The Impact of Mutational Hotspots on Cancer Survival
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Cancer Data
2.2. Detection of Hypermutated Samples
2.3. Statistical Analyses
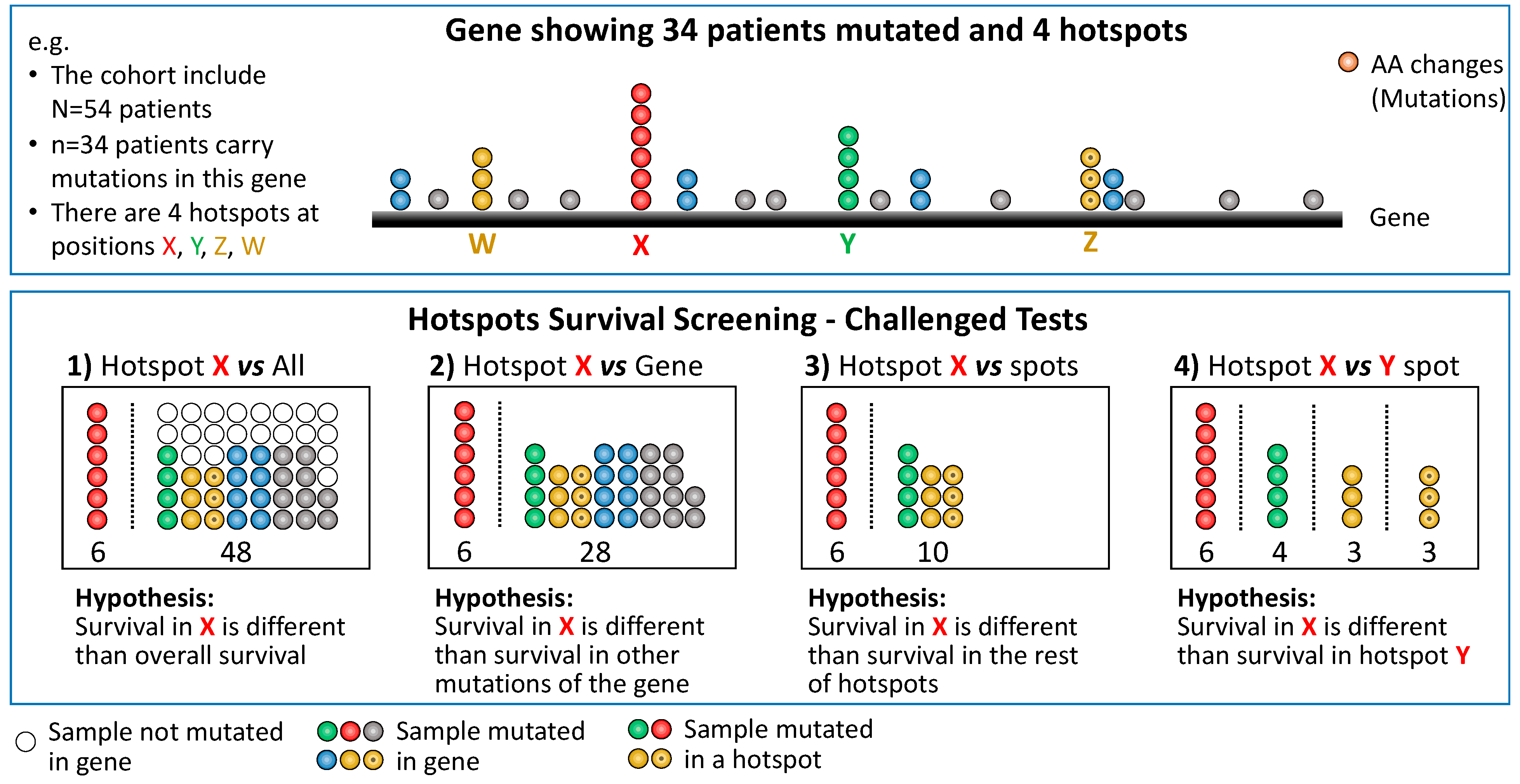
2.4. Test Strategies
3. Results
3.1. Hypermutated Samples Bias Hotspots Associated with Cancer Survival
3.2. Many Hotspots Are Potentially Associated with Cancer Survival
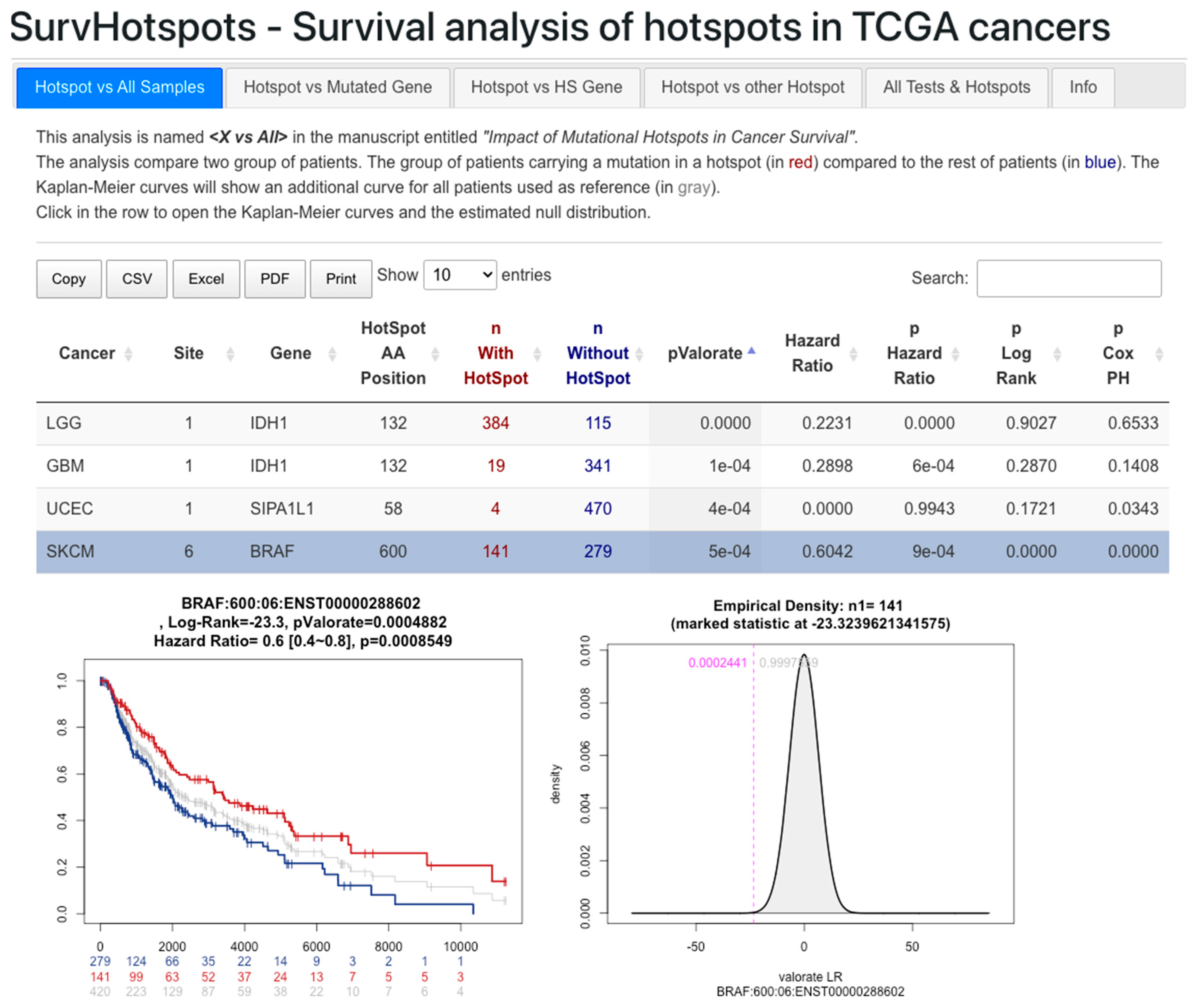
3.3. Web Resource
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, X.; Shao, Y.; Qin, H.F.; Tai, Y.H.; Gao, H.J. ALK-Rearrangement in Non-Small-Cell Lung Cancer (NSCLC). Thorac. Cancer 2018, 9, 423–430. [Google Scholar] [CrossRef]
- van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.M.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Sugio, K.; Uramoto, H.; Ono, K.; Oyama, T.; Hanagiri, T.; Sugaya, M.; Ichiki, Y.; So, T.; Nakata, S.; Morita, M.; et al. Mutations within the Tyrosine Kinase Domain of EGFR Gene Specifically Occur in Lung Adenocarcinoma Patients with a Low Exposure of Tobacco Smoking. Br. J. Cancer 2006, 94, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.A.; Mooij, T.M.; Roos-Blom, M.J.; Jervis, S.; Van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA-J. Am. Med. Assoc. 2017, 317, 2402–2416. [Google Scholar] [CrossRef] [PubMed]
- Holderfield, M.; Deuker, M.M.; McCormick, F.; McMahon, M. Targeting RAF Kinases for Cancer Therapy: BRAF-Mutated Melanoma and Beyond. Nat. Rev. Cancer 2014, 14, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Ledesma, E.; Flores, D.; Trevino, V. Computational Methods for Detecting Cancer Hotspots. Comput. Struct. Biotechnol. J. 2020, 18, 3567–3576. [Google Scholar] [CrossRef]
- Miller, M.L.; Reznik, E.; Gauthier, N.P.; Aksoy, B.A.; Korkut, A.; Gao, J.; Ciriello, G.; Schultz, N.; Sander, C. Pan-Cancer Analysis of Mutation Hotspots in Protein Domains. Cell Syst. 2015, 1, 197–209. [Google Scholar] [CrossRef]
- Li, V.D.; Li, K.H.; Li, J.T. TP53 Mutations as Potential Prognostic Markers for Specific Cancers: Analysis of Data from The Cancer Genome Atlas and the International Agency for Research on Cancer TP53 Database. J. Cancer Res. Clin. Oncol. 2019, 145, 625–636. [Google Scholar] [CrossRef]
- Tuna, M.; Ju, Z.; Yoshihara, K.; Amos, C.I.; Tanyi, J.L.; Mills, G.B. Clinical Relevance of TP53 Hotspot Mutations in High-Grade Serous Ovarian Cancers. Br. J. Cancer 2020, 122, 405–412. [Google Scholar] [CrossRef]
- Munch-Petersen, H.D.; Asmar, F.; Dimopoulos, K.; Areškevičiūtė, A.; Brown, P.; Girkov, M.S.; Pedersen, A.; Sjö, L.D.; Heegaard, S.; Broholm, H.; et al. TP53 Hotspot Mutations Are Predictive of Survival in Primary Central Nervous System Lymphoma Patients Treated with Combination Chemotherapy. Acta Neuropathol. Commun. 2016, 4, 40. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Sullivan, R.; LoRusso, P.; Boerner, S.; Dummer, R. Achievements and Challenges of Molecular Targeted Therapy in Melanoma. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, 177–186. [Google Scholar] [CrossRef]
- Supek, F.; Miñana, B.; Valcárcel, J.; Gabaldón, T.; Lehner, B. Synonymous Mutations Frequently Act as Driver Mutations in Human Cancers. Cell 2014, 156, 1324–1335. [Google Scholar] [CrossRef]
- Kikutake, C.; Suyama, M. Possible Involvement of Silent Mutations in Cancer Pathogenesis and Evolution. Sci. Rep. 2023, 13, 7593. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.A.; Gordenin, D.A. Hypermutation in Human Cancer Genomes: Footprints and Mechanisms. Nat. Rev. Cancer 2014, 14, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Treviño, V.; Martínez-Ledesma, E.; Tamez-Peña, J. Identification of Outcome-Related Driver Mutations in Cancer Using Conditional Co-Occurrence Distributions. Sci. Rep. 2017, 7, srep43350. [Google Scholar] [CrossRef] [PubMed]
- Vandin, F.; Papoutsaki, A.; Raphael, B.J.; Upfal, E. Accurate Computation of Survival Statistics in Genome-Wide Studies. PLoS Comput. Biol. 2015, 11, e1004071. [Google Scholar] [CrossRef] [PubMed]
- Treviño, V.; Tamez-Pena, J. VALORATE: Fast and Accurate Log-Rank Test in Balanced and Unbalanced Comparisons of Survival Curves and Cancer Genomics. Bioinformatics 2017, 33, 1900–1901. [Google Scholar] [CrossRef]
- Trevino, V. HotSpotAnnotations-A Database for Hotspot Mutations and Annotations in Cancer. Database 2020, 2020, baaa025. [Google Scholar] [CrossRef]
- Lin, K.W.; Yakymovych, I.; Jia, M.; Yakymovych, M.; Souchelnytskyi, S. Phosphorylation of EEF1A1 at Ser300 by TβR-I Results in Inhibition of MRNA Translation. Curr. Biol. 2010, 20, 1615–1625. [Google Scholar] [CrossRef] [PubMed]
- Soares, D.C.; Barlow, P.N.; Newbery, H.J.; Porteous, D.J.; Abbott, C.M. Structural Models of Human EEF1A1 and EEF1A2 Reveal Two Distinct Surface Clusters of Sequence Variation and Potential Differences in Phosphorylation. PLoS ONE 2009, 4, e6315. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Lu, S.X.; Liu, L.L.; Wang, C.H.; Yang, X.; Zhang, Z.Y.; Zhang, H.Z.; Yun, J. ping EEF1A1 Overexpression Enhances Tumor Progression and Indicates Poor Prognosis in Hepatocellular Carcinoma. Transl. Oncol. 2018, 11, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Nambiar, D.K.; Cao, H.; Viswanathan, V.; Kwok, S.; Hui, A.B.; Hou, Y.; Hildebrand, R.; von Eyben, R.; Holmes, B.J.; et al. NFE2L2 Mutations Enhance Radioresistance in Head and Neck Cancer by Modulating Intratumoral Myeloid Cells. Cancer Res. 2023, 83, 861–874. [Google Scholar] [CrossRef]
- Guerrero, S.; Casanova, I.; Farre, L.; Mazo, A.; Capella, G.; Mangues, R. K-Ras Codon 12 Mutation Induces Higher Level of Resistance to Apoptosis and Predisposition to Anchorage-Independent Growth than Codon 13 Mutation or Proto-Oncogene Overexpression. Cancer Res. 2000, 60, 6750–6756. [Google Scholar]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S.; et al. A Transforming Mutation in the Pleckstrin Homology Domain of AKT1 in Cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef]
- Taylor, S.E.; O’Connor, C.M.; Wang, Z.; Shen, G.; Song, H.; Leonard, D.; Sangodkar, J.; LaVasseur, C.; Avril, S.; Waggoner, S.; et al. The Highly Recurrent PP2A Aa-Subunit Mutation P179R Alters Protein Structure and Impairs PP2A Enzyme Function to Promote Endometrial Tumorigenesis. Cancer Res. 2019, 79, 4242–4257. [Google Scholar] [CrossRef]
- Christensen, E.; Birkenkamp-Demtröder, K.; Nordentoft, I.; Høyer, S.; van der Keur, K.; van Kessel, K.; Zwarthoff, E.; Agerbæk, M.; Ørntoft, T.F.; Jensen, J.B.; et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur. Urol. 2017, 71, 961–969. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent Somatic Mutations of GNAQ in Uveal Melanoma and Blue Naevi. Nature 2009, 457, 599–602. [Google Scholar] [CrossRef]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef]
- Wang, Y.; Chiappetta, G.; Guérois, R.; Liu, Y.; Romero, S.; Boesch, D.J.; Krause, M.; Dessalles, C.A.; Babataheri, A.; Barakat, A.I.; et al. PPP2R1A Regulates Migration Persistence through the NHSL1-Containing WAVE Shell Complex. Nat. Commun. 2023, 14, 3541. [Google Scholar] [CrossRef] [PubMed]
- Kilian, M.; Friedrich, M.; Sanghvi, K.; Green, E.; Pusch, S.; Kawauchi, D.; Löwer, M.; Sonner, J.K.; Krämer, C.; Zaman, J.; et al. T-Cell Receptor Therapy Targeting Mutant Capicua Transcriptional Repressor in Experimental Gliomas. Clin. Cancer Res. 2022, 28, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Razafinjatovo, C.F.; Stiehl, D.; Deininger, E.; Rechsteiner, M.; Moch, H.; Schraml, P. VHL Missense Mutations in the P53 Binding Domain Show Different Effects on P53 Signaling and HIFα Degradation in Clear Cell Renal Cell Carcinoma. Oncotarget 2017, 8, 10199–10212. [Google Scholar] [CrossRef] [PubMed]
- Maskin, C.R.; Raman, R.; Houvras, Y. PPP6C, a Serine-Threonine Phosphatase, Regulates Melanocyte Differentiation and Contributes to Melanoma Tumorigenesis through Modulation of MITF Activity. Sci. Rep. 2022, 12, 5573. [Google Scholar] [CrossRef] [PubMed]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-Associated SF3B1 Mutations Affect Alternative Splicing by Promoting Alternative Branchpoint Usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef]
- Brown, A.L.; Arts, P.; Carmichael, C.L.; Babic, M.; Dobbins, J.; Chong, C.E.; Schreiber, A.W.; Feng, J.; Phillips, K.; Wang, P.P.S.; et al. RUNX1-Mutated Families Show Phenotype Heterogeneity and a Somatic Mutation Profile Unique to Germline Predisposed AML. Blood Adv. 2020, 4, 1131–1144. [Google Scholar] [CrossRef]
- Furukawa, M.; Nagatomo, I.; Kumagai, T.; Yamadori, T.; Takahashi, R.; Yoshimura, M.; Yoneda, T.; Takeda, Y.; Goya, S.; Matsuoka, H.; et al. Gefitinib-Sensitive EGFR Lacking Residues 746-750 Exhibits Hypophosphorylation at Tyrosine Residue 1045, Hypoubiquitination, and Impaired Endocytosis. DNA Cell Biol. 2007, 26, 178–185. [Google Scholar] [CrossRef]
- Nogueira, G.; Fernandes, R.; García-Moreno, J.F.; Romão, L. Nonsense-Mediated RNA Decay and Its Bipolar Function in Cancer. Mol. Cancer 2021, 20, 72. [Google Scholar] [CrossRef]
- Bonjoch, L.; Fernandez-Rozadilla, C.; Alvarez-Barona, M.; Lopez-Novo, A.; Herrera-Pariente, C.; Amigo, J.; Bujanda, L.; Remedios, D.; Dacal, A.; Cubiella, J.; et al. BMPR2 as a Novel Predisposition Gene for Hereditary Colorectal Polyposis. Gastroenterology 2023, 165, 162–172.e5. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Khodadadi-Jamayran, A.; Dolgalev, I.; Cho, H.; Badri, S.; Chiriboga, L.A.; Zeck, B.; de Rodas Gregorio, M.L.; Dowling, C.M.; Labbe, K.; et al. Targeting the Atf7ip–Setdb1 Complex Augments Antitumor Immunity by Boosting Tumor Immunogenicity. Cancer Immunol. Res. 2021, 9, 1298–1315. [Google Scholar] [CrossRef] [PubMed]
- Javitt, A.; Shmueli, M.D.; Kramer, M.P.; Kolodziejczyk, A.A.; Cohen, I.J.; Radomir, L.; Sheban, D.; Kamer, I.; Litchfield, K.; Bab-Dinitz, E.; et al. The Proteasome Regulator PSME4 Modulates Proteasome Activity and Antigen Diversity to Abrogate Antitumor Immunity in NSCLC. Nat. Cancer 2023, 4, 629–647. [Google Scholar] [CrossRef]
- Trevino, V. Modeling and Analysis of Site-Specific Mutations in Cancer Identifies Known plus Putative Novel Hotspots and Bias Due to Contextual Sequences. Comput. Struct. Biotechnol. J. 2020, 18, 1664–1675. [Google Scholar] [CrossRef]
- Gutiontov, S.I.; Turchan, W.T.; Spurr, L.F.; Rouhani, S.J.; Chervin, C.S.; Steinhardt, G.; Lager, A.M.; Wanjari, P.; Malik, R.; Connell, P.P.; et al. CDKN2A Loss-of-Function Predicts Immunotherapy Resistance in Non-Small Cell Lung Cancer. Sci. Rep. 2021, 11, 20059. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Duan, X.; Zhang, C.; Yuan, J.; Wen, J.; Zheng, C.; Shi, J.; Yuan, M. KAT6B May Be Applied as a Potential Therapeutic Target for Glioma. J. Oncol. 2022, 2022, 2500092. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, Y.; Broaddus, R.; Sun, L.; Xue, F.; Zhang, W. Exon 3 Mutations of CTNNB1 Drive Tumorigenesis: A Review. Oncotarget 2018, 9, 5492–5508. [Google Scholar] [CrossRef]
- Ruz-Caracuel, I.; López-Janeiro, Á.; Heredia-Soto, V.; Ramón-Patino, J.L.; Yébenes, L.; Berjón, A.; Hernández, A.; Gallego, A.; Ruiz, P.; Redondo, A.; et al. Clinicopathological Features and Prognostic Significance of CTNNB1 Mutation in Low-Grade, Early-Stage Endometrial Endometrioid Carcinoma. Virchows Arch. 2021, 479, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Binkley, M.S.; Jeon, Y.J.; Nesselbush, M.; Moding, E.J.; Nabet, B.Y.; Almanza, D.; Kunder, C.; Stehr, H.; Yoo, C.H.; Rhee, S.; et al. KEAP1/NFE2L2 Mutations Predict Lung Cancer Radiation Resistance That Can Be Targeted by Glutaminase Inhibition. Cancer Discov. 2020, 10, 1826–1841. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Zhang, Y.; Cieślik, M.; Guo, J.; Tan, M.; Green, M.D.; Wang, W.; Lin, H.; Li, W.; et al. Epigenetic Driver Mutations in ARID1A Shape Cancer Immune Phenotype and Immunotherapy. J. Clin. Investig. 2020, 130, 2712–2726. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Mandelker, D.; Schmidt-Kittler, O.; Samuels, Y.; Velculescu, V.E.; Kinzler, K.W.; Vogelstein, B.; Gabelli, S.B.; Amzel, L.M. The Structure of a Human P110α/P85α Complex Elucidates the Effects of Oncogenic PI3Kα Mutations. Science 2007, 318, 1744–1748. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, R.; Xiu, J.; Goldberg, R.M.; Philip, P.A.; Seeber, A.; Battaglin, F.; Arai, H.; Lo, J.H.; Naseem, M.; Puccini, A.; et al. The Impact of ARID1A Mutation on Molecular Characteristics in Colorectal Cancer. Eur. J. Cancer 2020, 140, 119–129. [Google Scholar] [CrossRef]
- Christie, M.; Jorissen, R.N.; Mouradov, D.; Sakthianandeswaren, A.; Li, S.; Day, F.; Tsui, C.; Lipton, L.; Desai, J.; Jones, I.T.; et al. Different APC Genotypes in Proximal and Distal Sporadic Colorectal Cancers Suggest Distinct WNT/β-Catenin Signalling Thresholds for Tumourigenesis. Oncogene 2013, 32, 4675–4682. [Google Scholar] [CrossRef]
- Zaman, G.J.R.; De Roos, J.A.D.M.; Libouban, M.A.A.; Prinsen, M.B.W.; De Man, J.; Buijsman, R.C.; Uitdehaag, J.C.M. TTK Inhibitors as a Targeted Therapy for CTNNB1 (β-Catenin) Mutant Cancers. Mol. Cancer Ther. 2017, 16, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- King, J.L.; Zhang, B.; Li, Y.; Li, K.P.; Ni, J.J.; Saavedra, H.I.; Dong, J.T. TTK Promotes Mesenchymal Signaling via Multiple Mechanisms in Triple Negative Breast Cancer. Oncogenesis 2018, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wu, P.; Hu, H.; Tian, D.; Jiang, N.; Wu, C. Protein Kinase TTK Promotes Proliferation and Migration and Mediates Epithelial-Mesenchymal Transition in Human Bladder Cancer Cells. Int. J. Clin. Exp. Pathol. 2018, 11, 4854–4861. [Google Scholar] [PubMed]
- Chen, S.; Wang, J.; Wang, L.; Peng, H.; Xiao, L.; Li, C.; Lin, D.; Yang, K. Silencing TTK Expression Inhibits the Proliferation and Progression of Prostate Cancer. Exp. Cell Res. 2019, 385, 111669. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Jang, S.J.; Kim, J.; Sohn, I.; Lee, J.Y.; Cho, E.J.; Chun, S.M.; Sung, C.O. Spontaneous Mutations in the Single TTN Gene Represent High Tumor Mutation Burden. NPJ Genom. Med. 2020, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in Uveal Melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef]
- Vader, M.J.C.; Madigan, M.C.; Versluis, M.; Suleiman, H.M.; Gezgin, G.; Gruis, N.A.; Out-Luiting, J.J.; Bergman, W.; Verdijk, R.M.; Jager, M.J.; et al. GNAQ and GNA11 Mutations and Downstream YAP Activation in Choroidal Nevi. Br. J. Cancer 2017, 117, 884–887. [Google Scholar] [CrossRef]
- Lloyd, J.T.; McLaughlin, K.; Lubula, M.Y.; Gay, J.C.; Dest, A.; Gao, C.; Phillips, M.; Tonelli, M.; Cornilescu, G.; Marunde, M.R.; et al. Structural Insights into the Recognition of Mono-and Diacetylated Histones by the ATAD2B Bromodomain. J. Med. Chem. 2020, 63, 12799–12813. [Google Scholar] [CrossRef]
- Menyhart, O.; Weltz, B.; Gyorffy, B. Multipletesting.Com: A Tool for Life Science Researchers for Multiple Hypothesis Testing Correction. PLoS ONE 2021, 16, e0245824. [Google Scholar] [CrossRef]
- Ignatiadis, N.; Klaus, B.; Zaugg, J.B.; Huber, W. Data-Driven Hypothesis Weighting Increases Detection Power in Genome-Scale Multiple Testing. Nat. Methods 2016, 13, 577–580. [Google Scholar] [CrossRef] [PubMed]


| Cancer Type | Potential Hotspots | X vs. All | X vs. Gene | X vs. HS | X vs. Y |
|---|---|---|---|---|---|
| ACC | 1 | - | - | - | - |
| BLCA | 50 | 2 | - | - | 4 |
| BRCA | 56 | 3 | 3 | 1 | 14 |
| CESC | 10 | 1 | - | - | - |
| CHOL | 1 | - | - | - | - |
| COAD | 108 | 10 | 6 | 3 | 4 |
| DLBC | 2 | - | - | - | - |
| ESCA | 15 | - | - | - | 2 |
| GBM | 23 | 3 | 1 | 1 | 2 |
| HNSC | 50 | 3 | 2 | 4 | 27 |
| KICH | 0 | - | - | - | - |
| KIRC | 7 | 1 | 1 | - | - |
| KIRP | 3 | - | - | - | - |
| LAML | 8 | 1 | - | - | - |
| LGG | 35 | 6 | 3 | 2 | 7 |
| LIHC | 15 | 1 | - | - | - |
| LUAD | 29 | 2 | 2 | 3 | 2 |
| LUSC | 49 | 3 | 4 | 4 | 45 |
| MESO | 0 | - | - | - | - |
| OV | 38 | 3 | 3 | 3 | 28 |
| PAAD | 13 | 2 | - | - | - |
| PCPG | 3 | - | - | - | - |
| PRAD | 8 | - | - | - | - |
| READ | 15 | 2 | 1 | 1 | 3 |
| SARC | 3 | - | - | - | - |
| SKCM | 178 | 18 | 6 | 2 | 1 |
| STAD | 95 | 2 | 2 | - | - |
| TGCT | 4 | - | - | - | - |
| THCA | 5 | - | - | - | - |
| THYM | 4 | - | - | - | - |
| UCEC | 631 | 31 | 10 | 5 | 7 |
| UCS | 7 | - | - | - | - |
| UVM | 3 | 1 | - | - | - |
| Sum | 1469 | 95 | 44 | 29 | 146 |
| Comparisons | 1469 | 1226 | 594 | 3162 |
| Test | Cancer | Gene | Hotspot Position | n (with/without) | p | HR |
|---|---|---|---|---|---|---|
| X vs. All | LGG | IDH1 | 132 | 384/114 | 0 | 0.2 |
| SKCM * | BRAF | 600 | 141/182 | 0.01 | 0.7 | |
| PAAD | KRAS | 12 | 128/48 | 0.04 | 1.6 | |
| SKCM | BRAF | 600 | 47/373 | 0.05 | 2.9 | |
| UVM | GNAQ | 209 | 37/42 | 0.053 | 0.4 | |
| BLCA | FGFR3 | 249 | 29/357 | 0.04 | 0.5 | |
| UCEC | PPP2R1A | 179 | 26/448 | 0.04 | 2.4 | |
| BRCA | AKT1 | 17 | 24/918 | 0.02 | <1 | |
| LUAD | EGFR | 858 | 21/457 | 0.03 | 2 | |
| LGG | IDH2 | 172 | 20/478 | 0.04 | 0.2 | |
| GBM | IDH1 | 132 | 19/340 | 0 | 0.3 | |
| UCEC | SLC3A2 | 300 | 17/457 | 0.03 | <1 | |
| COAD | PIK3CA | 1047 | 16/326 | 0.05 | 0.2 | |
| UCEC | KRAS | 13 | 14/460 | 0.04 | <1 | |
| UVM | SF3B1 | 625 | 13/66 | 0 | <1 | |
| UCEC | OR14K1 | 14 | 13/461 | 0.01 | <1 | |
| LGG | CIC | 215 | 12/486 | 0 | <1 | |
| GBM | TP53 | 248 | 12/347 | 0.01 | 0.5 | |
| COAD | SETD1B | 8 | 12/330 | 0.04 | <1 | |
| BLCA | KRAS | 12 | 10/376 | 0 | 3.8 | |
| LIHC | TP53 | 249 | 10/339 | 0.054 | 2.6 | |
| HNSC | TP53 | 193 | 9/466 | 0.02 | 3.1 | |
| BRCA | TP53 | 196 | 8/934 | 0.01 | <1 | |
| READ | APC | 876 | 8/110 | 0.01 | <1 | |
| BRCA | RUNX1 | 96 | 7/935 | 0.04 | <1 | |
| UCEC | PTCH1 | 1203 | 7/467 | 0.03 | <1 | |
| UCEC | ZFP37 | 161 | 7/467 | 0.03 | <1 | |
| KIRC | VHL | 158 | 7/323 | 0.02 | 2.9 | |
| SKCM * | PPP6C | 264 | 7/316 | 0.04 | 0.4 | |
| PAAD | KRAS | 61 | 7/166 | 0.04 | 2.8 | |
| X vs. Gene | LUAD | EGFR | 746 | 15/43 | 0.01 | 0.2 |
| UCEC | UPF3A | 267 | 12/7 | 0 | >1 | |
| COAD | BMPR2 | 583 | 9/8 | 0.04 | >1 | |
| UCEC | ATF7IP | 320 | 7/14 | 0.01 | >1 | |
| COAD | DOCK3 | 1852 | 7/12 | 0.04 | <1 | |
| STAD | PSME4 | 1805 | 6/7 | 0.03 | <1 | |
| LUSC | CDKN2A | 108 | 6/64 | 0.01 | 3.4 | |
| LGG | KAT6B | 1203 | 6/5 | 0.04 | >1 | |
| LUSC | TP53 | 126 | 6/361 | 0.05 | 4.5 | |
| HNSC | TP53 | 306 | 6/310 | 0.02 | 4.5 | |
| UCEC | ZMYND8 | 635 | 6/18 | 0.04 | <1 | |
| COAD | FBXW7 | 505 | 5/40 | 0.04 | 4.4 | |
| SKCM * | SALL1 | 675 | 5/37 | 0.01 | <1 | |
| LUSC | TP53 | 176 | 5/362 | 0.01 | 4.1 | |
| OV | TP53 | 179 | 5/359 | 0.02 | 0.2 | |
| OV | TP53 | 244 | 5/359 | 0.03 | 0.2 | |
| OV | TP53 | 266 | 5/359 | 0.02 | 3.2 | |
| X vs. Other HS | UCEC | KRAS | 12 | 65/17 | 0.04 | >1 |
| UCEC | CTNNB1 | 37 | 19/69 | 0.04 | 3.8 | |
| LUSC | NFE2L2 | 29 | 14/35 | 0.05 | 2.8 | |
| LGG | TP53 | 248 | 14/128 | 0.04 | 0.3 | |
| UCEC | PIK3CA | 38 | 10/172 | 0.03 | 3.9 | |
| LUAD | TP53 | 125 | 10/144 | 0.01 | 3.9 | |
| UCEC | ARID1A | 1989 | 5/72 | 0.04 | <1 | |
| HNSC | NFE2L2 | 79 | 5/4 | 0.05 | 2.6 | |
| X vs. Y | UCEC | ARID1A | 1850 vs. 1989 | 17/5 | 0.03 | >1 |
| LUAD | EGFR | 746 vs. 858 | 15/21 | < 0.01 | 0.2 | |
| UCEC | FBXW7 | 505 vs. 545 | 12/4 | 0.04 | <1 | |
| UCEC | PIK3CA | 38 vs. 545 | 10/26 | 0.04 | 5.5 | |
| UCEC | PIK3CA | 38 vs. 542 | 10/22 | 0.03 | 9.1 | |
| UCEC | PIK3CA | 118 vs. 93 | 9/8 | 0.03 | >1 | |
| READ | APC | 213 vs. 876 | 6/8 | 0.04 | >1 | |
| READ | APC | 1114 vs. 213 | 5/6 | 0.02 | <1 | |
| READ | APC | 1114 vs. 1450 | 5/4 | 0.05 | <1 | |
| UCEC | PIK3CA | 111 vs. 118 | 4/9 | 0.04 | <1 | |
| HNSC | NFE2L2 | 29 vs. 79 | 4/5 | 0.05 | <1 | |
| LGG | CIC | 201 vs. 215 | 4/12 | 0.03 | >1 | |
| LGG | CIC | 202 vs. 215 | 4/12 | 0.04 | >1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Cárdenas, M.; Treviño, V. The Impact of Mutational Hotspots on Cancer Survival. Cancers 2024, 16, 1072. https://doi.org/10.3390/cancers16051072
Gonzalez-Cárdenas M, Treviño V. The Impact of Mutational Hotspots on Cancer Survival. Cancers. 2024; 16(5):1072. https://doi.org/10.3390/cancers16051072
Chicago/Turabian StyleGonzalez-Cárdenas, Melissa, and Víctor Treviño. 2024. "The Impact of Mutational Hotspots on Cancer Survival" Cancers 16, no. 5: 1072. https://doi.org/10.3390/cancers16051072







