Impact of Physical Activity on DNA Methylation Signatures in Breast Cancer Patients: A Systematic Review with Bioinformatic Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods and Materials
2.1. Study Selection
2.2. Data Collection Process
2.3. Functional Profiling and Tissue Enrichment Analysis
3. Results
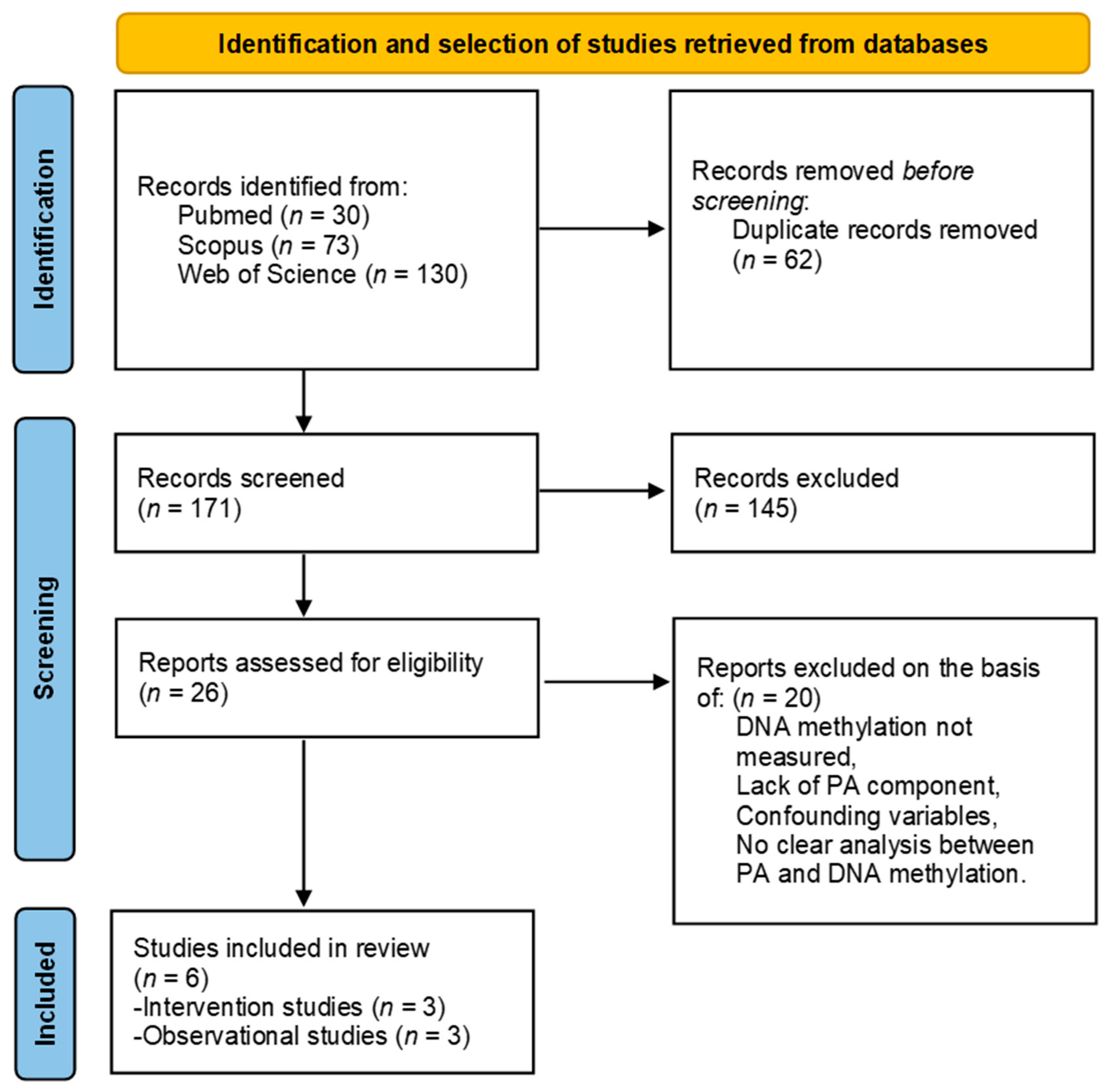
3.1. Search Results
3.2. Studies Characteristics
3.3. Study Populations and Treatment Descriptions
3.4. Results of Studies with Exercise Interventions
3.5. Results of Studies with Self-Reported PA Levels
3.6. Effects of Physical Activity on Recovery and Survival Outcomes
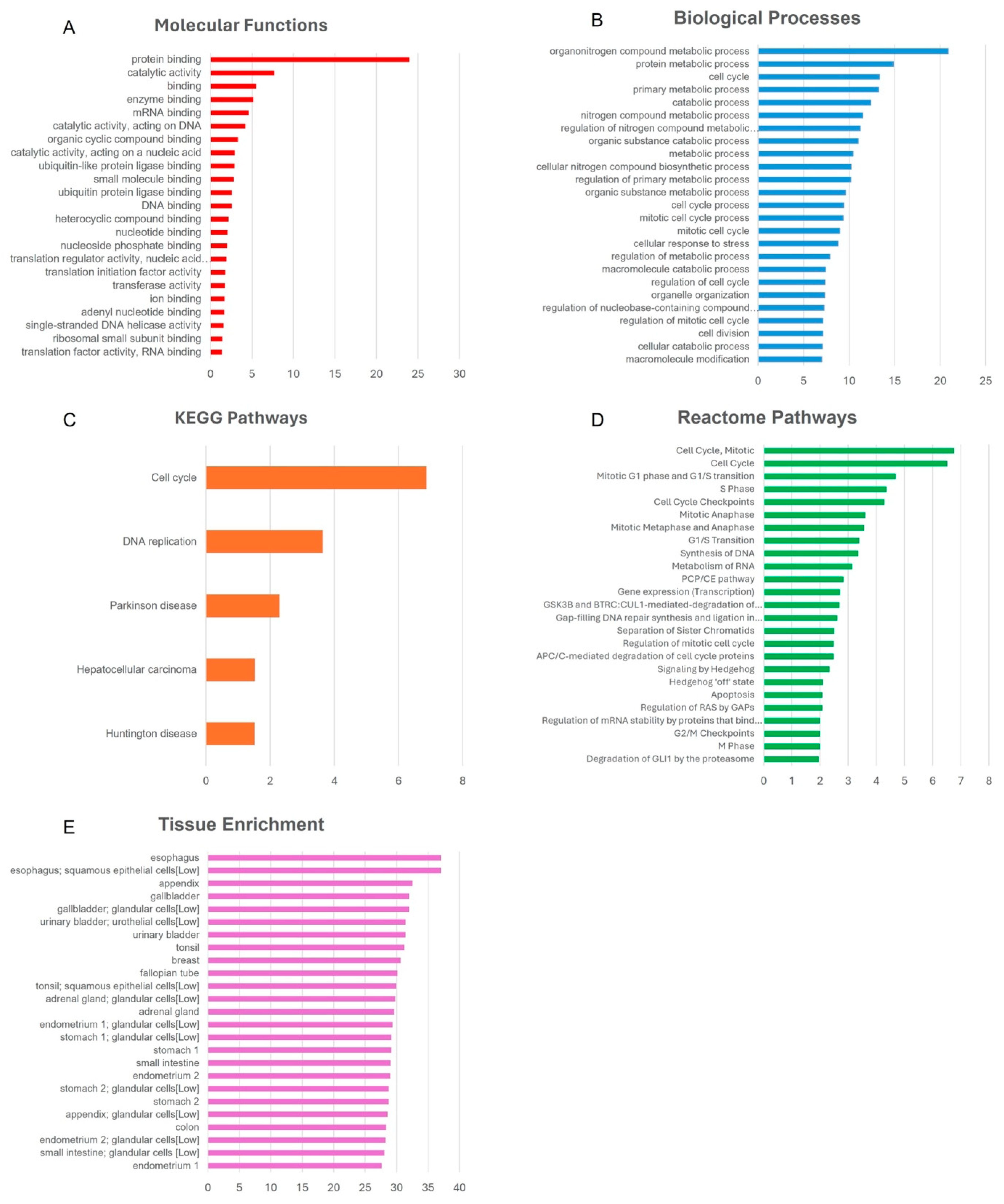
3.7. Bioinformatics Pathway Analysis
4. Discussion
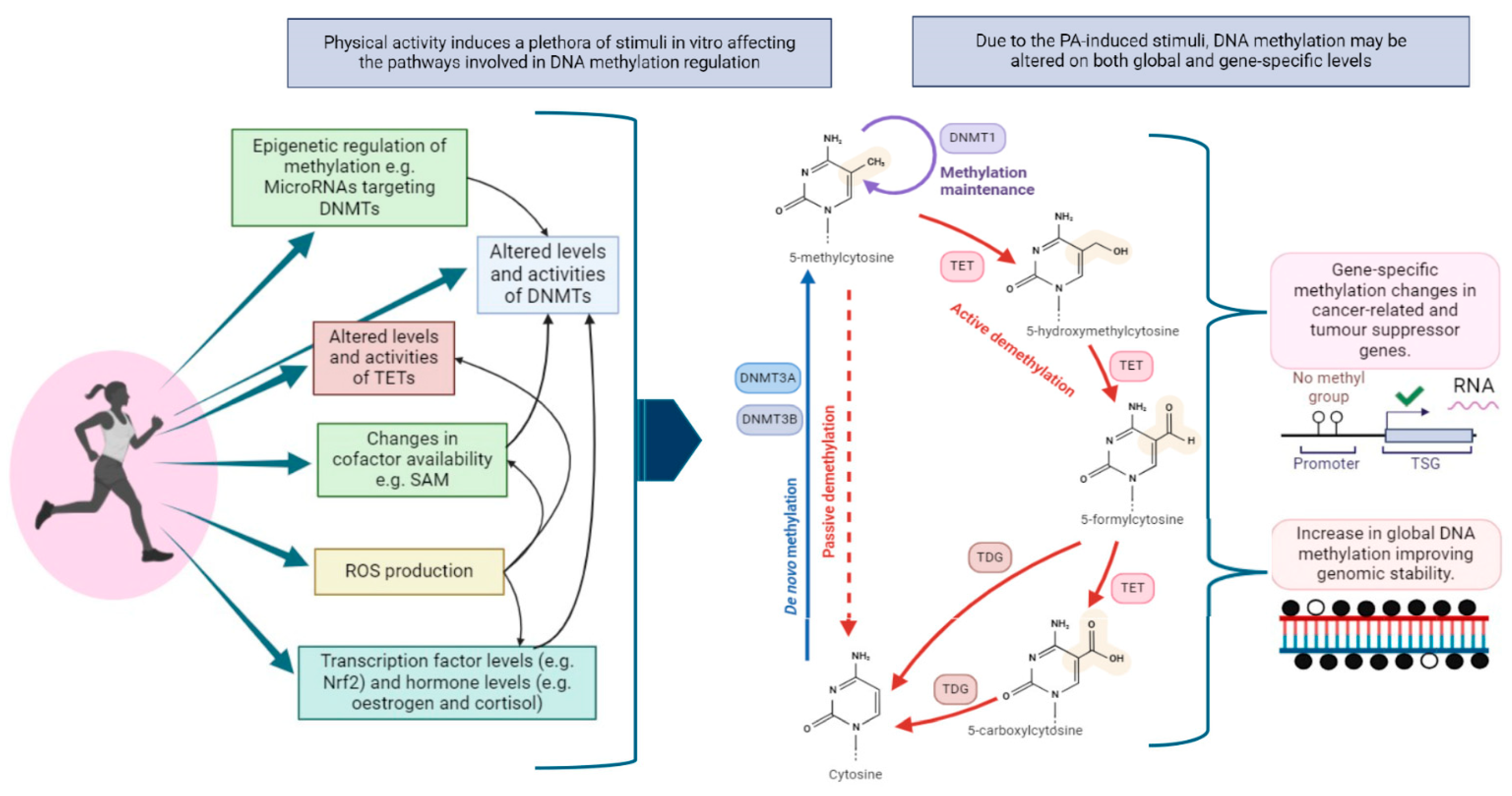
4.1. Global DNA Methylation
4.2. Gene-Specific DNA Methylation
4.3. Long-Term Effects of PA on DNA Methylation
4.4. Functional Impact of PA-Induced DNA Methylation Changes
4.5. Mechanisms Underlying PA-Induced DNA Methylation Changes
5. Study Limitations
6. Future Implications for Research
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cao, X.; Tang, Q.; Holland-Letz, T.; Gündert, M.; Ćuk, K.; Schott, S.; Heil, J.; Golatta, M.; Sohn, C.; Schneeweiss, A.; et al. Evaluation of Promoter Methylation of RASSF1A and ATM in Peripheral Blood of Breast Cancer Patients and Healthy Control Individuals. Int. J. Mol. Sci. 2018, 19, 900. [Google Scholar] [CrossRef]
- Zhu, G.; Zhang, X.; Wang, Y.; Xiong, H.; Zhao, Y.; Sun, F. Effects of Exercise Intervention in Breast Cancer Survivors: A Meta-Analysis of 33 Randomized Controlled Trails. OTT 2016, 9, 2153–2168. [Google Scholar] [CrossRef]
- Ahmad, A. Breast Cancer Statistics: Recent Trends. In Breast Cancer Metastasis and Drug Resistance; Ahmad, A., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2019; Volume 1152, pp. 1–7. ISBN 978-3-030-20300-9. [Google Scholar]
- Brennan, S.F.; Cantwell, M.M.; Cardwell, C.R.; Velentzis, L.S.; Woodside, J.V. Dietary Patterns and Breast Cancer Risk: A Systematic Review and Meta-Analysis. Am. J. Clin. Nutr. 2010, 91, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, M.M.; Gapstur, S.M.; Sun, J.; Diver, W.R.; Hannan, L.M.; Thun, M.J. Active Smoking and Breast Cancer Risk: Original Cohort Data and Meta-Analysis. JNCI J. Natl. Cancer Inst. 2013, 105, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Friedenreich, C.M.; Neilson, H.K.; Lynch, B.M. State of the Epidemiological Evidence on Physical Activity and Cancer Prevention. Eur. J. Cancer 2010, 46, 2593–2604. [Google Scholar] [CrossRef] [PubMed]
- Lynch, B.M.; Neilson, H.K.; Friedenreich, C.M. Physical Activity and Breast Cancer Prevention. In Physical Activity and Cancer; Courneya, K.S., Friedenreich, C.M., Eds.; Recent Results in Cancer Research; Springer: Berlin/Heidelberg, Germany, 2010; Volume 186, pp. 13–42. ISBN 978-3-642-04230-0. [Google Scholar]
- Friedenreich, C.M.; Woolcott, C.G.; McTiernan, A.; Ballard-Barbash, R.; Brant, R.F.; Stanczyk, F.Z.; Terry, T.; Boyd, N.F.; Yaffe, M.J.; Irwin, M.L.; et al. Alberta Physical Activity and Breast Cancer Prevention Trial: Sex Hormone Changes in a Year-Long Exercise Intervention Among Postmenopausal Women. JCO 2010, 28, 1458–1466. [Google Scholar] [CrossRef]
- Irwin, M.L.; Cadmus, L.; Alvarez-Reeves, M.; O’Neil, M.; Mierzejewski, E.; Latka, R.; Yu, H.; DiPietro, L.; Jones, B.; Knobf, M.T.; et al. Recruiting and Retaining Breast Cancer Survivors into a Randomized Controlled Exercise Trial: The Yale Exercise and Survivorship Study. Cancer 2008, 112, 2593–2606. [Google Scholar] [CrossRef]
- Zeng, H.; Irwin, M.L.; Lu, L.; Risch, H.; Mayne, S.; Mu, L.; Deng, Q.; Scarampi, L.; Mitidieri, M.; Katsaros, D.; et al. Physical Activity and Breast Cancer Survival: An Epigenetic Link through Reduced Methylation of a Tumor Suppressor Gene L3MBTL1. Breast Cancer Res Treat 2012, 133, 127–135. [Google Scholar] [CrossRef]
- Serra, M.C.; Ryan, A.S.; Ortmeyer, H.K.; Addison, O.; Goldberg, A.P. Resistance Training Reduces Inflammation and Fatigue and Improves Physical Function in Older Breast Cancer Survivors. Menopause 2018, 25, 211–216. [Google Scholar] [CrossRef]
- Grazioli, E.; Cerulli, C.; Dimauro, I.; Moretti, E.; Murri, A.; Parisi, A. New Strategy of Home-Based Exercise during Pandemic COVID-19 in Breast Cancer Patients: A Case Study. Sustainability 2020, 12, 6940. [Google Scholar] [CrossRef]
- Paronetto, M.P.; Dimauro, I.; Grazioli, E.; Palombo, R.; Guidotti, F.; Fantini, C.; Sgrò, P.; De Francesco, D.; Di Luigi, L.; Capranica, L.; et al. Exercise-Mediated Downregulation of MALAT1 Expression and Implications in Primary and Secondary Cancer Prevention. Free. Radic. Biol. Med. 2020, 160, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Dimauro, I.; Grazioli, E.; Antinozzi, C.; Duranti, G.; Arminio, A.; Mancini, A.; Greco, E.A.; Caporossi, D.; Parisi, A.; Di Luigi, L. Estrogen-Receptor-Positive Breast Cancer in Postmenopausal Women: The Role of Body Composition and Physical Exercise. Int. J. Environ. Res. Public Health 2021, 18, 9834. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, S.; Kaaks, R.; Friedenreich, C.M.; Key, T.J.; Travis, R.; Biessy, C.; Slimani, N.; Overvad, K.; Østergaard, J.N.; Tjønneland, A.; et al. Physical Activity, Sex Steroid, and Growth Factor Concentrations in Pre- and Post-Menopausal Women: A Cross-Sectional Study within the EPIC Cohort. Cancer Causes Control 2014, 25, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Grazioli, E.; Dimauro, I.; Mercatelli, N.; Wang, G.; Pitsiladis, Y.; Di Luigi, L.; Caporossi, D. Physical Activity in the Prevention of Human Diseases: Role of Epigenetic Modifications. BMC Genom. 2017, 18, 802. [Google Scholar] [CrossRef]
- Ligibel, J.A.; Dillon, D.; Giobbie-Hurder, A.; McTiernan, A.; Frank, E.; Cornwell, M.; Pun, M.; Campbell, N.; Dowling, R.J.O.; Chang, M.C.; et al. Impact of a Pre-Operative Exercise Intervention on Breast Cancer Proliferation and Gene Expression: Results from the Pre-Operative Health and Body (PreHAB) Study. Clin. Cancer Res. 2019, 25, 5398–5406. [Google Scholar] [CrossRef]
- Dimauro, I.; Paronetto, M.P.; Caporossi, D. Epigenomic Adaptations of Exercise in the Control of Metabolic Disease and Cancer. In Nutritional Epigenomics; Elsevier: Amsterdam, The Netherlands, 2019; pp. 289–316. ISBN 978-0-12-816843-1. [Google Scholar]
- Moulton, C.; Grazioli, E.; Antinozzi, C.; Fantini, C.; Cerulli, C.; Murri, A.; Duranti, G.; Ceci, R.; Vulpiani, M.C.; Pellegrini, P.; et al. Online Home-Based Physical Activity Counteracts Changes of Redox-Status Biomarkers and Fitness Profiles during Treatment Programs in Postsurgery Female Breast Cancer Patients. Antioxidants 2023, 12, 1138. [Google Scholar] [CrossRef]
- Van De Voorde, L.; Speeckaert, R.; Van Gestel, D.; Bracke, M.; De Neve, W.; Delanghe, J.; Speeckaert, M. DNA Methylation-Based Biomarkers in Serum of Patients with Breast Cancer. Mutat. Res./Rev. Mutat. Res. 2012, 751, 304–325. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, L.; Shu, X.-O.; Cai, Q.; Shu, X.; Li, B.; Guo, X.; Ye, F.; Michailidou, K.; Bolla, M.K.; et al. Genetically Predicted Levels of DNA Methylation Biomarkers and Breast Cancer Risk: Data From 228 951 Women of European Descent. JNCI J. Natl. Cancer Inst. 2020, 112, 295–304. [Google Scholar] [CrossRef]
- Győrffy, B.; Bottai, G.; Fleischer, T.; Munkácsy, G.; Budczies, J.; Paladini, L.; Børresen-Dale, A.-L.; Kristensen, V.N.; Santarpia, L. Aberrant DNA Methylation Impacts Gene Expression and Prognosis in Breast Cancer Subtypes: Prognostic Value of Methylation in Breast Cancer Subtypes. Int. J. Cancer 2016, 138, 87–97. [Google Scholar] [CrossRef]
- Lim, U.; Song, M.-A. Dietary and Lifestyle Factors of DNA Methylation. In Cancer Epigenetics; Dumitrescu, R.G., Verma, M., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 863, pp. 359–376. ISBN 978-1-61779-611-1. [Google Scholar]
- Boyne, D.J.; King, W.D.; Brenner, D.R.; McIntyre, J.B.; Courneya, K.S.; Friedenreich, C.M. Aerobic Exercise and DNA Methylation in Postmenopausal Women: An Ancillary Analysis of the Alberta Physical Activity and Breast Cancer Prevention (ALPHA) Trial. PLoS ONE 2018, 13, e0198641. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in Cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Terry, M.B.; Delgado-Cruzata, L.; Vin-Raviv, N.; Wu, H.C.; Santella, R.M. DNA Methylation in White Blood Cells: Association with Risk Factors in Epidemiologic Studies. Epigenetics 2011, 6, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Jones, P.A. DNA Methylation and Breast Carcinogenesis. Oncogene 2002, 21, 5462–5482. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Nakayama, T.; Kajita, M.; Miyake, T.; Iwamoto, T.; Kim, S.J.; Sakai, A.; Ishihara, H.; Tamaki, Y.; Noguchi, S. Detection of Aberrant Promoter Methylation of GSTP1, RASSF1A, and RARβ2 in Serum DNA of Patients with Breast Cancer by a Newly Established One-Step Methylation-Specific PCR Assay. Breast Cancer Res. Treat 2012, 132, 165–173. [Google Scholar] [CrossRef]
- Jin, Z.; Tamura, G.; Tsuchiya, T.; Sakata, K.; Kashiwaba, M.; Osakabe, M.; Motoyama, T. Adenomatous Polyposis Coli (APC) Gene Promoter Hypermethylation in Primary Breast Cancers. Br. J. Cancer 2001, 85, 69–73. [Google Scholar] [CrossRef]
- Burbee, D.G.; Forgacs, E.; Zochbauer-Muller, S.; Shivakumar, L.; Fong, K.; Gao, B.; Randle, D.; Kondo, M.; Virmani, A.; Bader, S.; et al. Epigenetic Inactivation of RASSF1A in Lung and Breast Cancers and Malignant Phenotype Suppression. JNCI J. Natl. Cancer Inst. 2001, 93, 691–699. [Google Scholar] [CrossRef]
- Dammann, R.; Yang, G.; Pfeifer, G.P. Hypermethylation of the cpG Island of Ras Association Domain Family 1A (RASSF1A), a Putative Tumor Suppressor Gene from the 3p21.3 Locus, Occurs in a Large Percentage of Human Breast Cancers. Cancer Res. 2001, 61, 3105–3109. [Google Scholar]
- Honorio, S.; Agathanggelou, A.; Schuermann, M.; Pankow, W.; Viacava, P.; Maher, E.R.; Latif, F. Detection of RASSF1A Aberrant Promoter Hypermethylation in Sputum from Chronic Smokers and Ductal Carcinoma in Situ from Breast Cancer Patients. Oncogene 2003, 22, 147–150. [Google Scholar] [CrossRef]
- Pu, R.T.; Laitala, L.E.; Alli, P.M.; Fackler, M.J.; Sukumar, S.; Clark, D.P. Methylation Profiling of Benign and Malignant Breast Lesions and Its Application to Cytopathology. Mod. Pathol. 2003, 16, 1095–1101. [Google Scholar] [CrossRef]
- Fackler, M.J.; McVeigh, M.; Mehrotra, J.; Blum, M.A.; Lange, J.; Lapides, A.; Garrett, E.; Argani, P.; Sukumar, S. Quantitative Multiplex Methylation-Specific PCR Assay for the Detection of Promoter Hypermethylation in Multiple Genes in Breast Cancer. Cancer Res. 2004, 64, 4442–4452. [Google Scholar] [CrossRef]
- Vasileva, F.; Hristovski, R.; Font-Lladó, R.; Georgiev, G.; Sacot, A.; López-Ros, V.; Calleja-González, J.; Barretina-Ginesta, J.; López-Bermejo, A.; Prats-Puig, A. Physical Exercise-Induced DNA Methylation in Disease-Related Genes in Healthy Adults-A Systematic Review With Bioinformatic Analysis. J. Strength Cond. Res. 2024, 38, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Etayo-Urtasun, P.; Sáez De Asteasu, M.L.; Izquierdo, M. Effects of Exercise on DNA Methylation: A Systematic Review of Randomized Controlled Trials. Sports Med. 2024, 54, 2059–2069. [Google Scholar] [CrossRef] [PubMed]
- Światowy, W.J.; Drzewiecka, H.; Kliber, M.; Sąsiadek, M.; Karpiński, P.; Pławski, A.; Jagodziński, P.P. Physical Activity and DNA Methylation in Humans. IJMS 2021, 22, 12989. [Google Scholar] [CrossRef]
- Li, S.; Tollefsbol, T.O. DNA Methylation Methods: Global DNA Methylation and Methylomic Analyses. Methods 2021, 187, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- King-Himmelreich, T.S.; Schramm, S.; Wolters, M.C.; Schmetzer, J.; Möser, C.V.; Knothe, C.; Resch, E.; Peil, J.; Geisslinger, G.; Niederberger, E. The Impact of Endurance Exercise on Global and AMPK Gene-Specific DNA Methylation. Biochem. Biophys. Res. Commun. 2016, 474, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.D.; Magnan, R.E.; Hooper, A.E.C.; Harlaar, N.; Hutchison, K.E. Physical Activity and Differential Methylation of Breast Cancer Genes Assayed from Saliva: A Preliminary Investigation. Ann. Behav. Med. 2013, 45, 89–98. [Google Scholar] [CrossRef]
- Gorski, P.P.; Raastad, T.; Ullrich, M.; Turner, D.C.; Hallén, J.; Savari, S.I.; Nilsen, T.S.; Sharples, A.P. Aerobic Exercise Training Resets the Human Skeletal Muscle Methylome 10 Years after Breast Cancer Treatment and Survival. FASEB J. 2023, 37, e22720. [Google Scholar] [CrossRef]
- McCullough, L.E.; Chen, J.; White, A.J.; Xu, X.; Cho, Y.H.; Bradshaw, P.T.; Eng, S.M.; Teitelbaum, S.L.; Terry, M.B.; Garbowski, G.; et al. Gene-Specific Promoter Methylation Status in Hormone-Receptor-Positive Breast Cancer Associates with Postmenopausal Body Size and Recreational Physical Activity. Int. J. Cancer Clin. Res. 2015, 2, 013. [Google Scholar] [CrossRef]
- Coyle, Y.M.; Xie, X.-J.; Lewis, C.M.; Bu, D.; Milchgrub, S.; Euhus, D.M. Role of Physical Activity in Modulating Breast Cancer Risk as Defined by APC and RASSF1A Promoter Hypermethylation in Nonmalignant Breast Tissue. Cancer Epidemiol. Biomark. Prev. 2007, 16, 192–196. [Google Scholar] [CrossRef]
- Moulton, C.; Murri, A.; Benotti, G.; Fantini, C.; Duranti, G.; Ceci, R.; Grazioli, E.; Cerulli, C.; Sgrò, P.; Rossi, C.; et al. The Impact of Physical Activity on Promoter-Specific Methylation of Genes Involved in the Redox-Status and Disease Progression: A Longitudinal Study on Post-Surgery Female Breast Cancer Patients Undergoing Medical Treatment. Redox Biol. 2024, 70, 103033. [Google Scholar] [CrossRef]
- McCullough, L.E.; Chen, J.; Cho, Y.H.; Khankari, N.K.; Bradshaw, P.T.; White, A.J.; Teitelbaum, S.L.; Terry, M.B.; Neugut, A.I.; Hibshoosh, H.; et al. Modification of the Association between Recreational Physical Activity and Survival after Breast Cancer by Promoter Methylation in Breast Cancer-Related Genes. Breast Cancer Res. 2017, 19, 19. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway Enrichment Analysis and Visualization of Omics Data Using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- McCullough, L.E.; Chen, J.; White, A.J.; Xu, X.; Cho, Y.H.; Bradshaw, P.T.; Eng, S.M.; Teitelbaum, S.L.; Terry, M.B.; Garbowski, G.; et al. Global DNA Methylation, Measured by the Luminometric Methylation Assay (LUMA), Associates with Postmenopausal Breast Cancer in Non-Obese and Physically Active Women. J. Cancer 2015, 6, 548–554. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines on Physical Activity and Sedentary Behaviour At a Glance; World Health Organization: Geneva, Switzerland, 2020; ISBN 978-92-4-001488-6. [Google Scholar]
- Bernstein, M.; Sloutskis, D.; Kumanyika, S.; Sparti, A.; Schutz, Y.; Morabia, A. Data-Based Approach for Developing a Physical Activity Frequency Questionnaire. Am. J. Epidemiol. 1998, 147, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, H.T.; Sigurdsson, M.I.; Fallin, M.D.; Irizarry, R.A.; Aspelund, T.; Cui, H.; Yu, W.; Rongione, M.A.; Ekström, T.J.; Harris, T.B.; et al. Intra-Individual Change over Time in DNA Methylation with Familial Clustering. JAMA 2008, 299, 2877–2883. [Google Scholar] [CrossRef]
- Xu, X.; Gammon, M.D.; Hernandez-Vargas, H.; Herceg, Z.; Wetmur, J.G.; Teitelbaum, S.L.; Bradshaw, P.T.; Neugut, A.I.; Santella, R.M.; Chen, J. DNA Methylation in Peripheral Blood Measured by LUMA Is Associated with Breast Cancer in a Population-Based Study. FASEB J. 2012, 26, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Kawasaki, T.; Brahmandam, M.; Cantor, M.; Kirkner, G.J.; Spiegelman, D.; Makrigiorgos, G.M.; Weisenberger, D.J.; Laird, P.W.; Loda, M.; et al. Precision and Performance Characteristics of Bisulfite Conversion and Real-Time PCR (MethyLight) for Quantitative DNA Methylation Analysis. J. Mol. Diagn. 2006, 8, 209–217. [Google Scholar] [CrossRef]
- Maksimovic, J.; Phipson, B.; Oshlack, A. A Cross-Package Bioconductor Workflow for Analysing Methylation Array Data. F1000Res 2017, 5, 1281. [Google Scholar] [CrossRef]
- Thiese, M.S. Observational and Interventional Study Design Types; an Overview. Biochem. Med. 2014, 24, 199–210. [Google Scholar] [CrossRef]
- Lee, E.-H.; Kang, E.H.; Kang, H.-J. Evaluation of Studies on the Measurement Properties of Self-Reported Instruments. Asian Nurs. Res. 2020, 14, 267–276. [Google Scholar] [CrossRef]
- Mohsen, K.; Johansson, S.; Ekström, T.J. Using LUMA: A Luminometric-Based Assay for Global DNA-Methylation. Epigenetics 2006, 1, 46–49. [Google Scholar] [CrossRef]
- Karimi, M.; Johansson, S.; Stach, D.; Corcoran, M.; Grandér, D.; Schalling, M.; Bakalkin, G.; Lyko, F.; Larsson, C.; Ekström, T.J. LUMA (LUminometric Methylation Assay)—A High Throughput Method to the Analysis of Genomic DNA Methylation. Exp. Cell Res. 2006, 312, 1989–1995. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.S.; Estécio, M.R.H.; Doshi, K.; Kondo, Y.; Tajara, E.H.; Issa, J.-P.J. A Simple Method for Estimating Global DNA Methylation Using Bisulfite PCR of Repetitive DNA Elements. Nucleic Acids Res. 2004, 32, e38. [Google Scholar] [CrossRef]
- Lisanti, S.; Omar, W.A.W.; Tomaszewski, B.; De Prins, S.; Jacobs, G.; Koppen, G.; Mathers, J.C.; Langie, S.A.S. Comparison of Methods for Quantification of Global DNA Methylation in Human Cells and Tissues. PLoS ONE 2013, 8, e79044. [Google Scholar] [CrossRef]
- Schulz, W.A.; Steinhoff, C.; Florl, A.R. Methylation of Endogenous Human Retroelements in Health and Disease. In DNA Methylation: Development, Genetic Disease and Cancer; Doerfler, W., Böhm, P., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2006; Volume 310, pp. 211–250. ISBN 978-3-540-31180-5. [Google Scholar]
- Kim, J.K.; Samaranayake, M.; Pradhan, S. Epigenetic Mechanisms in Mammals. Cell. Mol. Life Sci. 2009, 66, 596. [Google Scholar] [CrossRef] [PubMed]
- White, A.J.; Sandler, D.P.; Bolick, S.C.E.; Xu, Z.; Taylor, J.A.; DeRoo, L.A. Recreational and Household Physical Activity at Different Time Points and DNA Global Methylation. Eur. J. Cancer 2013, 49, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, Z.; Cui, D.; Liu, H.; Hao, X. Glutathione S-Transferase P1 Ile105Val Polymorphism and Breast Cancer Risk: A Meta-Analysis Involving 34,658 Subjects. Breast Cancer Res. Treat 2011, 125, 253–259. [Google Scholar] [CrossRef]
- Bonasio, R.; Lecona, E.; Reinberg, D. MBT Domain Proteins in Development and Disease. Semin. Cell Dev. Biol. 2010, 21, 221–230. [Google Scholar] [CrossRef]
- Satoh, K.; Ginsburg, E.; Vonderhaar, B.K. Msx-1 and Msx-2 in Mammary Gland Development. J. Mammary Gland Biol. Neoplasia 2004, 9, 195–205. [Google Scholar] [CrossRef]
- Panis, C.; Herrera, A.C.S.A.; Victorino, V.J.; Campos, F.C.; Freitas, L.F.; De Rossi, T.; Colado Simão, A.N.; Cecchini, A.L.; Cecchini, R. Oxidative Stress and Hematological Profiles of Advanced Breast Cancer Patients Subjected to Paclitaxel or Doxorubicin Chemotherapy. Breast Cancer Res. Treat 2012, 133, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Delrieu, L.; Touillaud, M.; Pérol, O.; Morelle, M.; Martin, A.; Friedenreich, C.M.; Mury, P.; Dufresne, A.; Bachelot, T.; Heudel, P.-E.; et al. Impact of Physical Activity on Oxidative Stress Markers in Patients with Metastatic Breast Cancer. Oxidative Med. Cell. Longev. 2021, 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Friedenreich, C.M.; Pialoux, V.; Wang, Q.; Shaw, E.; Brenner, D.R.; Waltz, X.; Conroy, S.M.; Johnson, R.; Woolcott, C.G.; Poulin, M.J.; et al. Effects of Exercise on Markers of Oxidative Stress: An Ancillary Analysis of the Alberta Physical Activity and Breast Cancer Prevention Trial. BMJ Open Sport Exerc. Med. 2016, 2, e000171. [Google Scholar] [CrossRef]
- Hojan, K.; Gerreth, K.; Procyk, D.; Mania, K.; Zalewska, A.; Maciejczyk, M. Redox Status Response of Physical Exercise Training in Women with Breast Cancer during Trastuzumab Therapy. Healthcare 2022, 10, 2039. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; Van Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018, 8, 1898. [Google Scholar] [CrossRef]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; van Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Methylome of Human Skeletal Muscle after Acute & Chronic Resistance Exercise Training, Detraining & Retraining. Sci. Data 2018, 5, 180213. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.C.; Seaborne, R.A.; Sharples, A.P. Comparative Transcriptome and Methylome Analysis in Human Skeletal Muscle Anabolism, Hypertrophy and Epigenetic Memory. Sci. Rep. 2019, 9, 4251. [Google Scholar] [CrossRef]
- Verschoor, N.; Deger, T.; Jager, A.; Sleijfer, S.; Wilting, S.M.; Martens, J.W.M. Validity and Utility of HER2/ERBB2 Copy Number Variation Assessed in Liquid Biopsies from Breast Cancer Patients: A Systematic Review. Cancer Treat. Rev. 2022, 106, 102384. [Google Scholar] [CrossRef]
- Maasar, M.-F.; Turner, D.C.; Gorski, P.P.; Seaborne, R.A.; Strauss, J.A.; Shepherd, S.O.; Cocks, M.; Pillon, N.J.; Zierath, J.R.; Hulton, A.T.; et al. The Comparative Methylome and Transcriptome After Change of Direction Compared to Straight Line Running Exercise in Human Skeletal Muscle. Front. Physiol. 2021, 12, 619447. [Google Scholar] [CrossRef]
- Molinari, M. Cell Cycle Checkpoints and Their Inactivation in Human Cancer. Cell Prolif. 2000, 33, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Ma, H.; Wang, J.; Huang, M.; Fu, D.; Qin, L.; Yin, Q. Energy Metabolism Pathways in Breast Cancer Progression: The Reprogramming, Crosstalk, and Potential Therapeutic Targets. Transl. Oncol. 2022, 26, 101534. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, S.; Wang, X. The Metabolic Mechanisms of Breast Cancer Metastasis. Front. Oncol. 2020, 10, 602416. [Google Scholar] [CrossRef]
- Voisin, S.; Eynon, N.; Yan, X.; Bishop, D.J. Exercise Training and DNA Methylation in Humans. Acta Physiol. 2015, 213, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, S.; Isozaki, M.; Yao, A.; Higashino, Y.; Yamauchi, T.; Kidoguchi, M.; Kawajiri, S.; Tsunetoshi, K.; Neish, H.; Imoto, H.; et al. Cross-Tissue Correlations of Genome-Wide DNA Methylation in Japanese Live Human Brain and Blood, Saliva, and Buccal Epithelial Tissues. Transl. Psychiatry 2023, 13, 72. [Google Scholar] [CrossRef]
- McGee, S.L.; Walder, K.R. Exercise and the Skeletal Muscle Epigenome. Cold Spring Harb. Perspect Med. 2017, 7, a029876. [Google Scholar] [CrossRef]
- Tsai, I.-H.; Su, Y.-J. Thyrotoxic Periodic Paralysis with Ventricular Tachycardia. J. Electrocardiol. 2019, 54, 93–95. [Google Scholar] [CrossRef]
- Kietzmann, T.; Petry, A.; Shvetsova, A.; Gerhold, J.M.; Görlach, A. The Epigenetic Landscape Related to Reactive Oxygen Species Formation in the Cardiovascular System. Br. J. Pharmacol. 2017, 174, 1533–1554. [Google Scholar] [CrossRef]
- Niu, Y.; DesMarais, T.L.; Tong, Z.; Yao, Y.; Costa, M. Oxidative Stress Alters Global Histone Modification and DNA Methylation. Free. Radic. Biol. Med. 2015, 82, 22–28. [Google Scholar] [CrossRef]
- Adam, S.; Anteneh, H.; Hornisch, M.; Wagner, V.; Lu, J.; Radde, N.E.; Bashtrykov, P.; Song, J.; Jeltsch, A. DNA Sequence-Dependent Activity and Base Flipping Mechanisms of DNMT1 Regulate Genome-Wide DNA Methylation. Nat. Commun. 2020, 11, 3723. [Google Scholar] [CrossRef]
- Jeltsch, A.; Adam, S.; Dukatz, M.; Emperle, M.; Bashtrykov, P. Deep Enzymology Studies on DNA Methyltransferases Reveal Novel Connections between Flanking Sequences and Enzyme Activity. J. Mol. Biol. 2021, 433, 167186. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.; Bräcker, J.; Klingel, V.; Osteresch, B.; Radde, N.E.; Brockmeyer, J.; Bashtrykov, P.; Jeltsch, A. Flanking Sequences Influence the Activity of TET1 and TET2 Methylcytosine Dioxygenases and Affect Genomic 5hmC Patterns. Commun. Biol. 2022, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Dakhlallah, D.A.; Wisler, J.; Gencheva, M.; Brown, C.M.; Leatherman, E.R.; Singh, K.; Brundage, K.; Karsies, T.; Dakhlallah, A.; Witwer, K.W.; et al. Circulating Extracellular Vesicle Content Reveals de Novo DNA Methyltransferase Expression as a Molecular Method to Predict Septic Shock. J. Extracell. Vesicles 2019, 8, 1669881. [Google Scholar] [CrossRef]
- Silva-Llanes, I.; Shin, C.H.; Jiménez-Villegas, J.; Gorospe, M.; Lastres-Becker, I. The Transcription Factor NRF2 Has Epigenetic Regulatory Functions Modulating HDACs, DNMTs, and miRNA Biogenesis. Antioxidants 2023, 12, 641. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, R.; Rivenbark, A.G.; Coleman, W.B. Loss of Post-Transcriptional Regulation of DNMT3b by microRNAs: A Possible Molecular Mechanism for the Hypermethylation Defect Observed in a Subset of Breast Cancer Cell Lines. Int. J. Oncol. 2012, 41, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Barber, J.L.; Zellars, K.N.; Barringhaus, K.G.; Bouchard, C.; Spinale, F.G.; Sarzynski, M.A. The Effects of Regular Exercise on Circulating Cardiovascular-Related MicroRNAs. Sci. Rep. 2019, 9, 7527. [Google Scholar] [CrossRef]
- Lisi, V.; Moulton, C.; Fantini, C.; Grazioli, E.; Guidotti, F.; Sgrò, P.; Dimauro, I.; Capranica, L.; Parisi, A.; Di Luigi, L.; et al. Steady-State Redox Status in Circulating Extracellular Vesicles: A Proof-of-Principle Study on the Role of Fitness Level and Short-Term Aerobic Training in Healthy Young Males. Free Radic. Biol. Med. 2023, 204, 266–275. [Google Scholar] [CrossRef]
| Database | Query |
|---|---|
| Pubmed | ((Breast cancer [Title/Abstract]) AND (exercise [Title/Abstract] OR Physical activity [Title/Abstract]) AND (DNA Methylation [Title/Abstract] OR DNMT [Title/Abstract] OR Epigenetics [Title/Abstract] OR methyltransferase [Title/Abstract] OR promoter methylation [Title/Abstract] OR methylome [Title/Abstract])) |
| Scopus | (TITLE-ABS-KEY (“breast cancer”)) AND (TITLE-ABS-KEY (exercise OR “physical activity”)) AND (TITLE-ABS-KEY (“dna methylation” OR epigenetics OR methyltransferase OR “promoter methylation” OR methylome OR dnmt)) AND (EXCLUDE (DOCTYPE, “re”)) |
| Web of Science | ((ALL = Breast cancer) AND (ALL = exercise OR ALL = Physical activity) AND (ALL = DNA Methylation OR ALL = Epigenetics OR ALL = methyltransferase OR ALL = promoter methylation OR ALL = methylome OR ALL = DNMT)) |
| Intervention Studies | ||||||
|---|---|---|---|---|---|---|
| Author | Study Design | Population Sample Size | Sample | Exercise Intervention | Methylation Detection Technique and Analysed Target | Significant Modulation(s) in DNA Methylation as a Result of PA/Exercise |
| Gorski et al., 2022 [43] | Two-armed randomised controlled exercise training trial | Breast cancer survivors diagnosed with stage II-III HER2-negative breast cancer Cancer trained (n = 7, 62.5 ± 4.2 age of years) Cancer-untrained (n = 6, 59.9 ± 5.7 age of years) Healthy age-matched, trained (n = 10, 57.6 ± 4.0 age of years) | Skeletal muscle | 5 months of three-times -per-week treadmill-based endurance training, aimed at increasing VO2peak | Infinium MethylationEPIC BeadChip array: Methylome DMPs and DMRs | DMPs: RAD51L3, ZFR, SIRT2, PPWD1, TMEM146, EIF4G3, UQCRC1, INO80C, KIAA0226, ROBLD3, CCDC28A, IFT80, JOSD1, AP1G2, PPP4R3B, CHMP5, COX7A2L, ZNF641, NXT1, GTF3C1, HIST1H4A, DNAJA3, COQ10B, C16orf46, CARHSP1, C11orf48, BUB1B, ALDH4A1, MYADM, DFFA, C14orf178, RNF145, CBR4, IMMP2L, INTS10, OAZ2, TRAIP, RBM26-AS1, R3HDM1, LZTR1, CYP2U1, WDR60, MTCH2, WDR19, GANC, SIDT2, C4orf14, PTRH2, SLC35A3, HACD3, RBL2, AP2A1, SHMT1, TGFB1, SNORA6, NDUFB11, YPEL1, ZNF394, CELSR2, CDCA7L, MYH10, TSPAN33, KIFC1, G3BP1, ING1, VKORC1, CHTF8, MDH1, DIDO1, ATF7, SLC25A37, SLC38A10, SIVA1, ATPIF1, NPHP4, LDHA, NAGK, MCM3, UBE3B, CLP1, RRM2B, ATXN1L, BANP, PRSS27, CDKN2D, TYMS, ZNF570, DHPS, GNE, HIST1H4B, SMCR7L, RTN4, CBARA1, ZKSCAN5, OTUD7B, KCTD21, CLNS1A, ATP5L, ZDHHC5, GSK3B, ELOVL6, DIS3, ACIN1, NUMB, FAM151B, PNPLA6, SGOL1, PRMT7, ZNF594, MTR, TOMM34, TMEM170A, SOD1, NDUFV2, MORN1, RHBDD2, MKL1, ZNF282, LOC100329108, ESD, DMAP1, TMPO, SNORD43, SURF6, ZFYVE1, LOC400027, NFYA, PELI3, PIP4K2C, TEX2, BUB1B, LMBRD1, NDUFS2, ST5, ARL8B, UBA52, UBL5, ZNF337, MYO9A, AFG3L1, ALG14, DMXL1, CCNC, KIFC1, MYBBP1A, ZFP62, C6orf147, DLG4, C21orf91, C9orf40, C11orf80, SARM1, PFDN4, SPATS2, ZNF222, TYMP, PEX5, NDEL1, PTGES3, TNFAIP3, PSMC3IP, PAF1, FANCG, PGM1, SCYL3, PIGY, ACTG1, TRA2A, PIGP, TCTN2, C3orf71, NAA15, SPG7, USE1, DPM3, IMPA2, ACAD10, HNRNPA0, C1orf112, RNASEN, SMC3, CAST, TAF15, FRMD8, TAF5, RPL36AL, COL4A3BP, C1orf35, GMPR, MTBP, ANKRD31, BBS2, PHF12, NDUFA9, CLNS1A, HACE1, CD320, OTUD3, AURKB, P4HTM, C15orf29, CORIN, EXOSC4, CKAP2, MAP3K8, URM1, STAT5B, SEL1L, KLC4, AMZ2, TOR2A, PODNL1, AGFG1, LOC284900, ESCO1, DNAJC28, SSR2, CENPV, MAK16, EXD2, USP21, ELMO2, PYROXD1, TRIM24, BUB1, REXO2, ZNF143, EIF4G1, GNRHR2, MCM3APAS, CASC1, SRBD1, PLBD2, TPCN1, CSRP1, HS3ST3B1, ANKRD9, GIGYF2, COL4A3BP, PGRMC2, ICMT, C19orf43, NIPA2, RNFT1, CCDC102B, DCPS, BCL2L2-PABPN1, TACO1, FAM46A, RB1CC1, KBTBD4, ARF4, BTRC, EWSR1, HAUS8, NDUFA2, CAPNS1, CASP6, SETD5, TSNAX-DISC1, ZNF391, ZNF74, OSCP1, ZNF501, ZDHHC14, UBTF, ATP6V0A1, TUBG2, SLC25A3, DCBLD2, CPSF4, OCIAD1, NPHP4, C21orf59, NCK2, NUSAP1, TPD52L2, C10orf84, FASN, PLCD3, SEC24A, C1orf101, UBL3, HYAL3, FARSA, ZNF136, ARNTL2, TMEM55B, PNKD, IFFO2, CDK5RAP3, TNNI3K, LRRCC1, GSTZ1, ATPGD1, MINK1, NOP58, DDX58, OCEL1, CMBL, ZNF222, RCBTB2, ASL, MKI67, UGDH, UTP23, MIF4GD, RAP2B, BAT3, HDAC7, FAM164A, SLU7, POLD2, G6PC3, CHMP1B, TTK, CASP6, AAK1, FRY, SUCLG1, PPM1F, TRIM27, CPEB3, CCDC59, DPYD, C16orf72, MTIF2, RUFY2, CCNB2, KBTBD7, SLC35A3, STMN1, TBCB, RBCK1, FTSJD1, C16orf42, RANBP3, NIT2, NAGK, ATRN, FICD, RPUSD3, C15orf61, GPATCH2, NUDT6, RSPH1, EHD4, ALDH9A1, UBE2H, MTR, HDAC11, ELP2, ST8SIA1, ELMO2, TSPAN3, RPAP1, SLMO2, IPO8, GUCD1, TUBB, FANCC, UBOX5, HELQ, PRKCA, USP33, GADD45B, SAMM50, CPEB2, LRPPRC, NSUN6, EIF4A3, PCIF1, ZNF707, SYNPO, HDLBP, DCUN1D4, AHDC1, CCDC93, RASA2, ECHS1, FXR1, ZNF570, FNIP1, GIN1, POLD2, PTCD3, ANKRD11, C14orf21, ELP2P, GSDMD, CDKN2D, CHMP5, SLC10A7, PJA2, C6orf1, TIPIN, DNAJC8, AEN, NEU1, SLC19A2, CDC123, C2orf42, PPP1R3E, CLSPN, PPPDE2, DCBLD1, PLEKHG2, SPAG5, DCP1A, STRAP, ARHGAP12, PSMG1, AMIGO2, ZNF318, MTMR4, ALG8, ZNF259, UBE2J1, DUS4L, MTIF2, CASP8AP2, SPATA6, TAPBP, C14orf80, ZBTB42, INTS5, PCYOX1L, ANAPC13, NUP50, NDUFB5, PLEKHG4, TERF2, RBM15B, APPL1, C19orf40, N6AMT1, KCTD7, UBA52, OSBPL2, PSMB1, PSMD9, MCM4, PDE4C, NME3, RPS6, BRD2, KCNK6, DCAF4, ACOT7, SYNRG, EIF4B, SELO, GPATCH3, BRIX1, TAPBP, RPSA, MEA1, PGAM5, KLHL7-AS1, NT5C3, SRSF7, CLK3, SAMHD1, ELAC2, KIAA0895, WSB1, GANAB, PNPLA8, UNC119B, CCAR1, TTC4, DAAM1, XPO4, MRPL12, NCL, CPPED1, CEP95, SRSF10, PDCD5, C11orf54, NSUN6, SLC35C1, BAT2, MED11, HS1BP3, SETMAR, ANXA4, C12orf10, FAM63A, HERPUD2, LNP1, FAF2, JOSD1, LOC100130987, PEF1, PPP2R2A, ZFP90, ZNF639, ZZEF1, SIX5, MLL5, AATF, VRK1, TM2D1, C20orf11, NARF, PPP1CC, SIDT2, CTNNBIP1, CHCHD2, HCN3, MAP3K6, FAM111A, KCNA3, ERBB2IP, ELL, RPLP2, SEC31A, TNK2, TPRKB, TRAPPC3, TFCP2, PEX7, C7orf30, WRNIP1, IER2, LSG1, GRK4, MOBKL3, TRAM1, KIAA0564, PHF20L1, MDH1B, CEP164, MUL1, PHB2, ZZZ3, BTBD7, SIX5, CAMK2D, NAA38, TRIM45, MRPL55, NEDD4L, C2orf42, MARS2, KIAA1949, MTHFD1, POLE3, EZH2, TSPAN4, SH2D3C, TRIM45, LAPTM4A, SNORA76, ANKRD52, NAGS, ZDHHC5, DENND6A, FTSJD1, DCBLD2, FBXO34, RPP25L, ZBTB7A, IQCH-AS1, LINC00899, MORF4L1, CPEB2, HIST1H2BJ, PPM1B, NDE1, QSER1, FBXO31, LRRC8A, BLCAP, BRI3BP, PRMT1, RPLP1, GNRHR2, METTL23, ZNF490, SRP14, NCAPG, TMEM149, GBAS, MKNK1, H3F3B, ANKRD27, LOC440356, VOPP1, KLHDC2, PODNL1, RAD17, FIS1, SSNA1, SGCE, FKBP3, CHP, ZNHIT6, MIF4GD, FDFT1, GPD1L, ZNF566, MLL, IGF1R, BANP, ZBTB41, MICA, CPSF2, ERO1LB, FOSL1, C19orf61, ABHD13, ATP1A1, SFPQ, C20orf30, ZNF546, YWHAZ, CYB5R3, MFN2, TP53, S100A6, LMAN2, FGFBP3, RFC3, CDC14A, C20orf27, ZNF775, ABHD4, SGSM3, ZNF295, COPB2, PEX26, LOC100506100, DCLRE1A, POLD4, ANKRD11, STX5, LAPTM4A, UNG, NDUFB3, BTG2, HSPA9, RTN4, TLCD1, WDR47, SLC37A4, CPNE8, FZD7, TMEM48, ZNF620, CENPM, C1orf51, STK10, MPHOSPH6, UEVLD, DNAJB12, MTERFD3, TBRG4, NDUFB5, MIR92B, OXNAD1, GPR180, BSCL2, SLC9A8, NDUFV2, RABIF, CD47, DDX46, RBMXL1, CDC7, CDK19, TMEM135, STRAP, ERGIC2, MGC16275, EHD3, ANKRD49, WIZ, DCP1B, KLHL22, PYCR2, PTCD3, ATXN3, ZFYVE19, SMIM14, NDUFA2, FAM55C, SMARCA5, PRDX5, SNHG4, IPO8, KANSL1L, NF2, ATP6V1H, POLD1, RBM15B, PPP6R1, ATG5, PSMB9, LOC400657, AP2M1, XRCC1, INIP, GATAD1, RPL36AL, VPS54, RCHY1, GCH1, ERCC2, MUS81, NDUFA7, SLC35A4, UBXN1, MED7, MYO15B, VPS45, EPB41, PRPF8, LSS, S100A6, BRF1, NAA35, ATP6V0D1, FAM175B, SLC10A7, TIGD1, C19orf61, CNOT8, RAPGEF6, CLN5, DAPK3, STT3A, CDC42EP4, MYH10, XYLB, CDKN2A, MRPL44, BTBD10, EIF4G2, CABIN1, CDR2, FUT8, PPP2R4, HMGN1, CDK1, EVI5, GK5, SNORA76, CPNE1, EDC4, SS18L2, PTBP1, NAA25, TCF12, EME1, GSS, HK1, DFFA, ZNF792, C1orf212, ATP5H, SNX27, KIAA0528, ANXA4, MYADM, GPR113, ATP5G2, NVL, FICD, ZNF582, POLR3H, ABCB6, ZCCHC24, GNS, TMEM18, MIR148A, EPB41, SPOPL, CDNF, ZNF688, KLHDC2, RAP2B, MSRB2, FAM98A, RPL29, ABI1, ZNF784, CDC6, MRPL18, SNRNP70, RAB3GAP2, RAP1A, C9orf163, PINX1, CNST, TRIM41, WEE1, ZNF805, ATAD2B, IMMP2L, WDR24, C6orf173, GOLGA1, MTCH1, PELP1, TMEM44-AS1, STK24, HYAL2, GLUD1, RAB2A, ENO2, POLR2E, TMEM131, RCBTB2, TULP3, MLL5, YOD1, C1orf43, EIF2AK2, CGGBP1, ZNF707, RSU1, GTF3C5, PTEN, TMEM80, ERMAP, CCT2, KIF21A, C6orf204, PSMA5, MXD1, SFT2D2, AACS, USP18, ZNF782, SNORD50B, BAT5, CAB39L, C7orf50, SMC3, E4F1, BAT5, BNIP2, SAPS3, C17orf62, BAG2, BBIP1, EIF6, ERCC4, ERLIN2, GFI1, SLC27A5, LTB, ZNF57, ERO1L, CCDC134, BTG2, LRSAM1, ZCCHC2, C1orf83, GATAD2B, SLTM, RPL15, IP6K1, DENR, ANKRD13A, ZBTB2, SUPT5H, EEF2, ERGIC2, USP21, ZSWIM7, DSCR3, ORC1L, ZNF77, C6orf1, ZFP36L2, TSTD2, SOX4, ETFDH, ANP32B, RAI14, PSMA7, BRUNOL6, ZFP36L2, PGLS, PHF12, MTHFD1L, IFT140, PHTF1, RNASEN, ZNF862, SPC25, SPRY2, ORAI1, ANKRD11, DDX12, TXNRD1, CISD1, EIF3A, NDRG3, CNIH4, MRPS9, TMEM106B, ZNF782, PRDX5, LOC282997, NPHP4, POLE4, LENG8, METTL13, ITPR1, ZNF764, MFAP1, RNFT1, PSMA2, SYNGAP1, TMEM214, KLF10, SCAMP1-AS1, DEPDC1, NAA40, MTA2, PIGT, ARRDC2, BAT5, HSPA13, IL4R, WDR87, ZDHHC14, NSUN3, CDC2, PSMA5, C8orf59, PLEKHF1, ACAD11, PEF1, TCF7, DCTPP1, HDAC1, BAT5, NANP, METAP2, KIAA0652, CLK3, ITCH, NUP160, CCDC150, PPPDE2, BCL2L1, MARS2, CENPK, CHIC2, FZD6, PSMD7, PRKAA1, TMEM194A, C6orf136, MTIF2, ARRB2, RAP1GDS1, SNX33, SCAI, SEC23IP, ARF6, ZZEF1, ATP1A1, PRR3, NBPF3, ZNF169, CBX6, TMEM143, APPBP2, ZNF573, DHDDS, AKAP13, SFRS13A, RPL26, IL12A, KLRAQ1, SERGEF, CCNF, PSMA5, GTF2A1, CPNE8, ZNF578, COX10, LCA5, FAM96B, TMEM14C, ENTPD5, JUNB, PDRG1, MED18, SMIM20, LRIG1, EIF4A2, C10orf4, FABP5, RNASEH2B, LZTFL1, ERMP1, UGDH, MAP1S, HMGN1, RB1, ECI1, ZHX2, CD55, GRSF1, IBTK, LSS, HINFP, PCNA, SH3YL1, STK36, CCDC21, MKL2, PTBP3, NUF2, TMEM68, C4orf19, OSCP1, HNRNPUL1, SYDE2, PDCD5, SAMHD1, NUDT18, PSMD1, MICALL1, NFE2L2, CAPN1, TMEM161A, PSME2, SLBP, ACIN1, MARS, TUBA1A, MTIF3, C22orf25, C20orf7, PDP2, ISM1, EIF1AD, MKL2, ZC3H12A, TMEM251, RAB11B, PPPDE2, MSH5, PDE7A, KLC2, TMEM218, PEX26, PARPBP, CLASP2, MUDENG, ZNF507, DVL1, CD82, YTHDF2, APPL1, AGXT2L2, FNTB, NR4A2, SLC25A28, NFU1, BTBD1, SLC16A13, DNAJB6, ADRB2, HSD17B8, XKR9, CISH, CETN3, THUMPD3, CHST12, CDON, TAPBP, TIMELESS, KIAA2018, NANS, GRPEL2, COQ3, PRR13, NEU3, SRPRB, Mar-08, VCPKMT, LENG1, RNF103, MLF1IP, STARD3, C14orf4, MEF2D, IDH3B, FAM115A, GPX4, HNRNPL, PDP2, PTCD2, LOC93622, IKZF1, ATP2C1, RPL18, TSGA14, RTTN, RPS16, ZNF793, NETO2, SUGT1, ANXA4, NAT14, OGDH, SSRP1, SGSM2, TIPIN, GGCX, KIAA1324, GNS, ACADS, NDUFS8, RNF40, CDK5, MRPL41, LYSMD4, HCFC2, CPEB2, B4GALT3, DNA2, ZNF395, WIPF1, AP1M1, MTURN, MAP4K5, NUP188, ARIH2, STT3B, LOC100272217, TWISTNB DMRs: BAG1, BTG2, CHP1, KIFC1, MKL2, MTR, PEX11B, POLD2, S100A6, SNORD104, SPG7 |
| Moulton et al., 2024 [46] | Two-armed randomised controlled trial | Female first primary BC patients undergoing medical treatment (training group: n = 10; control group: n = 10) (45–65 years of age) | Blood sample | 16 weeks of mixed-modality exercise training 2 times/week | MSP (SOD1, SOD2, Catalase, RASSF1A, L3MBTL1, RASSF1A) | SOD1, SOD2, L3MBTL1 |
| Zeng et al., 2012 [10] | Randomised clinical trial | Physically inactive, postmenopausal female BC patients (n = 12) (56.5 ± 9.5 years of age) Breast tumour samples (n = 348) (111 ≤ 50, 139 = 50–65, 98 = ~65 years of age) | Blood sample Breast tumour sample | 6 months of moderate-intensity exercise (150 min/week) | Infinium HumanMethylation27 BeadChip: Panel of 14 495 genes MSP: (L3MBTL1) | EPS15, DYDC1, WNT7A, SULF1, KPNA5, AQP5, ALG1, C1R, PARP11, INSRR, CDC26, ZNF222, PPP2R3A, TMEM100, IFT172, C8orf53, CXCL10, NALP11, HINT2, OSTF1, ERVK6, DC-UbP, RASA1, DCC RP11-450P7.3, KIAA0980, RBM10, PLAGL1, MEG3, ORM2, DYNC1I1, GAB1, ABCB1, SLC9A7, LRRC14, L3MBTL1, MSX1, PCTK3, BCL2L11, WNK3, GLUD1, MGC39633, PLCZ1 L3MBTL1 |
| Self-reported PA studies | ||||||
| Author | Study design | Population | Sample | Exercise/PA behaviour measured | Methylation detection technique and analysed target | Significant modulation(s) in DNA methylation as a Result of PA/exercise |
| McCullough et al., 2015 (specific) [44] | Population-based-case–control study | Female first primary BC patients (n = 532) (20–98 years of age, mean age 59.6) | Breast tumour sample | RPA | -MSP: ESR1, PR, BRCA1 -MethyLight assay: APC, CDH1, CCND2, DAPK, GSTP1, HIN, P16, RARB, RASSF1A, TWIST1 | GSTP1 |
| McCullough et al., 2015 (global) [49] | Population-based-case–control study | Postmenopausal female first primary BC (n = 1300) (20–98 years of age) | Blood sample | Postmenopausal RPA | LUMA global DNA methylation assay: LUMA Pyrosequencing-based methylation assay: LINE-1 | Global DNA methylation |
| McCullough et al., 2017 [47] | Population-based-case–control study | Female first primary BC patients (n = 807) (20–98 years of age) | Breast tumour sample Blood sample | RPA | Gene-specific: -MSP: ESR1, PR, BRCA1 -MethyLight assay: APC, CDH1, CCND2, DAPK, GSTP1, HIN, P16, RARB, RASSF1A, TWIST1 Global: -LUMA global DNA methylation assay: LUMA -Pyrosequencing-based methylation assay: LINE-1 | APC, CCND2, HIN, TWIST1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moulton, C.; Lisi, V.; Silvestri, M.; Ceci, R.; Grazioli, E.; Sgrò, P.; Caporossi, D.; Dimauro, I. Impact of Physical Activity on DNA Methylation Signatures in Breast Cancer Patients: A Systematic Review with Bioinformatic Analysis. Cancers 2024, 16, 3067. https://doi.org/10.3390/cancers16173067
Moulton C, Lisi V, Silvestri M, Ceci R, Grazioli E, Sgrò P, Caporossi D, Dimauro I. Impact of Physical Activity on DNA Methylation Signatures in Breast Cancer Patients: A Systematic Review with Bioinformatic Analysis. Cancers. 2024; 16(17):3067. https://doi.org/10.3390/cancers16173067
Chicago/Turabian StyleMoulton, Chantalle, Veronica Lisi, Monica Silvestri, Roberta Ceci, Elisa Grazioli, Paolo Sgrò, Daniela Caporossi, and Ivan Dimauro. 2024. "Impact of Physical Activity on DNA Methylation Signatures in Breast Cancer Patients: A Systematic Review with Bioinformatic Analysis" Cancers 16, no. 17: 3067. https://doi.org/10.3390/cancers16173067
APA StyleMoulton, C., Lisi, V., Silvestri, M., Ceci, R., Grazioli, E., Sgrò, P., Caporossi, D., & Dimauro, I. (2024). Impact of Physical Activity on DNA Methylation Signatures in Breast Cancer Patients: A Systematic Review with Bioinformatic Analysis. Cancers, 16(17), 3067. https://doi.org/10.3390/cancers16173067










