Biomolecular Classification in Endometrial Cancer: Onset, Evolution, and Further Perspectives: A Critical Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature and Search Strategy
2.2. Outcome Measures
2.2.1. Histotype Specificity
2.2.2. Follow-Up
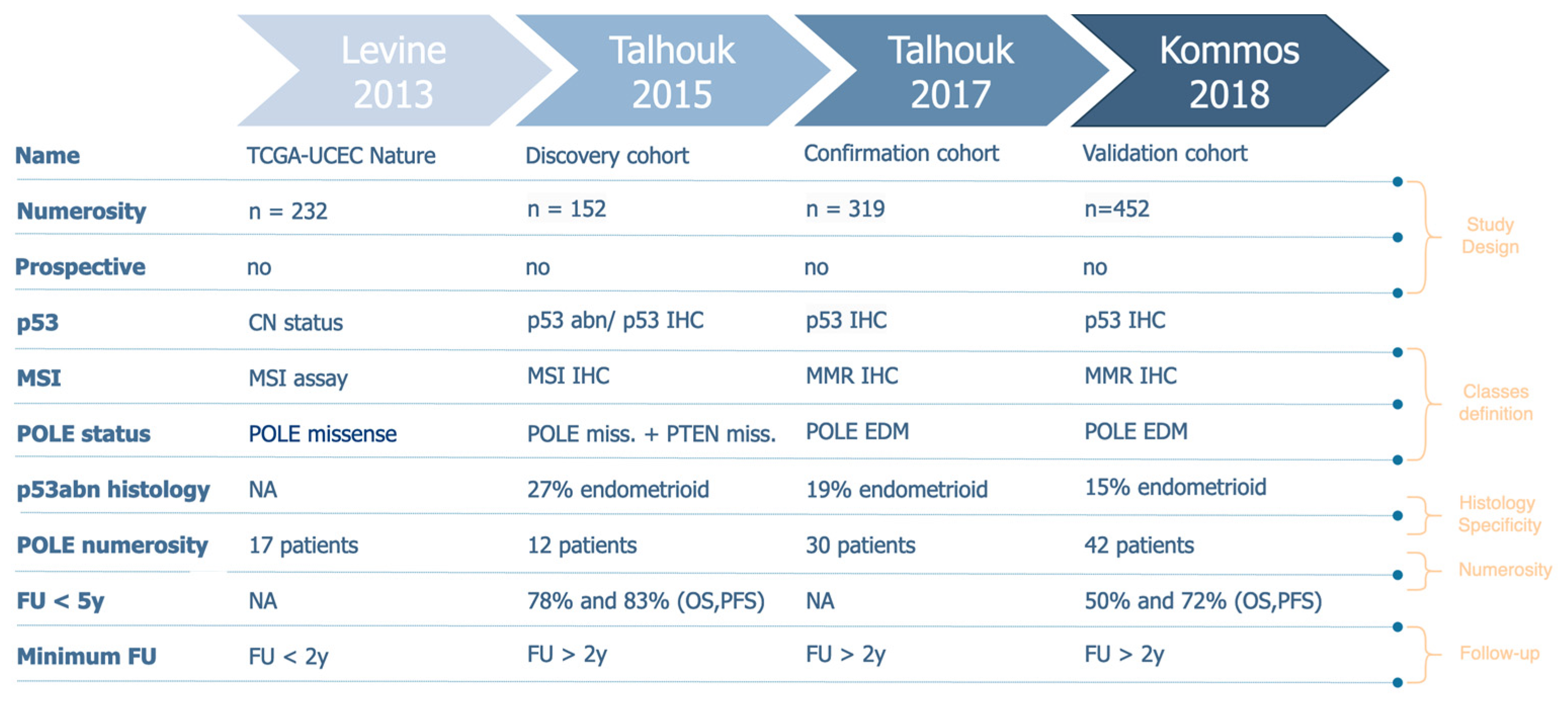
2.2.3. Molecular Profiles Definition and Numerosity
2.2.4. Study Design
3. Results
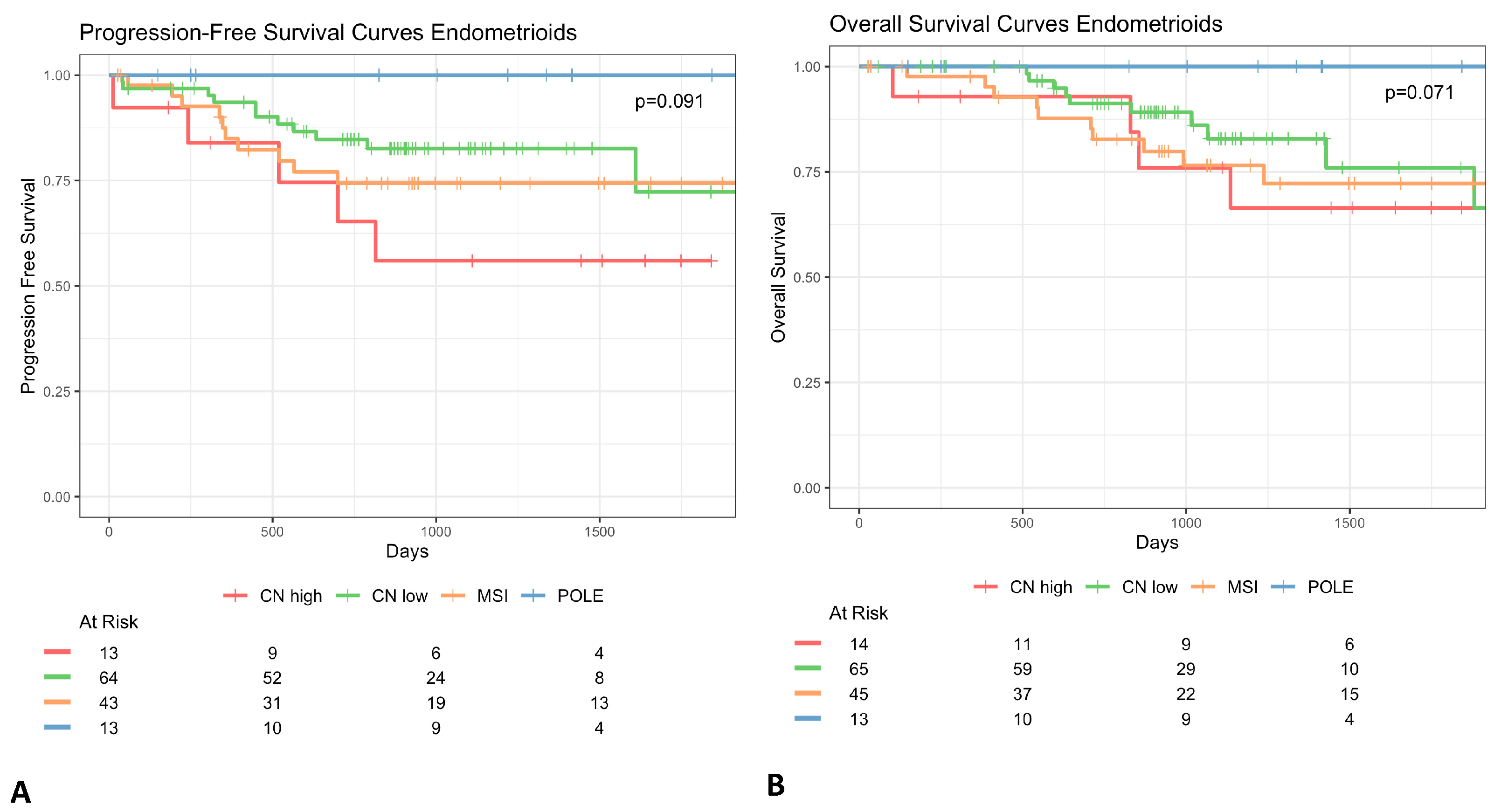
3.1. Histotype Specificity
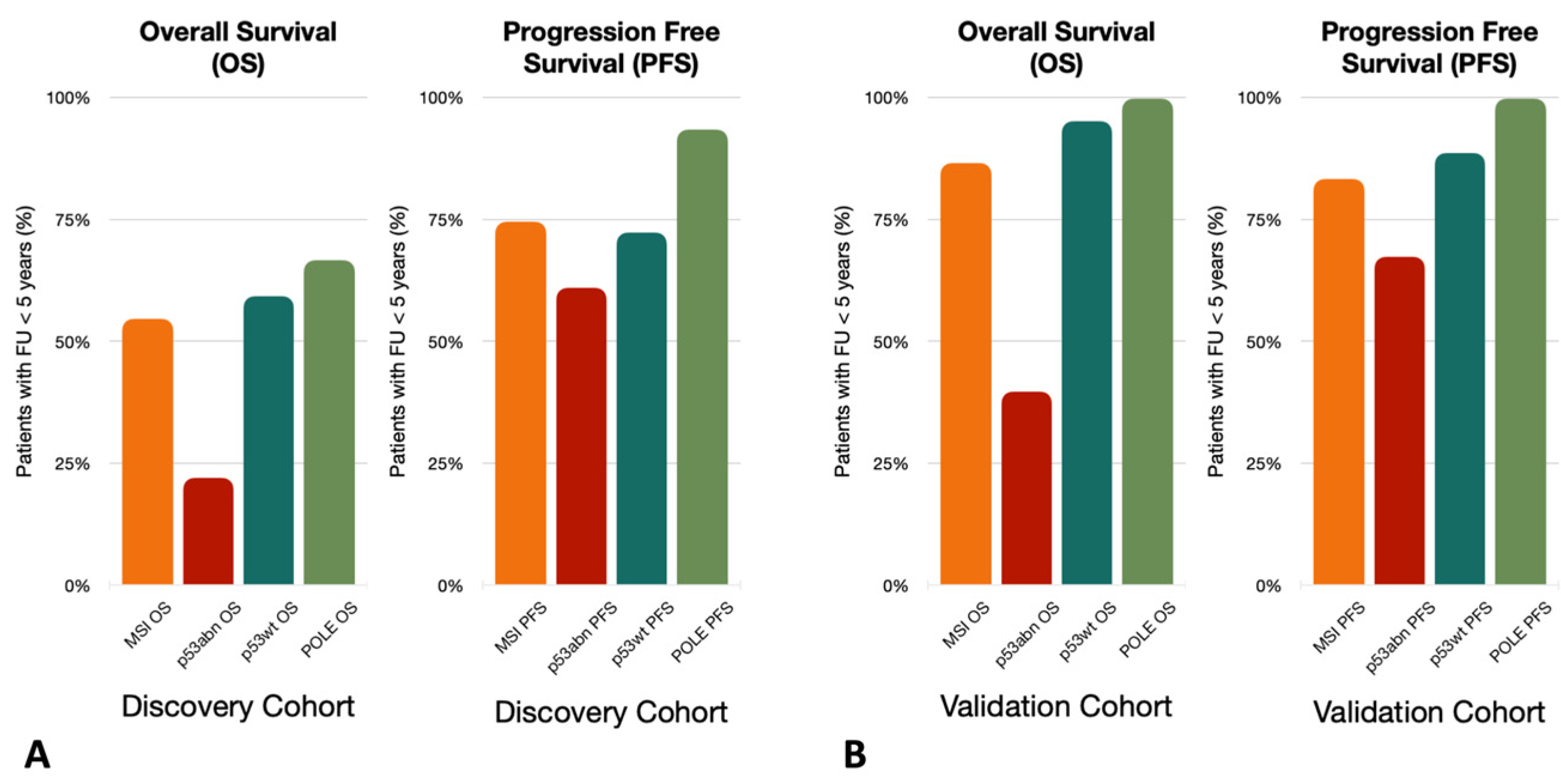
3.2. Follow-Up
3.3. Molecular Profiles Definition and Numerosity
3.4. Study Design
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
List of Abbreviations
| CN | copy-number |
| CN-high | Copy Number high |
| EC | Endometrial Cancer |
| ESGO | European Society of Gynaecological Oncology |
| ESMO | European Society of Medical Oncology |
| ESTRO | European SocieTy for Radiotherapy & Oncology |
| FIGO | International Federation of Gynaecology and Obstetrics |
| IHC | immunohistochemistry |
| KM | Kaplan-Meier |
| LoH | Loss of Heterozygosity |
| MMR | mismatch repair |
| MSI | Micro Satellite Instability |
| NSMP | no specific molecular profile |
| POLE | polymerase-E |
| ProMisE | Proactive Molecular Risk Classifier for Endometrial Cancer |
| TCGA | The Cancer Genome Atlas |
References
- Levine, D.; The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, E.; Bosse, T.; Nout, R.A.; Mackay, H.J.; Church, D.N.; Nijman, H.W.; Leary, A.; Edmondson, R.J.; Powell, M.E.; Crosbie, E.J.; et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod. Pathol. 2015, 28, 836–844. [Google Scholar] [CrossRef]
- León-Castillo, A.; de Boer, S.M.; Powell, M.E.; Mileshkin, L.R.; Mackay, H.J.; Leary, A.; Nijman, H.W.; Singh, N.; Pollock, P.M.; Bessette, P.; et al. Molecular Classification of the PORTEC-3 Trial for High-Risk Endometrial Cancer: Impact on Prognosis and Benefit From Adjuvant Therapy. J. Clin. Oncol. 2020, 38, 3388–3397. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- van der Putten, L.J.; Visser, N.C.; van de Vijver, K.; Santacana, M.; Bronsert, P.; Bulten, J.; Hirschfeld, M.; Colas, E.; Gil-Moreno, A.; Garcia, A.; et al. L1CAM expression in endometrial carcinomas: An ENITEC collaboration study. Br. J. Cancer 2016, 115, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Van Gool, I.C.; Stelloo, E.; Nout, R.A.; Nijman, H.W.; Edmondson, R.J.; Church, D.N.; MacKay, H.J.; Leary, A.; Powell, M.E.; Mileshkin, L.; et al. Prognostic significance of L1CAM expression and its association with mutant p53 expression in high-risk endometrial cancer. Mod. Pathol. 2016, 29, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef]
- León-Castillo, A.; Britton, H.; McConechy, M.K.; McAlpine, J.N.; Nout, R.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; Rau, T.T.; et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J. Pathol. 2019, 250, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, S.; McConechy, M.K.; Kommoss, F.; Leung, S.; Bunz, A.; Magrill, J.; Britton, H.; Grevenkamp, F.; Karnezis, A.; Yang, W.; et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann. Oncol. 2018, 29, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Colombo, N.; Creutzberg, C.; Amant, F.; Bosse, T.; González-Martín, A.; Ledermann, J.; Marth, C.; Nout, R.; Querleu, D.; Mirza, M.R.; et al. ESMO-ESGO-ESTRO consensus con-ference on endometrial cancer: Diagnosis, treatment and follow-up. Ann. Oncol. 2016, 26, 2–30. [Google Scholar]
- Cosgrove, C.M.; Tritchler, D.L.; Cohn, D.E.; Mutch, D.G.; Rush, C.M.; Lankes, H.A.; Creasman, W.T.; Miller, D.S.; Ramirez, N.C.; Geller, M.A.; et al. An NRG Oncology/GOG study of molecular classification for risk prediction in endometrioid endometrial cancer. Gynecol. Oncol. 2018, 148, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef] [PubMed]
- Berek, J.S.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lindemann, K.; Mutch, D.; Concin, N.; Endometrial Cancer Staging Subcommittee, FIGO Women's Cancer Committee; et al. FIGO staging of endometrial cancer: 2023. Int. J. Gynecol. Obstet. 2023, 162, 383–394. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, J.; Leon-Castillo, A.; Bosse, T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J. Pathol. 2018, 244, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Piulats, J.M.; Guerra, E.; Gil-Martín, M.; Roman-Canal, B.; Gatius, S.; Sanz-Pamplona, R.; Velasco, A.; Vidal, A.; Matias-Guiu, X. Molecular approaches for classifying endometrial carcinoma. Gynecol. Oncol. 2016, 145, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Vermij, L.; Smit, V.; Nout, R.; Bosse, T. Incorporation of molecular characteristics into endometrial cancer management. Histopathology 2019, 76, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Köbel, M.; Nelson, G.S. Letter in response to: McAlpine J, Leon-Castillo A, Bosse T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J Pathol 2018; 244: 538–549. J. Pathol. 2018, 245, 249–250. [Google Scholar] [CrossRef] [PubMed]
- McCluggage, W.G.; Bosse, T.; Gilks, C.B.; Howitt, B.E.; McAlpine, J.N.; Nucci, M.R.; Rabban, J.T.; Singh, N.; Talia, K.L.; Parra-Herran, C. FIGO 2023 endometrial cancer staging: Too much, too soon? Int. J. Gynecol. Cancer 2024, 34, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Church, D.N.; Stelloo, E.; Nout, R.A.; Valtcheva, N.; Depreeuw, J.; ter Haar, N.; Noske, A.; Amant, F.; Tomlinson, I.P.M.; Wild, P.J.; et al. Prognostic Significance of POLE Proofreading Mutations in Endometrial Cancer. JNCI J. Natl. Cancer Inst. 2014, 107, 402. [Google Scholar] [CrossRef] [PubMed]
- León-Castillo, A.; Gilvazquez, E.; Nout, R.; Smit, V.T.; McAlpine, J.N.; McConechy, M.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; et al. Clinicopathological and molecular characterisation of ‘multiple-classifier’ endometrial carcinomas. J. Pathol. 2020, 250, 312–322. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruno, V.; Betti, M.; Mauro, J.; Buda, A.; Vizza, E. Biomolecular Classification in Endometrial Cancer: Onset, Evolution, and Further Perspectives: A Critical Review. Cancers 2024, 16, 2959. https://doi.org/10.3390/cancers16172959
Bruno V, Betti M, Mauro J, Buda A, Vizza E. Biomolecular Classification in Endometrial Cancer: Onset, Evolution, and Further Perspectives: A Critical Review. Cancers. 2024; 16(17):2959. https://doi.org/10.3390/cancers16172959
Chicago/Turabian StyleBruno, Valentina, Martina Betti, Jessica Mauro, Alessandro Buda, and Enrico Vizza. 2024. "Biomolecular Classification in Endometrial Cancer: Onset, Evolution, and Further Perspectives: A Critical Review" Cancers 16, no. 17: 2959. https://doi.org/10.3390/cancers16172959
APA StyleBruno, V., Betti, M., Mauro, J., Buda, A., & Vizza, E. (2024). Biomolecular Classification in Endometrial Cancer: Onset, Evolution, and Further Perspectives: A Critical Review. Cancers, 16(17), 2959. https://doi.org/10.3390/cancers16172959







