
| 4 | DWIglszmGrayLevelNonUniformityNormalized | 0.0592 | −2.85 × 10−13 | |
| 5 | DWIgldmSmallDependenceEmphasis | 0.0507 | 9.34 × 10−15 | |
| 6 | DWIfirstorderMedian | 0.0481 | −2.17 × 10−11 | |
| 7 | DWIglrlmGrayLevelNonUniformityNormalized | 0.0445 | −2.95 × 10−13 | |
| 8 | DWIglcmImc1 | 0.0429 | 5.94 × 10−14 | |
| 9 | DWIglcmIdmn | 0.0423 | −3.91 × 10−13 | |
| 10 | DWIglcmCorrelation | 0.0410 | −1.85 × 10−13 | |
| (c) | ||||
| # | Feature | RF Importance | LogRegCoef | |
| 1 | ADCglszmGrayLevelNonUniformityNormalized | 0.0339 | 1.25 × 10−33 | |
| 2 | ADCglcmSumAverage | 0.0320 | −3.62 × 10−15 | |
| 3 | ADCshapeMeshVolume | 0.0297 | −1.07 × 10−15 | |
| 4 | ADCfirstorderUniformity | 0.0274 | −7.88 × 10−33 | |
| 5 | ADCshapeMajorAisLength | 0.0251 | 3.17 × 10−15 | |
| 6 | ADCfirstorderMeanAbsoluteDeviation | 0.0249 | 1.27 × 10−15 | |
| 7 | ADCshapeMaimum2DDiameterSlice | 0.0231 | 2.15 × 10−15 | |
| 8 | ADCglszmSmallAreaLowGrayLevelEmphasis | 0.0228 | 1.56 × 10−32 | |
| 9 | ADCglrlmRunVariance | 0.0222 | 9.95 × 10−32 | |
| 10 | ADCglcmImc1 | 0.0222 | 7.52 × 10−32 | |
| SEQUENCES | MODEL | Accuracy | AUC |
|---|---|---|---|
| T2 nod | Random Forest | 0.78 | 0.78 |
| Logistic Regression | 0.78 | 0.78 | |
| Support Vector Machine | 0.78 | 0.17 | |
| DWI | Random Forest | 0.86 | 0.89 |
| Logistic Regression | 0.78 | 0.67 | |
| Support Vector Machine | 0.89 | 0.28 | |
| ADC | Random Forest | 0.89 | 0.67 |
| Logistic Regression | 0.67 | 0.67 | |
| Support Vector Machine | 0.78 | 0.67 |
| (a) | ||||
| Model Comparison | AUC Radiomics Model | AUC Nomogram | Z-Score | p-Value |
| LR vs. Briganti | 0.89 | 0.79 | 0.833 | 0.405 |
| LR vs. Partin | 0.89 | 0.78 | 1.117 | 0.264 |
| LR vs. MSKCC | 0.89 | 0.78 | 1.132 | 0.258 |
| LR vs. YALE | 0.89 | 0.78 | 1.123 | 0.262 |
| RF vs. Briganti | 0.78 | 0.79 | −0.345 | 0.730 |
| RF vs. Partin | 0.78 | 0.78 | 0 | 1 |
| RF vs. MSKCC | 0.78 | 0.78 | 0 | 1 |
| RF vs. YALE | 0.78 | 0.78 | 0 | 1 |
| SVM vs. Briganti | 0.17 | 0.79 | 1248.94 | <0.05 |
| SVM vs. Partin | 0.17 | 0.78 | 1221.28 | <0.05 |
| SVM vs. MSKCC | 0.17 | 0.78 | 1221.64 | <0.05 |
| SVM vs. YALE | 0.17 | 0.78 | 1221.46 | <0.05 |
| (b) | ||||
| Model Comparison | AUC radiomics model | AUC nomogram | Z-Score | p-Value |
| RF vs. Briganti | 0.89 | 0.79 | 2.00 | 0.0455 |
| RF vs. Partin | 0.89 | 0.78 | 2.20 | 0.0278 |
| RF vs. MSKCC | 0.89 | 0.78 | 2.20 | 0.0278 |
| RF vs. YALE | 0.89 | 0.78 | 2.20 | 0.0278 |
| LR vs. Briganti | 0.671 | 0.79 | 1.733 | 0.083 |
| LR vs. Partin | 0.671 | 0.78 | 1.546 | 0.122 |
| LR vs. MSKCC | 0.671 | 0.78 | 1.529 | 0.126 |
| LR vs. YALE | 0.671 | 0.78 | 1.507 | 0.132 |
| (c) | ||||
| Model Comparison | AUC radiomics model | AUC nomogram | Z-score | p-value |
| LR vs. Briganti | 0.67 | 0.79 | −0.439 | 0.661 |
| LR vs. Partin | 0.67 | 0.78 | 0.039 | 0.969 |
| LR vs. MSKCC | 0.67 | 0.78 | −0.028 | 0.978 |
| LR vs. YALE | 0.67 | 0.78 | 0.065 | 0.948 |
| RF vs. Briganti | 0.67 | 0.80 | −1.27 | 0.205 |
| RF vs. Partin | 0.67 | 0.78 | −1.11 | 0.268 |
| RF vs. MSKCC | 0.67 | 0.78 | −1.06 | 0.290 |
| RF vs. YALE | 0.67 | 0.78 | −1.05 | 0.295 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santucci, D.; Ragone, R.; Vergantino, E.; Vaccarino, F.; Esperto, F.; Prata, F.; Scarpa, R.M.; Papalia, R.; Beomonte Zobel, B.; Grasso, F.R.; et al. Comparison between Three Radiomics Models and Clinical Nomograms for Prediction of Lymph Node Involvement in PCa Patients Combining Clinical and Radiomic Features. Cancers 2024, 16, 2731. https://doi.org/10.3390/cancers16152731
Santucci D, Ragone R, Vergantino E, Vaccarino F, Esperto F, Prata F, Scarpa RM, Papalia R, Beomonte Zobel B, Grasso FR, et al. Comparison between Three Radiomics Models and Clinical Nomograms for Prediction of Lymph Node Involvement in PCa Patients Combining Clinical and Radiomic Features. Cancers. 2024; 16(15):2731. https://doi.org/10.3390/cancers16152731
Chicago/Turabian StyleSantucci, Domiziana, Raffaele Ragone, Elva Vergantino, Federica Vaccarino, Francesco Esperto, Francesco Prata, Roberto Mario Scarpa, Rocco Papalia, Bruno Beomonte Zobel, Francesco Rosario Grasso, and et al. 2024. "Comparison between Three Radiomics Models and Clinical Nomograms for Prediction of Lymph Node Involvement in PCa Patients Combining Clinical and Radiomic Features" Cancers 16, no. 15: 2731. https://doi.org/10.3390/cancers16152731
APA StyleSantucci, D., Ragone, R., Vergantino, E., Vaccarino, F., Esperto, F., Prata, F., Scarpa, R. M., Papalia, R., Beomonte Zobel, B., Grasso, F. R., & Faiella, E. (2024). Comparison between Three Radiomics Models and Clinical Nomograms for Prediction of Lymph Node Involvement in PCa Patients Combining Clinical and Radiomic Features. Cancers, 16(15), 2731. https://doi.org/10.3390/cancers16152731







