Multiomics-Based Outcome Prediction in Personalized Ultra-Fractionated Stereotactic Adaptive Radiotherapy (PULSAR)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
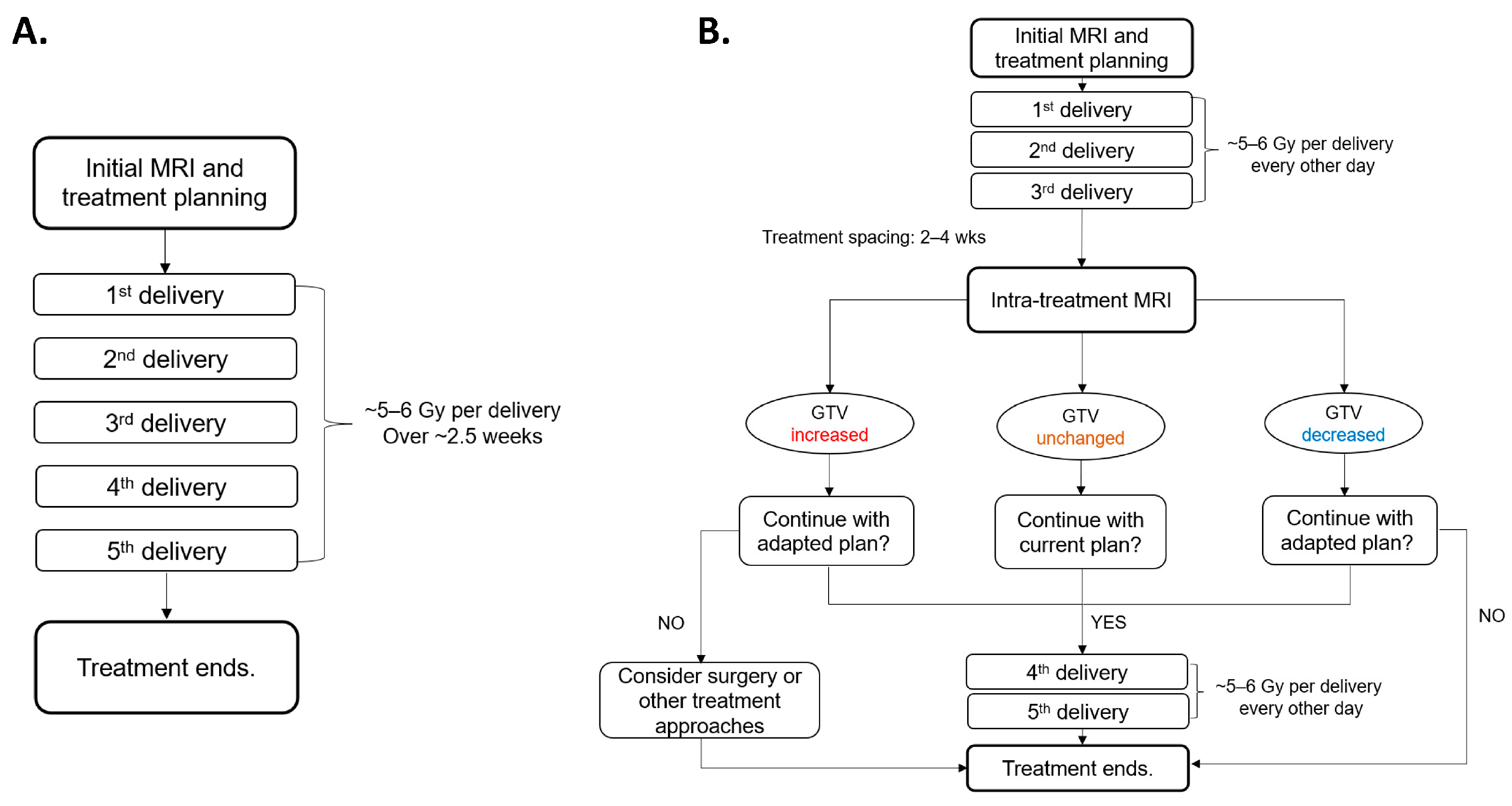
2.1. PULSAR Treatment, Study Population, and Data Acquisition
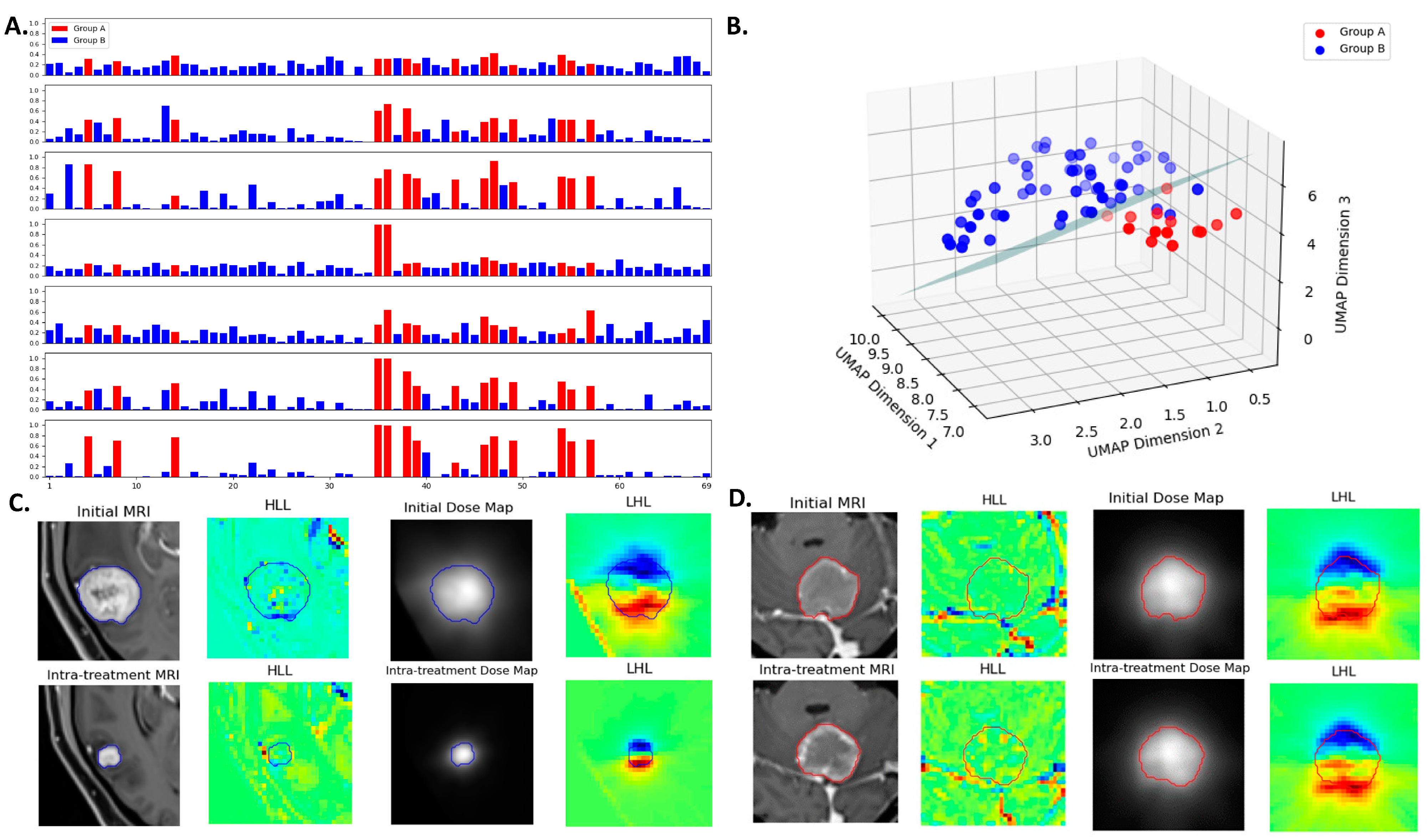
2.2. Data Processing and Multiomics Feature Extraction
2.3. Multi-Level Feature Selection
2.4. Predictive Model and Performance Evaluation
2.5. Ensemble Feature Selection (EFS)
3. Results
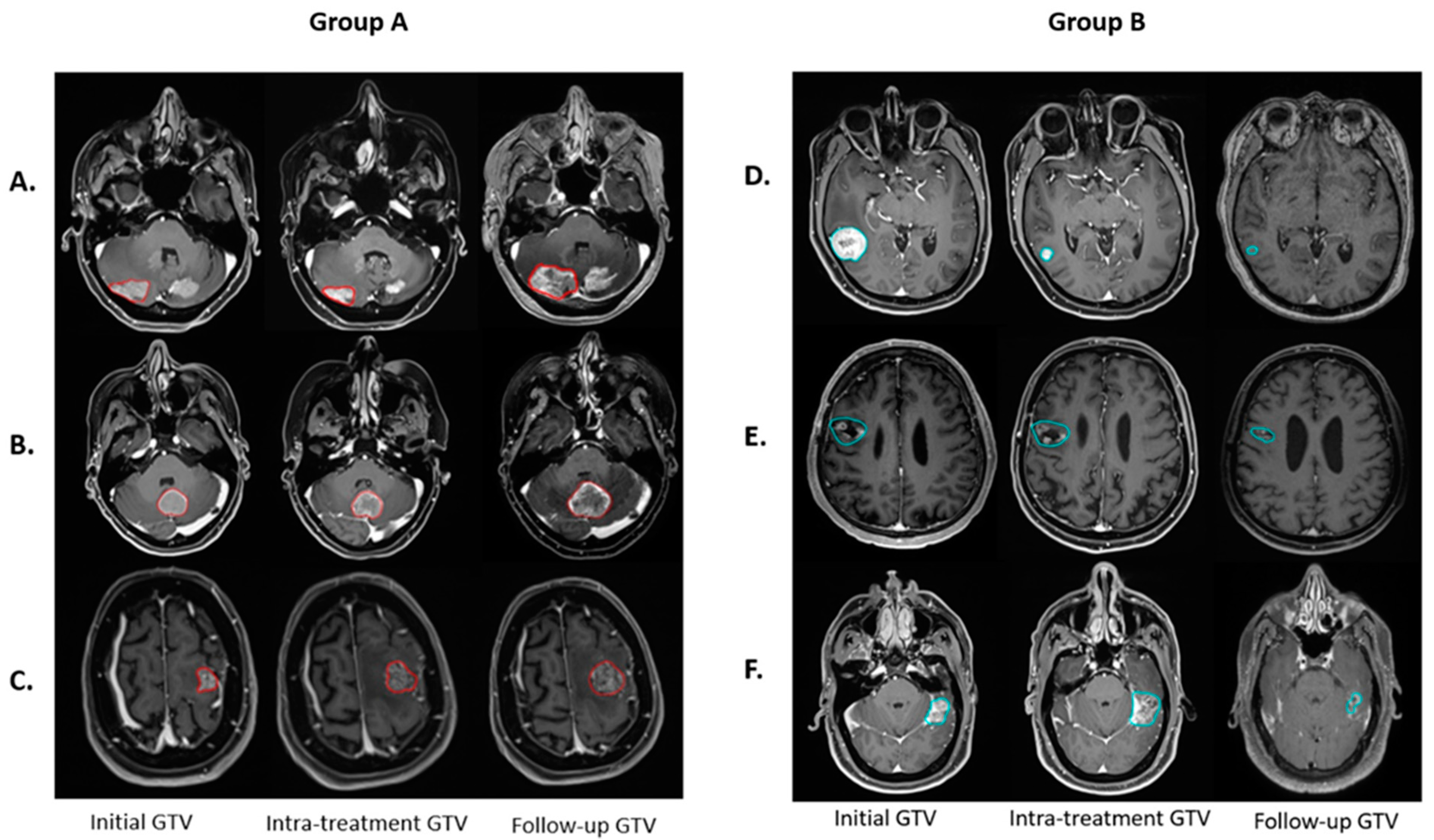
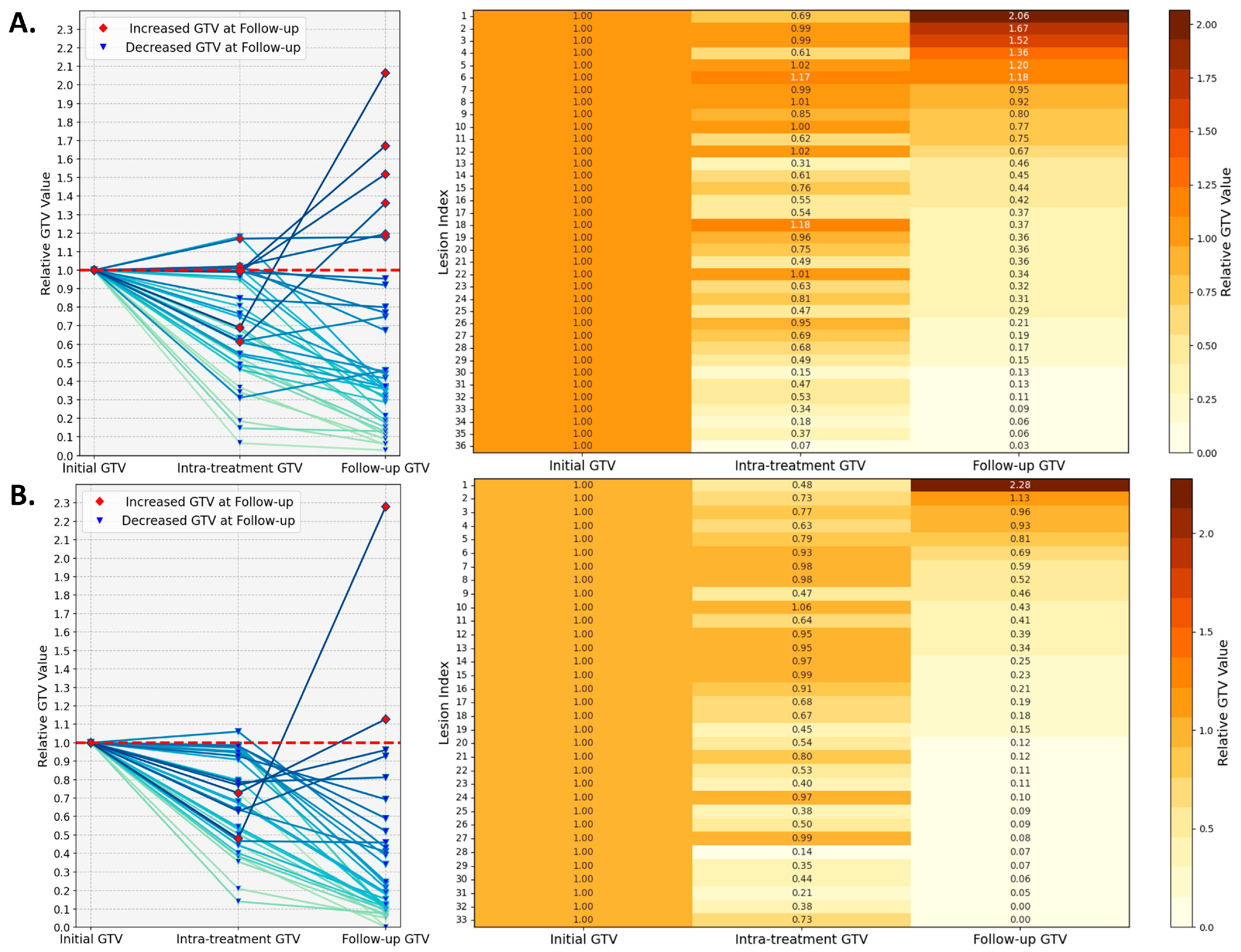
3.1. Treatment Response
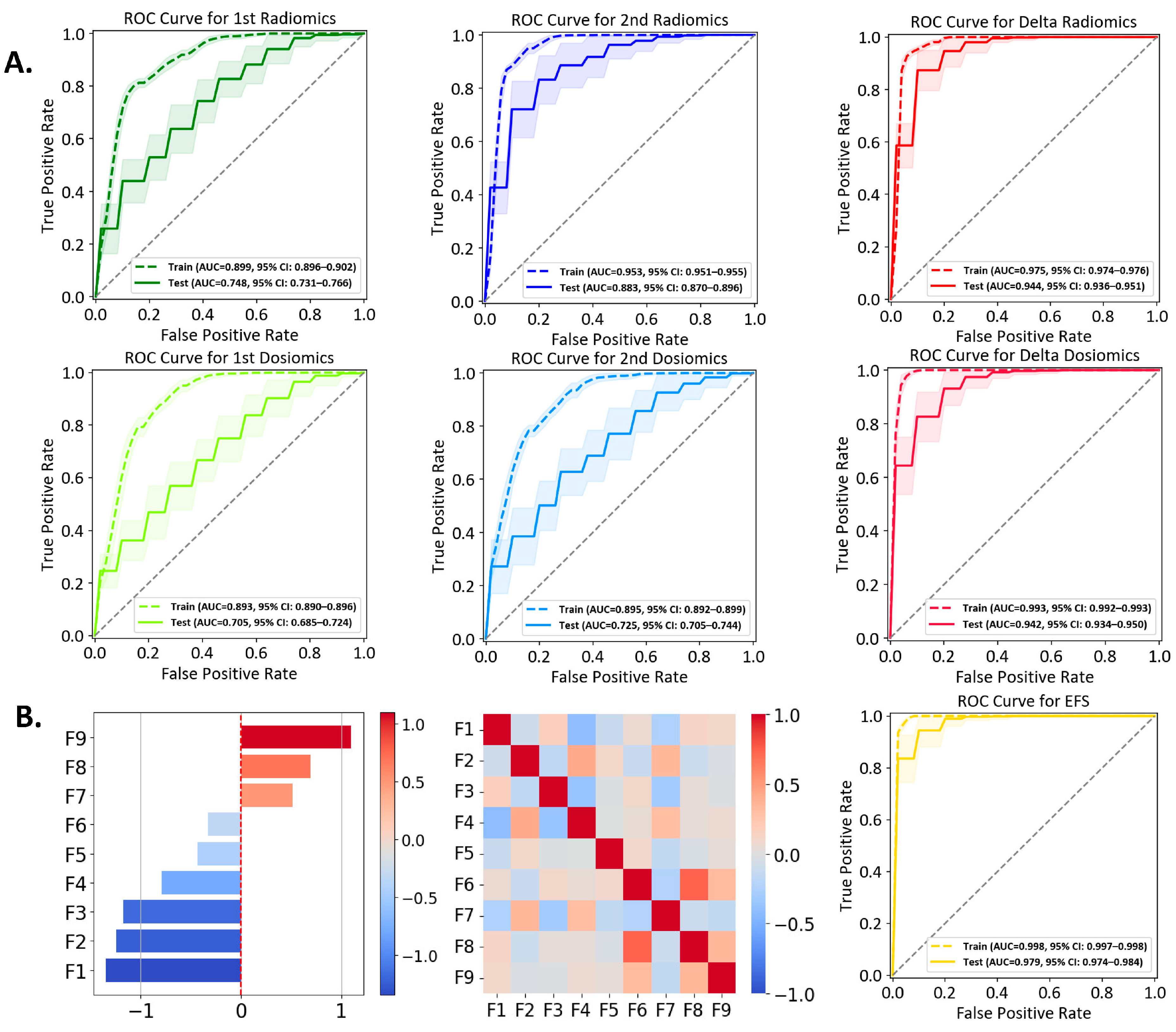
3.2. Performances of Individual Models
3.3. Ensemble Feature Selection (EFS) Model
3.4. Multiomics Feature Interpretation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ippen, F.M.; Mahadevan, A.; Wong, E.T.; Uhlmann, E.J.; Sengupta, S.; Kasper, E.M. Stereotactic Radiosurgery for Renal Cancer Brain Metastasis: Prognostic Factors and the Role of Whole-Brain Radiation and Surgical Resection. J. Oncol. 2015, 2015, 636918. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Loeffler, J. Management of brain metastases. Oncology 1999, 13, 941–954, 957–961. [Google Scholar] [PubMed]
- de la Pinta, C.; Fernández-Lizarbe, E.; Sevillano, D.; Capúz, A.; Martín, M.; Hernanz, R.; Vallejo, C.; Sancho, S. Brain metastases: Single-dose radiosurgery versus hypofractionated stereotactic radiotherapy: A retrospective study. J. Clin. Transl. Res. 2020, 6, 6. [Google Scholar]
- Sarmey, N.; Kaisman-Elbaz, T.; Mohammadi, A.M. Management strategies for large brain metastases. Front. Oncol. 2022, 12, 827304. [Google Scholar] [CrossRef]
- Higuchi, Y.; Serizawa, T.; Nagano, O.; Matsuda, S.; Ono, J.; Sato, M.; Iwadate, Y.; Saeki, N. Three-staged stereotactic radiotherapy without whole brain irradiation for large metastatic brain tumors. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1543–1548. [Google Scholar] [CrossRef]
- Sinclair, G.; Stenman, M.; Benmakhlouf, H.; Johnstone, P.; Wersäll, P.; Lindskog, M.; Hatiboglu, M.; Harmenberg, U. Adaptive radiosurgery based on two simultaneous dose prescriptions in the management of large renal cell carcinoma brain metastases in critical areas: Towards customization. Surg. Neurol. Int. 2020, 11, 21. [Google Scholar] [CrossRef]
- Minniti, G.; Scaringi, C.; Paolini, S.; Lanzetta, G.; Romano, A.; Cicone, F.; Osti, M.; Enrici, R.M.; Esposito, V. Single-fraction versus multifraction (3× 9 Gy) stereotactic radiosurgery for large (> 2 cm) brain metastases: A comparative analysis of local control and risk of radiation-induced brain necrosis. Int. J. Radiat. Oncol. Biol. Phys. 2016, 95, 1142–1148. [Google Scholar] [CrossRef]
- Oermann, E.K.; Kress, M.-A.S.; Todd, J.V.; Collins, B.T.; Hoffman, R.; Chaudhry, H.; Collins, S.P.; Morris, D.; Ewend, M.G. The impact of radiosurgery fractionation and tumor radiobiology on the local control of brain metastases. J. Neurosurg. 2013, 119, 1131–1138. [Google Scholar] [CrossRef]
- Navarria, P.; Pessina, F.; Cozzi, L.; Ascolese, A.M.; De Rose, F.; Fogliata, A.; Franzese, C.; Franceschini, D.; Tozzi, A.; D’Agostino, G. Hypo-fractionated stereotactic radiotherapy alone using volumetric modulated arc therapy for patients with single, large brain metastases unsuitable for surgical resection. Radiat. Oncol. 2016, 11, 76. [Google Scholar] [CrossRef]
- Angelov, L.; Mohammadi, A.M.; Bennett, E.E.; Abbassy, M.; Elson, P.; Chao, S.T.; Montgomery, J.S.; Habboub, G.; Vogelbaum, M.A.; Suh, J.H. Impact of 2-staged stereotactic radiosurgery for treatment of brain metastases ≥ 2 cm. J. Neurosurg. 2017, 129, 366–382. [Google Scholar] [CrossRef]
- Moore, C.; Hsu, C.-C.; Chen, W.-M.; Chen, B.P.; Han, C.; Story, M.; Aguilera, T.; Pop, L.M.; Hannan, R.; Fu, Y.-X. Personalized ultrafractionated stereotactic adaptive radiotherapy (PULSAR) in preclinical models enhances single-agent immune checkpoint blockade. Int. J. Radiat. Oncol. Biol. Phys. 2021, 110, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Filatenkov, A.; Baker, J.; Mueller, A.M.; Kenkel, J.; Ahn, G.-O.; Dutt, S.; Zhang, N.; Kohrt, H.; Jensen, K.; Dejbakhsh-Jones, S. Ablative tumor radiation can change the tumor immune cell microenvironment to induce durable complete remissions. Clin. Cancer Res. 2015, 21, 3727–3739. [Google Scholar] [CrossRef]
- Patel, T.R.; McHugh, B.J.; Bi, W.L.; Minja, F.J.; Knisely, J.P.; Chiang, V.L. A comprehensive review of MR imaging changes following radiosurgery to 500 brain metastases. Am. J. Neuroradiol. 2011, 32, 1885–1892. [Google Scholar] [CrossRef]
- Oft, D.; Schmidt, M.A.; Weissmann, T.; Roesch, J.; Mengling, V.; Masitho, S.; Bert, C.; Lettmaier, S.; Frey, B.; Distel, L.V. Volumetric regression in brain metastases after stereotactic radiotherapy: Time course, predictors, and significance. Front. Oncol. 2021, 10, 590980. [Google Scholar] [CrossRef]
- Lin, N.U.; Lee, E.Q.; Aoyama, H.; Barani, I.J.; Barboriak, D.P.; Baumert, B.G.; Bendszus, M.; Brown, P.D.; Camidge, D.R.; Chang, S.M. Response assessment criteria for brain metastases: Proposal from the RANO group. Lancet Oncol. 2015, 16, e270–e278. [Google Scholar] [CrossRef] [PubMed]
- DuMontier, C.; Loh, K.P.; Bain, P.A.; Silliman, R.A.; Hshieh, T.; Abel, G.A.; Djulbegovic, B.; Driver, J.A.; Dale, W. Defining undertreatment and overtreatment in older adults with cancer: A scoping literature review. J. Clin. Oncol. 2020, 38, 2558. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Zanfardino, M.; Franzese, M.; Pane, K.; Cavaliere, C.; Monti, S.; Esposito, G.; Salvatore, M.; Aiello, M. Bringing radiomics into a multi-omics framework for a comprehensive genotype–phenotype characterization of oncological diseases. J. Transl. Med. 2019, 17, 1–21. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Liang, B.; Yan, H.; Tian, Y.; Chen, X.; Yan, L.; Zhang, T.; Zhou, Z.; Wang, L.; Dai, J. Dosiomics: Extracting 3D spatial features from dose distribution to predict incidence of radiation pneumonitis. Front. Oncol. 2019, 9, 269. [Google Scholar] [CrossRef]
- Murakami, Y.; Soyano, T.; Kozuka, T.; Ushijima, M.; Koizumi, Y.; Miyauchi, H.; Kaneko, M.; Nakano, M.; Kamima, T.; Hashimoto, T. Dose-based radiomic analysis (dosiomics) for intensity modulated radiation therapy in patients with prostate cancer: Correlation between planned dose distribution and biochemical failure. Int. J. Radiat. Oncol. Biol. Phys. 2022, 112, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Reginelli, A.; Grassi, R.; Boldrini, L.; Vacca, G.; D’Ippolito, E.; Annunziata, S.; Farchione, A.; Belfiore, M.P.; Desideri, I. Delta radiomics: A systematic review. Radiol. Medica 2021, 126, 1571–1583. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Karalis, J.D.; Elamir, A.; Bifolco, A.; Wachsmann, M.; Capretti, G.; Spaggiari, P.; Enrico, S.; Balasubramanian, K.; Fatimah, N. Delta radiomic features predict resection margin status and overall survival in neoadjuvant-treated pancreatic cancer patients. Ann. Surg. Oncol. 2024, 31, 2608–2620. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Kalbasi, A.; Hsu, W.; Ruan, D.; Fu, J.; Shao, J.; Cao, M.; Wang, C.; Eilber, F.C.; Bernthal, N. Treatment effect prediction for sarcoma patients treated with preoperative radiotherapy using radiomics features from longitudinal diffusion-weighted MRIs. Phys. Med. Biol. 2020, 65, 175006. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Kukreja, S.L.; Löfberg, J.; Brenner, M.J. A least absolute shrinkage and selection operator (LASSO) for nonlinear system identification. IFAC Proc. Vol. 2006, 39, 814–819. [Google Scholar] [CrossRef]
- Yan, K.; Zhang, D. Feature selection and analysis on correlated gas sensor data with recursive feature elimination. Sens. Actuators B Chem. 2015, 212, 353–363. [Google Scholar] [CrossRef]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Gautam, A.K.; Bansal, A. Email-based cyberstalking detection on textual data using multi-model soft voting technique of machine learning approach. J. Comput. Inf. Syst. 2023, 63, 1362–1381. [Google Scholar] [CrossRef]
- Zhou, J.; Lu, J.; Gao, C.; Zeng, J.; Zhou, C.; Lai, X.; Cai, W.; Xu, M. Predicting the response to neoadjuvant chemotherapy for breast cancer: Wavelet transforming radiomics in MRI. BMC Cancer 2020, 20, 100. [Google Scholar] [CrossRef]
- Jiang, W.; Song, Y.; Sun, Z.; Qiu, J.; Shi, L. Dosimetric factors and radiomics features within different regions of interest in planning CT images for improving the prediction of radiation pneumonitis. Int. J. Radiat. Oncol. Biol. Phys. 2021, 110, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Vaios, E.; Wang, Y.; Yang, Z.; Cui, Y.; Reitman, Z.J.; Lafata, K.J.; Fecci, P.; Kirkpatrick, J.; Yin, F.-F. Dose-Incorporated Deep Ensemble Learning for Improving Brain Metastasis Stereotactic Radiosurgery Outcome Prediction. Int. J. Radiat. Oncol. Biol. Phys. 2024, 120, 603–613. [Google Scholar] [CrossRef] [PubMed]
- MacCallum, R.C.; Widaman, K.F.; Preacher, K.J.; Hong, S. Sample size in factor analysis: The role of model error. Multivar. Behav. Res. 2001, 36, 611–637. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-Y.; Lee, C.-C.; Yang, H.-C.; Chen, C.-J.; Chung, W.-Y.; Wu, H.-M.; Guo, W.-Y.; Liu, R.-S.; Lu, C.-F. Enhancement of radiosurgical treatment outcome prediction using MRI radiomics in patients with non-small cell lung cancer brain metastases. Cancers 2021, 13, 4030. [Google Scholar] [CrossRef]
- Wang, K.; An, Y.; Zhou, J.; Long, Y.; Chen, X. A novel multi-level feature selection method for radiomics. Alex. Eng. J. 2023, 66, 993–999. [Google Scholar] [CrossRef]
- Moradmand, H.; Aghamiri, S.M.R.; Ghaderi, R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J. Appl. Clin. Med. Phys. 2020, 21, 179–190. [Google Scholar] [CrossRef]
- Peng, H.; Moore, C.; Zhang, Y.; Saha, D.; Jiang, S.; Timmerman, R. An AI-based approach for modeling the synergy between radiotherapy and immunotherapy. Sci. Rep. 2024, 14, 8250. [Google Scholar] [CrossRef]
- Young, S.; Goldberg, D.; Hannallah, J.; Struycken, L.; Woodhead, G. Advancing Radioembolization through Personalized Dosimetry. Adv. Clin. Radiol. 2024, 6, 55–64. [Google Scholar] [CrossRef]
- Tunali, I.; Stringfield, O.; Guvenis, A.; Wang, H.; Liu, Y.; Balagurunathan, Y.; Lambin, P.; Gillies, R.J.; Schabath, M.B. Radial gradient and radial deviation radiomic features from pre-surgical CT scans are associated with survival among lung adenocarcinoma patients. Oncotarget 2017, 8, 96013. [Google Scholar] [CrossRef]
- Caballo, M.; Pangallo, D.R.; Sanderink, W.; Hernandez, A.M.; Lyu, S.H.; Molinari, F.; Boone, J.M.; Mann, R.M.; Sechopoulos, I. Multi-marker quantitative radiomics for mass characterization in dedicated breast CT imaging. Med. Phys. 2021, 48, 313–328. [Google Scholar] [CrossRef]
- Sharpton, S.R.; Oermann, E.K.; Moore, D.T.; Schreiber, E.; Hoffman, R.; Morris, D.E.; Ewend, M.G. The volumetric response of brain metastases after stereotactic radiosurgery and its post-treatment implications. Neurosurgery 2014, 74, 9–16. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Value |
|---|---|
| Patient Information | |
| Age (range) | 61 (28–84) |
| Total Patients (Male: Female) | 39 (14:25) |
| Lesion Information | |
| Single brain metastases | 26 |
| Multiple brain metastases | 43 |
| Total Lesions (Decreased: Non-Decreased) * | 69 (55:14) |
| Lesion size | |
| Initial Tumor Volume (mm3) | 3666.3 (14.5–37607.6) |
| 3D Diameter (mm) | 25.5 (3.7–79.8) |
| Delivered Dose and Fractionation by Lesion Count (Gy, Fx) | |
| 30 Gy, 5 Fx | 57 |
| 27.5 Gy, 5 Fx | 2 |
| 25 Gy, 5 Fx | 9 |
| 24 Gy, 4 Fx | 1 |
| 1st Radiomics | 2nd Radiomics | Delta Radiomics | 1st Dosiomics | 2nd Dosiomics | Delta Dosiomics | Ensemble Feature Selection (EFS) | |
|---|---|---|---|---|---|---|---|
| Sensitivity | 0.503 ± 0.271 (95% CI: 0.469–0.537) | 0.755 ± 0.266 (95% CI: 0.722–0.789) | 0.903 ± 0.174 (95% CI: 0.882–0.925) | 0.559 ± 0.271 (95% CI: 0.525–0.592) | 0.538 ± 0.309 (95% CI: 0.499–0.577) | 0.802 ± 0.242 (95% CI: 0.772–0.832) | 0.907 ± 0.169 (95% CI: 0.886–0.928) |
| Specificity | 0.745 ± 0.128 (95% CI: 0.729–0.761) | 0.854 ± 0.102 (95% CI: 0.841–0.867) | 0.836 ± 0.117 (95% CI: 0.822–0.851) | 0.636 ± 0.141 (95% CI: 0.618–0.654) | 0.713 ± 0.129 (95% CI: 0.697–0.729) | 0.872 ± 0.102 (95% CI: 0.860–0.885) | 0.920 ± 0.085 (95% CI: 0.909–0.931) |
| Accuracy | 0.696 ± 0.107 (95% CI: 0.682–0.709) | 0.833 ± 0.090 (95% CI: 0.822–0.844) | 0.850 ± 0.094 (95% CI: 0.838–0.862) | 0.621 ± 0.112 (95% CI: 0.607–0.635) | 0.677 ± 0.111 (95% CI: 0.664–0.691) | 0.858 ± 0.086 (95% CI: 0.847–0.869) | 0.917 ± 0.072 (95% CI: 0.908–0.926) |
| AUC | 0.748 ± 0.143 (95% CI: 0.731–0.766) | 0.883 ± 0.107 (95% CI: 0.870–0.896) | 0.944 ± 0.061 (95% CI: 0.936–0.951) | 0.705 ± 0.157 (95% CI: 0.685–0.724) | 0.725 ± 0.158 (95% CI: 0.705–0.744) | 0.942 ± 0.062 (95% CI: 0.934–0.950) | 0.979 ± 0.039 (95% CI: 0.974–0.984) |
| Precision | 0.344 ± 0.197 (95% CI: 0.319–0.368) | 0.605 ± 0.232 (95% CI: 0.577–0.634) | 0.632 ± 0.197 (95% CI: 0.607–0.657) | 0.285 ± 0.140 (95% CI: 0.268–0.302) | 0.319 ± 0.182 (95% CI: 0.296 –0.341) | 0.661 ± 0.224 (95% CI: 0.633–0.689) | 0.786 ± 0.201 (95% CI: 0.761–0.811) |
| F1 Score | 0.389 ± 0.193 (95% CI: 0.365–0.413) | 0.640 ± 0.199 (95% CI: 0.615–0.665) | 0.720 ± 0.151 (95% CI: 0.701–0.739) | 0.366 ± 0.162 (95% CI: 0.346–0.386) | 0.386 ± 0.205 (95% CI: 0.361–0.412) | 0.694 ± 0.186 (95% CI: 0.671–0.718) | 0.821 ± 0.147 (95% CI: 0.803–0.839) |
| 1st Radiomics vs. 2nd Radiomics | 1st Radiomics vs. Delta Radiomics | 1st Radiomics vs. 1st Dosiomics | 1st Radiomics vs. 2nd Dosiomics | 1st Radiomics vs. Delta Dosiomics | 1st Radiomics vs. EFS | 2nd Radiomics vs. Delta Radiomics | 2nd Radiomics vs. 1st Dosiomics | 2nd Radiomics vs. 2nd Dosiomics | 2nd Radiomics vs. Delta Dosiomics | 2nd Radiomics vs. EFS | Delta Radiomics vs. 1st Dosiomics | Delta Radiomics vs. 2nd Dosiomics | Delta Radiomics vs. Delta Dosiomics | Delta Radiomics vs. EFS | 1st Dosiomics vs. 2nd Dosiomics | 1st Dosiomics vs. Delta Dosiomics | 1st Dosiomics vs. EFS | 2nd Dosiomics vs. Delta Dosiomics | 2nd Dosiomics vs. EFS | Delta Dosiomics vs. EFS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | <0.002 * | <0.002 * | 0.022 | 0.176 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | 0.041 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | 0.829 | 0.428 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * |
| Specificity | <0.002 * | <0.002 * | <0.002 * | 0.006 | <0.002 * | <0.002 * | 0.070 | <0.002 * | <0.002 * | 0.047 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * |
| Accuracy | <0.002 * | <0.002 * | <0.002 * | 0.063 | <0.002 * | <0.002 * | 0.045 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | 0.328 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * |
| AUC | <0.002 * | <0.002 * | <0.002 * | 0.079 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | 0.758 | <0.002 * | 0.158 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * |
| Precision | <0.002 * | <0.002 * | <0.002 * | 0.140 | <0.002 * | <0.002 * | 0.170 | <0.002 * | <0.002 * | 0.007 | <0.002 * | <0.002 * | <0.002 * | 0.127 | <0.002 * | 0.022 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * |
| F1 Score | <0.002 * | <0.002 * | 0.152 | 0.882 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * | 0.088 | <0.002 * | 0.221 | <0.002 * | <0.002 * | <0.002 * | <0.002 * | <0.002 * |
| Abbreviation | Multi-Omics Type | Wavelet Filtering/Original | Feature Class | Feature Name | Feature Weight Coefficient | Feature Values (Z-Score) | ||
|---|---|---|---|---|---|---|---|---|
| Group A | Group B | p-Value | ||||||
| F1 | Delta Radiomics | Original | shape | Least Axis Length | −1.34382 | −0.231 ± 0.233 | 0.059 ± 0.468 | <0.05 * |
| F2 | 1st Dosiomics | Wavelet-LHL | glszm | Low Gray Level Zone Emphasis | −1.23960 | −0.58 ± 0.867 | 0.148 ± 0.994 | <0.05 * |
| F3 | 2nd Radiomics | Wavelet-LHH | glcm | Correlation | −1.17479 | −0.577 ± 0.584 | 0.147 ± 1.043 | <0.05 * |
| F4 | 2nd Radiomics | Original | glrlm | Long Run Low Gray Level Emphasis | −0.78866 | −0.444 ± 0.195 | 0.113 ± 1.098 | <0.05 * |
| F5 | Delta Radiomics | Wavelet-HHH | gldm | Large Dependence High Gray Level Emphasis | −0.32663 | −0.597 ± 0.951 | 0.152 ± 0.765 | <0.05 * |
| F6 | Delta Radiomics | Wavelet-HHH | glszm | Gray Level NonUniformity | −0.43211 | −0.379 ± 0.674 | 0.096 ± 0.497 | <0.05 * |
| F7 | 1st Radiomics | original | glcm | MCC | 0.51466 | 0.146 ± 0.812 | −0.037 ± 1.054 | 0.486 |
| F8 | Delta Radiomics | Wavelet-LLH | firstorder | Median | 0.69508 | 0.558 ± 1.214 | −0.142 ± 1.086 | 0.064 |
| F9 | Delta Radiomics | Wavelet-HLL | firstorder | Kurtosis | 1.09794 | 0.254 ± 0.831 | −0.065 ± 0.326 | 0.181 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Dohopolski, M.; Stojadinovic, S.; Schmitt, L.G.; Anand, S.; Kim, H.; Pompos, A.; Godley, A.; Jiang, S.; Dan, T.; et al. Multiomics-Based Outcome Prediction in Personalized Ultra-Fractionated Stereotactic Adaptive Radiotherapy (PULSAR). Cancers 2024, 16, 3425. https://doi.org/10.3390/cancers16193425
Zhang H, Dohopolski M, Stojadinovic S, Schmitt LG, Anand S, Kim H, Pompos A, Godley A, Jiang S, Dan T, et al. Multiomics-Based Outcome Prediction in Personalized Ultra-Fractionated Stereotactic Adaptive Radiotherapy (PULSAR). Cancers. 2024; 16(19):3425. https://doi.org/10.3390/cancers16193425
Chicago/Turabian StyleZhang, Haozhao, Michael Dohopolski, Strahinja Stojadinovic, Luiza Giuliani Schmitt, Soummitra Anand, Heejung Kim, Arnold Pompos, Andrew Godley, Steve Jiang, Tu Dan, and et al. 2024. "Multiomics-Based Outcome Prediction in Personalized Ultra-Fractionated Stereotactic Adaptive Radiotherapy (PULSAR)" Cancers 16, no. 19: 3425. https://doi.org/10.3390/cancers16193425
APA StyleZhang, H., Dohopolski, M., Stojadinovic, S., Schmitt, L. G., Anand, S., Kim, H., Pompos, A., Godley, A., Jiang, S., Dan, T., Wardak, Z., Timmerman, R., & Peng, H. (2024). Multiomics-Based Outcome Prediction in Personalized Ultra-Fractionated Stereotactic Adaptive Radiotherapy (PULSAR). Cancers, 16(19), 3425. https://doi.org/10.3390/cancers16193425





