Pathological Insights: Enhanced Vision Transformers for the Early Detection of Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
- Inclusion of other pathological findings in addition to polyp for early CRC diagnosis;
- ViTCol: Boosted ViT endoscopic pathological findings classifier for early CRC detection;
- PUTS: A novel Swin-Unet transformer-based polyp segmentation model;
- Comparison of ViTCol and PUTS with state-of-the-art CNN and ViT methods.
2. Related Works
3. Materials and Methods
3.1. ViTCol: Vision Transformer-Based Classifier for CRC Detection
3.1.1. Motivation
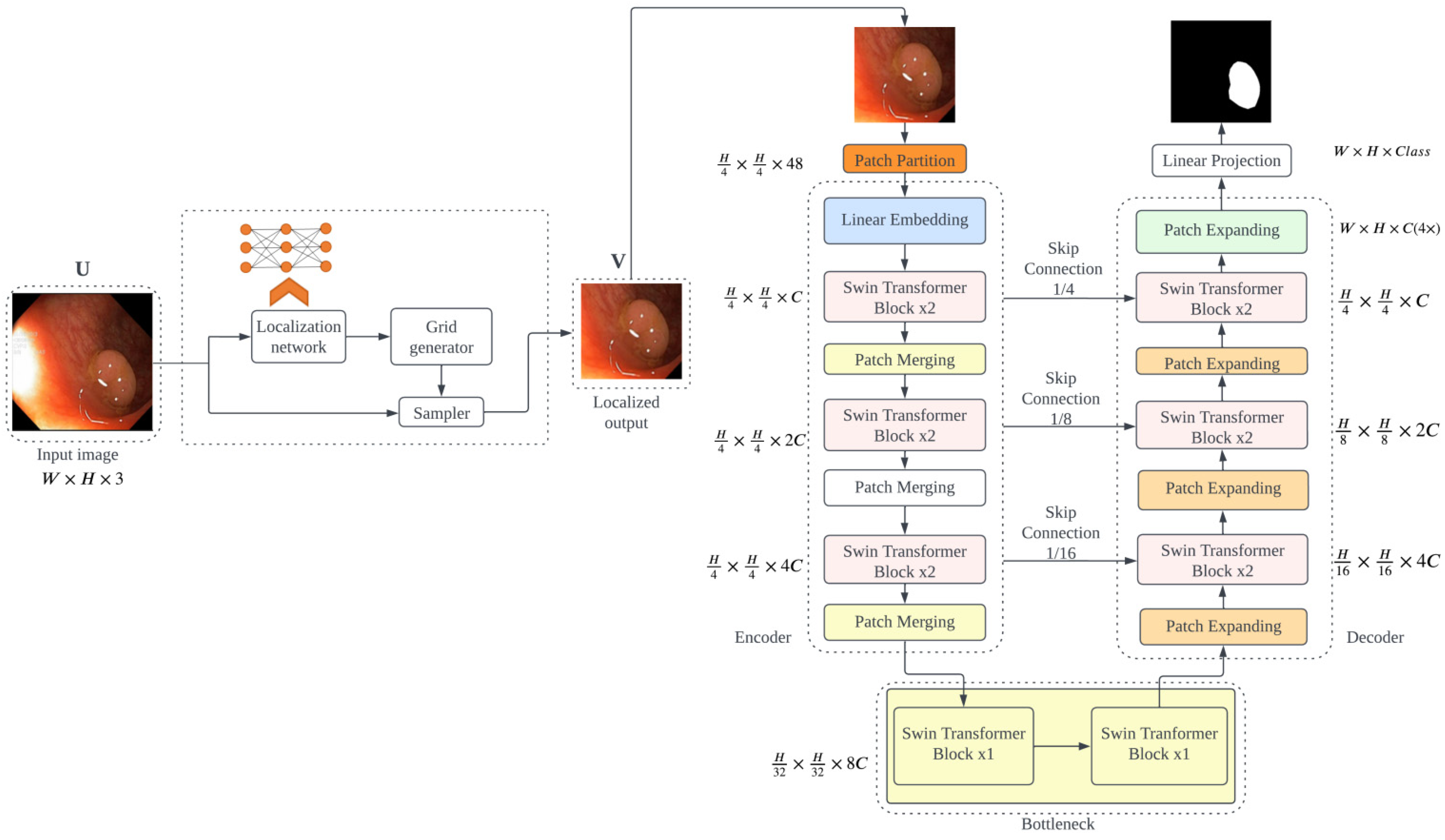
3.1.2. ViTCol Architecture
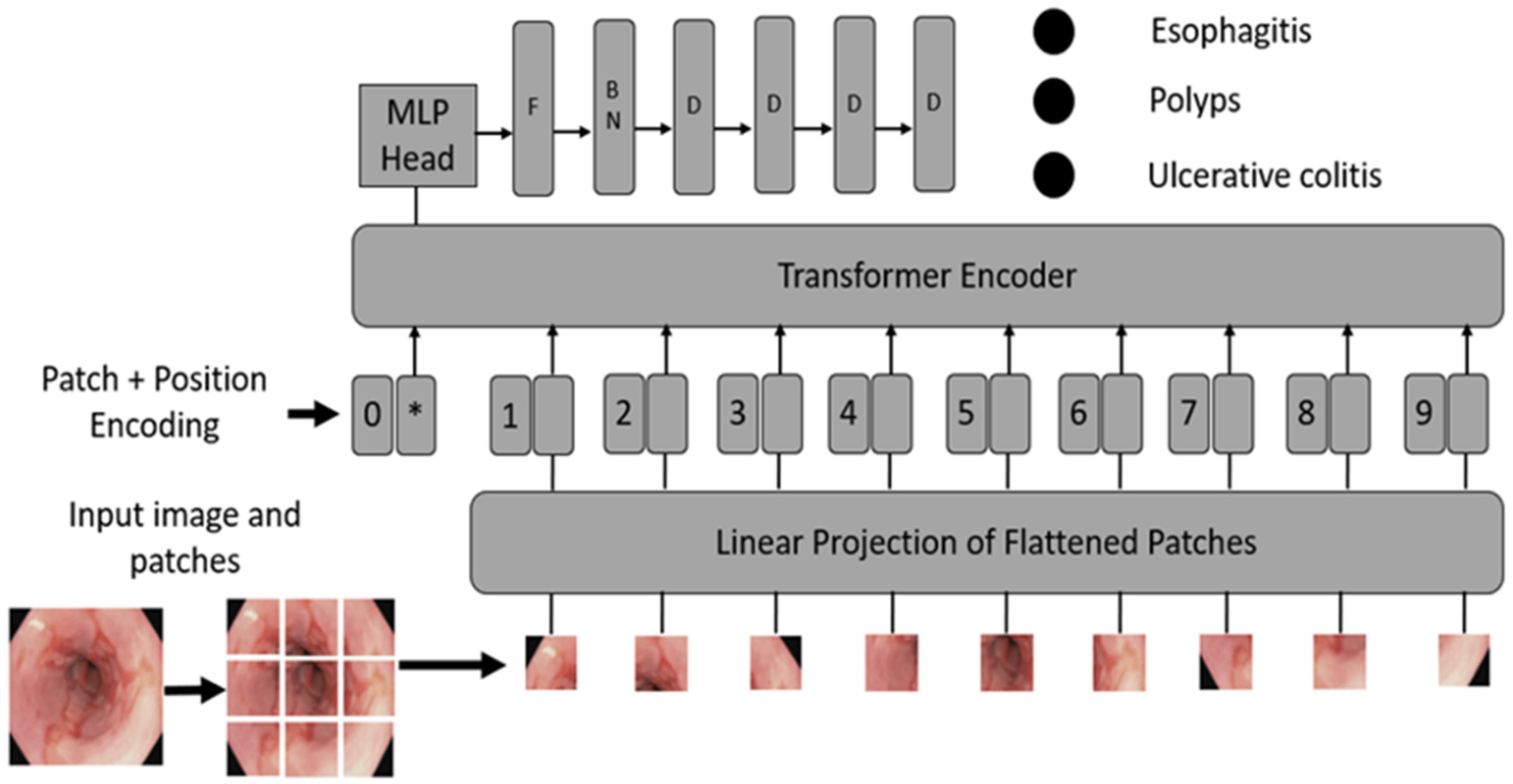
- Convert images into patches: Image patches are used as tokens of words by vision transformers, as in the original paper by Vaswani et al. [52]. For images, it is possible to take pixels; however, if pixels are considered, the computational cost will be high, and it is challenging to find hardware to process high-resolution images, as in the case of medical images. Thus, converting input images into patches has been proposed by Dosovitskiy et al. [36], as shown in Figure 2A. An image with is converted into patches of size .
- Learnable and positional embeddings: Patch projection is concatenated together with learnable embeddings, which are used later for classification. Transformers use positional embeddings to construct a certain order in the patches since, unlike time sequence models, patches do not spontaneously form into sequences. Similar to patch embedding, the positional encoding matrix is produced at random.
- Multilayer perceptron (MLP) head: The MLP head receives the transformer encoder unit’s outputs for classification. Despite the multiple outputs of the transformer encoder, the MLP takes only one output related to the class embedding, while the other outputs are ignored. The probability distribution of the labels that the associated images belong to is output by MLP. Figure 3 shows the ViT architecture used in ViTCol.
3.1.3. Model Definition
3.1.4. Implementation
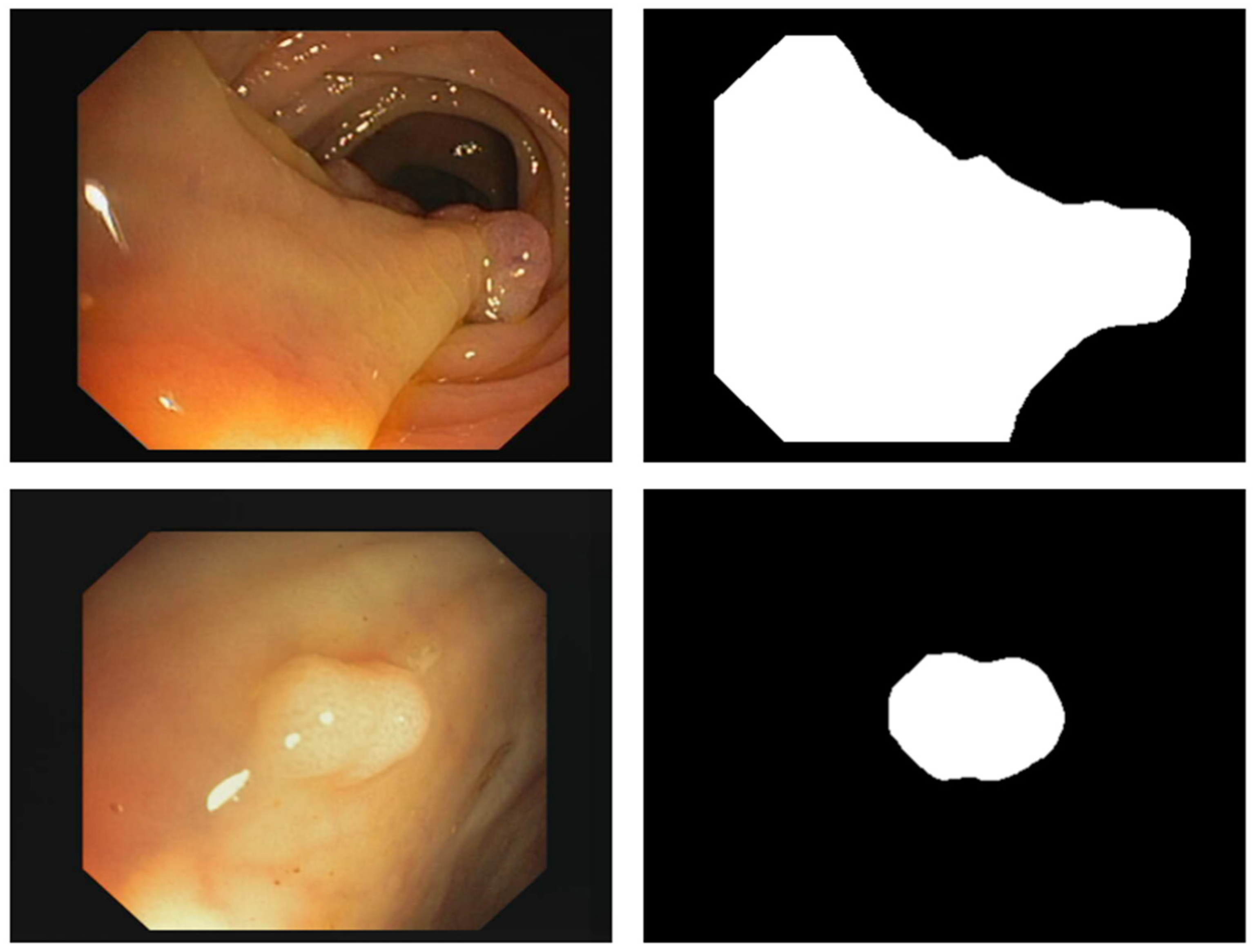
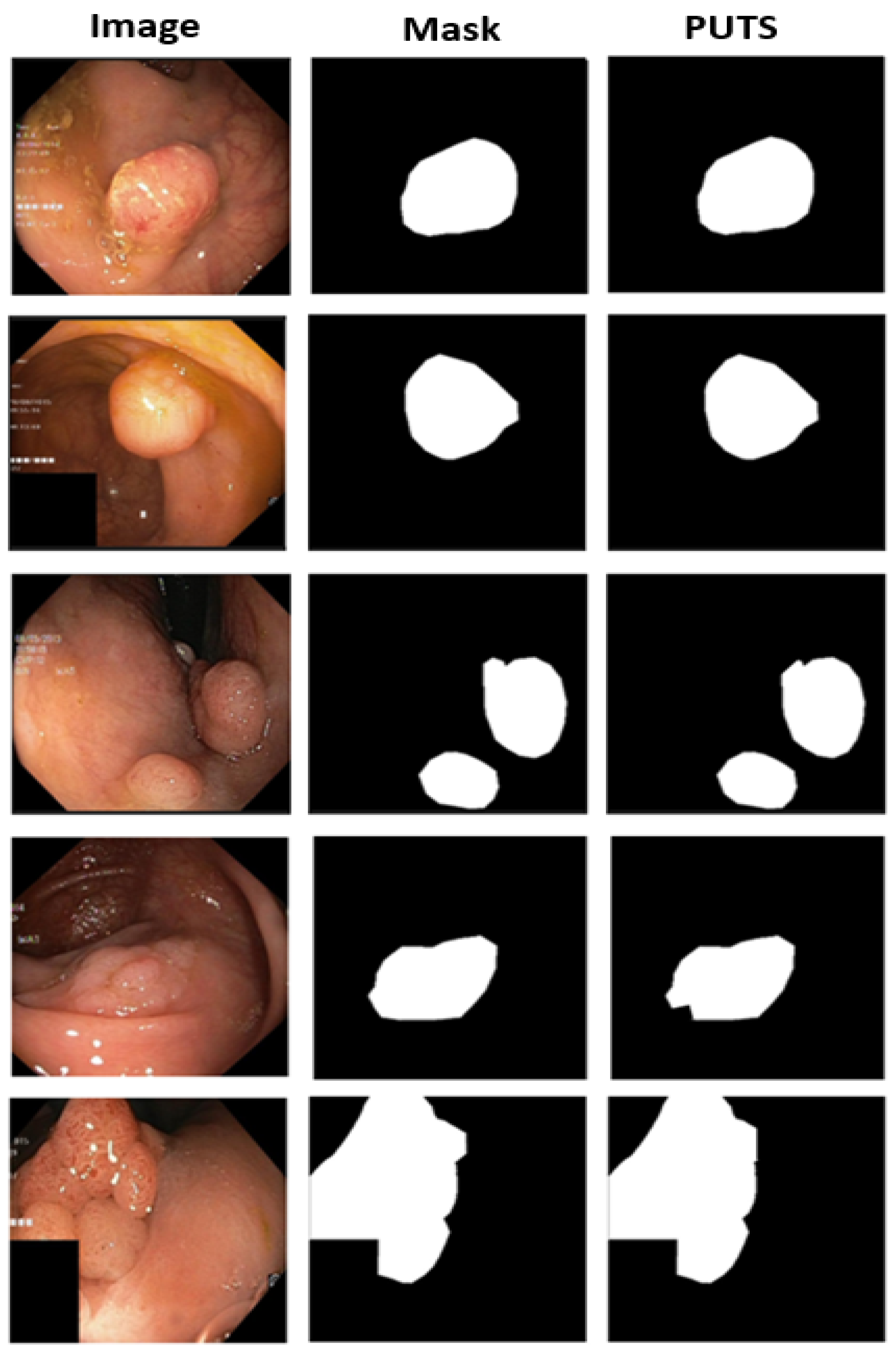
3.2. PUTS: Vision Transformer-Based Polyp Segmentation
3.2.1. Motivation
3.2.2. PUTS Architecture
3.2.3. Model Definition
3.2.4. Implementation
3.3. Parameters Selection
4. Results
4.1. Pathological Findings Classification Performance Results
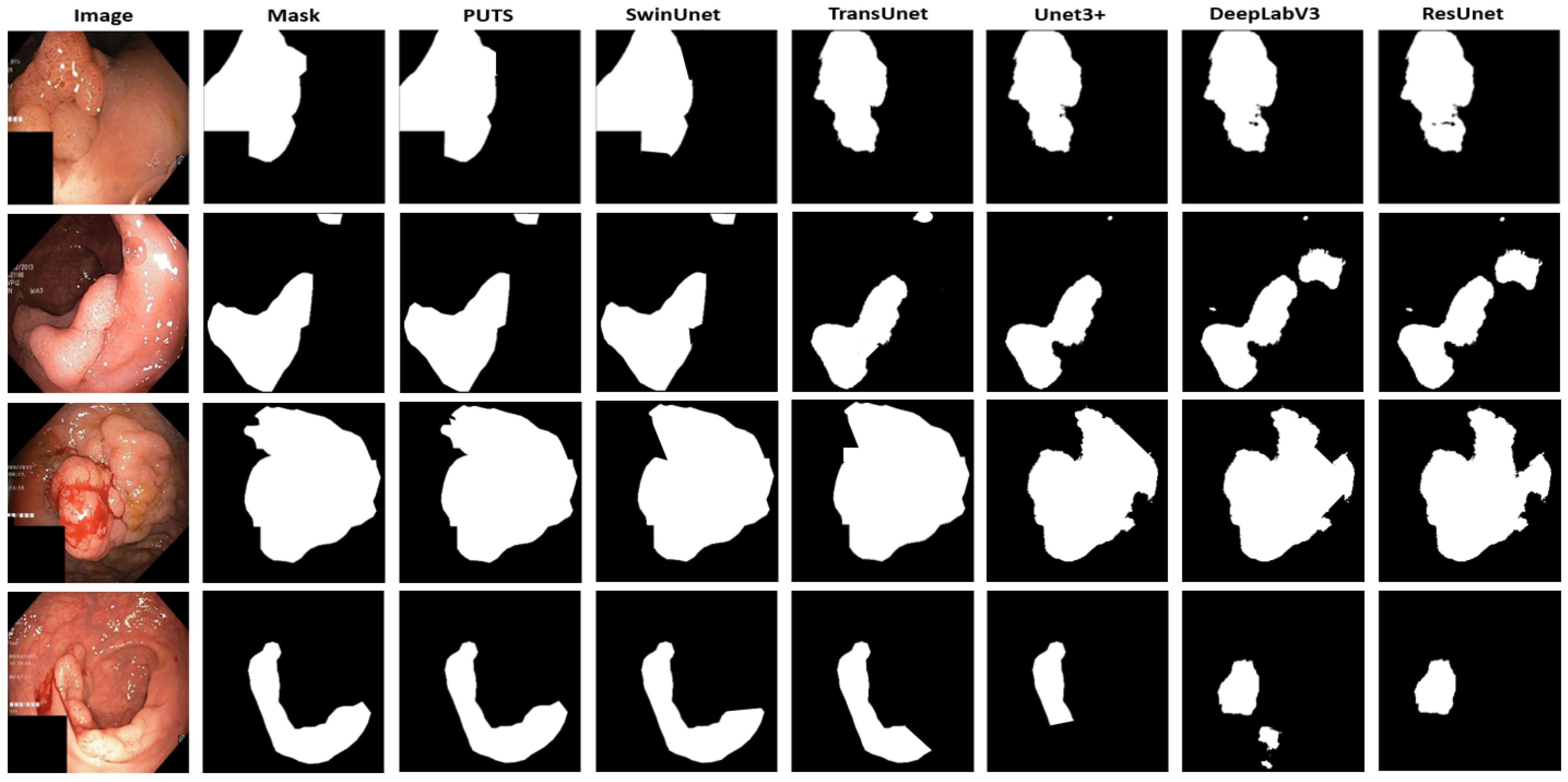
4.2. Polyp Segmentation Performance Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of Colorectal Cancer: Incidence, Mortality, Survival, and Risk Factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Khil, H.; Kim, S.M.; Hong, S.; Gil, H.M.; Cheon, E.; Lee, D.H.; Kim, Y.A.; Keum, N. Time Trends of Colorectal Cancer Incidence and Associated Lifestyle Factors in South Korea. Sci. Rep. 2021, 11, 2413. [Google Scholar] [CrossRef] [PubMed]
- Ayana, G.; Ryu, J.; Choe, S. Ultrasound-Responsive Nanocarriers for Breast Cancer Chemotherapy. Micromachines 2022, 13, 1508. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Translational Oncology Global Colorectal Cancer Burden in 2020 and Projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Helsingen, L.M.; Kalager, M. Colorectal Cancer Screening—Approach, Evidence, and Future Directions. NEJM Evid. 2022, 1, EVIDra2100035. [Google Scholar] [CrossRef]
- Shahmoradi, M.K.; Soleimaninejad, M.; Sharifian, M. Evaluation of Colonoscopy Data for Colorectal Polyps and Associated Histopathological Findings. Ann. Med. Surg. 2020, 57, 7–10. [Google Scholar] [CrossRef]
- Hao, Y.; Wang, Y.; Qi, M.; He, X.; Zhu, Y.; Hong, J. Risk Factors for Recurrent Colorectal Polyps. Gut Liver 2020, 14, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.U.; Kim, H.O.; Kim, H. Epidemiology, Risk Factors, and Prevention of Colorectal Cancer-An English Version. J. Anus Rectum Colon 2022, 6, 2022–2050. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, Y.; Fang, J. Comprehensive Review of Targeted Therapy for Colorectal Cancer. Signal Transduct. Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef]
- Baeg, M.K.; Choi, M.G.; Jung, Y.D.; Ko, S.H.; Lim, C.H.; Kim, H.H.; Kim, J.S.; Cho, Y.K.; Park, J.M.; Lee, I.S.; et al. Esophageal Squamous Cell Carcinoma Patients Have an Increased Risk of Coexisting Colorectal Neoplasms. Gut Liver 2016, 10, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, O.; Nishizawa, T.; Yoshida, S.; Matsuno, T.; Odawara, N.; Toyoshima, A.; Sakitani, K.; Watanabe, H.; Fujishiro, M.; Suzuki, H. Consistency between the Endoscopic Kyoto Classification and Pathological Updated Sydney System for Gastritis: A Cross-Sectional Study. J. Gastroenterol. Hepatol. 2022, 37, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Logan, R.F.A.; Skelly, M.M. Barrett’s Oesophagus and Colorectal Neoplasia: Scope for Screening? Gut 1999, 44, 775–776. [Google Scholar] [CrossRef]
- Solaymani-Dodaran, M.; Logan, R.F.A.; West, J.; Card, T.; Coupland, C. Risk of Extra-Oesophageal Malignancies and Colorectal Cancer in Barrett’s Oesophagus and Gastro-Oesophageal Reflux. Scand. J. Gastroenterol. 2004, 39, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Dese, K.; Ayana, G.; Lamesgin Simegn, G. Low Cost, Non-Invasive, and Continuous Vital Signs Monitoring Device for Pregnant Women in Low Resource Settings (Lvital Device). HardwareX 2022, 11, e00276. [Google Scholar] [CrossRef]
- del Amor, R.; Meseguer, P.; Parigi, T.L.; Villanacci, V.; Colomer, A.; Launet, L.; Bazarova, A.; Tontini, G.E.; Bisschops, R.; de Hertogh, G.; et al. Constrained Multiple Instance Learning for Ulcerative Colitis Prediction Using Histological Images. Comput. Methods Programs Biomed. 2022, 224, 107012. [Google Scholar] [CrossRef]
- Sutton, R.T.; Zaiane, O.R.; Goebel, R.; Baumgart, D.C. Artificial Intelligence Enabled Automated Diagnosis and Grading of Ulcerative Colitis Endoscopy Images. Sci. Rep. 2022, 12, 2748. [Google Scholar] [CrossRef]
- Marabotto, E.; Kayali, S.; Buccilli, S.; Levo, F.; Bodini, G.; Giannini, E.G.; Savarino, V.; Savarino, E.V. Colorectal Cancer in Inflammatory Bowel Diseases: Epidemiology and Prevention: A Review. Cancers 2022, 14, 4254. [Google Scholar] [CrossRef]
- Lucafò, M.; Curci, D.; Franzin, M.; Decorti, G. In Fl Ammatory Bowel Disease and Risk of Colorectal Cancer: An Overview From Pathophysiology to Pharmacological Prevention. Front. Pharmacol. 2021, 12, 772101. [Google Scholar] [CrossRef]
- Stoffel, E.M.; Murphy, C.C. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology 2020, 158, 341–353. [Google Scholar] [CrossRef]
- Kővári, B.; Pai, R.K. Upper Gastrointestinal Tract Involvement in Inflammatory Bowel Diseases: Histologic Clues and Pitfalls. Adv. Anat. Pathol. 2022, 29, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; De Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.T.; Lux, M.; Schmidt, P.T.; et al. Kvasir: A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection. In Proceedings of the 8th ACM on Multimedia Systems Conference, Taipei, Taiwan, 20–23 June 2017; pp. 164–169. [Google Scholar] [CrossRef]
- Jha, D.; Ali, S.; Hicks, S.; Thambawita, V.; Borgli, H.; Smedsrud, P.H.; de Lange, T.; Pogorelov, K.; Wang, X.; Harzig, P.; et al. A Comprehensive Analysis of Classification Methods in Gastrointestinal Endoscopy Imaging. Med. Image Anal. 2021, 70, 102007. [Google Scholar] [CrossRef] [PubMed]
- Dalju, H.B.; Rushdi, M.A.; Morsy, A. Superpixel-Based Segmentation and Classification of Gastrointestinal Landmarks and Diseases. In Proceedings of the 2021 IEEE Biomedical Circuits and Systems Conference (BioCAS), Berlin, Germany, 7–9 October 2021; pp. 1–5. [Google Scholar]
- Zhang, R.; Zheng, Y.; Mak, T.W.C.; Yu, R.; Wong, S.H.; Lau, J.Y.W.; Poon, C.C.Y. Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features from Nonmedical Domain. IEEE J. Biomed. Health Inform. 2017, 21, 41–47. [Google Scholar] [CrossRef]
- Ali, S.; Dmitrieva, M.; Ghatwary, N.; Bano, S.; Polat, G.; Temizel, A.; Krenzer, A.; Hekalo, A.; Guo, Y.B.; Matuszewski, B.; et al. Deep Learning for Detection and Segmentation of Artefact and Disease Instances in Gastrointestinal Endoscopy. Med. Image Anal. 2021, 70, 102002. [Google Scholar] [CrossRef]
- Younas, F.; Usman, M.; Yan, W.Q. A Deep Ensemble Learning Method for Colorectal Polyp Classification with Optimized Network Parameters. Appl. Intell. 2022, 53, 2410–2433. [Google Scholar] [CrossRef]
- Nogueira-Rodríguez, A.; Domínguez-Carbajales, R.; Campos-Tato, F.; Herrero, J.; Puga, M.; Remedios, D.; Rivas, L.; Sánchez, E.; Iglesias, Á.; Cubiella, J.; et al. Real-Time Polyp Detection Model Using Convolutional Neural Networks. Neural Comput. Appl. 2022, 34, 10375–10396. [Google Scholar] [CrossRef]
- Pacal, I.; Karaboga, D. A Robust Real-Time Deep Learning Based Automatic Polyp Detection System. Comput. Biol. Med. 2021, 134, 104519. [Google Scholar] [CrossRef]
- Keshtkar, K. Convolutional Neural Networks in Computer-Aided Diagnosis of Colorectal Polyps and Cancer: A Review. Preprint 2021. [Google Scholar] [CrossRef]
- Id, K.P.; Li, K.; Tao, K.; Wang, Q.; Bansal, A.; Rastogi, A.; Wang, G. A Comparative Study on Polyp Classification Using Convolutional Neural Networks. PLoS ONE 2020, 15, e0236452. [Google Scholar] [CrossRef]
- Ayana, G.; Park, J.; Choe, S. Patchless Multi-Stage Transfer Learning for Improved Mammographic Breast Mass Classification. Cancers 2022, 14, 1280. [Google Scholar] [CrossRef]
- Dese, K.; Raj, H.; Ayana, G.; Yemane, T.; Adissu, W.; Krishnamoorthy, J.; Kwa, T. Accurate Machine-Learning-Based Classification of Leukemia from Blood Smear Images. Clin. Lymphoma Myeloma Leuk. 2021, 21, e903–e914. [Google Scholar] [CrossRef]
- Ayana, G.; Dese, K.; Raj, H.; Krishnamoorthy, J.; Kwa, T. De-Speckling Breast Cancer Ultrasound Images Using a Rotationally Invariant Block Matching Based Non-Local Means (RIBM-NLM) Method. Diagnostics 2022, 12, 862. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 9992–10002. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. In European Conference on Computer Vision; Springer Nature: Cham, Switzerland, 2021; pp. 1–14. [Google Scholar]
- Park, K.-B.; Lee, J.Y. SwinE-Net: Hybrid Deep Learning Approach to Novel Polyp Segmentation Using Convolutional Neural Network and Swin Transformer. J. Comput. Des. Eng. 2022, 9, 616–632. [Google Scholar] [CrossRef]
- Ayana, G.; Choe, S. BUViTNet: Breast Ultrasound Detection via Vision Transformers. Diagnostics 2022, 12, 2654. [Google Scholar] [CrossRef] [PubMed]
- Ayana, G.; Dese, K.; Dereje, Y.; Kebede, Y.; Barki, H.; Amdissa, D.; Husen, N.; Mulugeta, F.; Habtamu, B.; Choe, S.-W. Vision-Transformer-Based Transfer Learning for Mammogram Classification. Diagnostics 2023, 13, 178. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhuo, L.; Zhang, H.; Yang, Y.; Li, X.; Zhang, J.; Wei, W. Polyp Segmentation Network with Hybrid Channel-Spatial Attention and Pyramid Global Context Guided Feature Fusion. Comput. Med. Imaging Graph. 2022, 98, 102072. [Google Scholar] [CrossRef]
- Hu, J.; Xu, Y.; Tang, Z. DAN-PD: Domain Adaptive Network with Parallel Decoder for Polyp Segmentation. Comput. Med. Imaging Graph. 2022, 101, 102124. [Google Scholar] [CrossRef]
- Yang, H.; Chen, Q.; Fu, K.; Zhu, L.; Jin, L.; Qiu, B.; Ren, Q.; Du, H.; Lu, Y. Boosting Medical Image Segmentation via Conditional-Synergistic Convolution and Lesion Decoupling. Comput. Med. Imaging Graph. 2022, 101, 102110. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhao, Y.; Li, F.; Wang, L. MIA-Net: Multi-Information Aggregation Network Combining Transformers and Convolutional Feature Learning for Polyp Segmentation. Knowl.-Based Syst. 2022, 247, 108824. [Google Scholar] [CrossRef]
- Pan, S.; Liu, X.; Xie, N.; Chong, Y. EG-TransUNet: A Transformer-Based U-Net with Enhanced and Guided Models for Biomedical Image Segmentation. BMC Bioinform. 2023, 24, 85. [Google Scholar] [CrossRef] [PubMed]
- Duc, N.T.; Oanh, N.T.; Thuy, N.T.; Triet, T.M.; Dinh, V.S. ColonFormer: An Efficient Transformer Based Method for Colon Polyp Segmentation. IEEE Access 2022, 10, 80575–80586. [Google Scholar] [CrossRef]
- Dong, B.; Wang, W.; Fan, D.-P.; Li, J.; Fu, H.; Shao, L. Polyp-PVT: Polyp Segmentation with Pyramid Vision Transformers. arXiv 2021, arXiv:2108.06932. [Google Scholar] [CrossRef]
- Patel, K.; Li, F.; Wang, G. FuzzyNet: A Fuzzy Attention Module for Polyp Segmentation. In Proceedings of the 36th Conference on Neural Information Processing Systems, NeurIPS 2022, New Orleans, LA, USA, 28 November–9 December 2022; pp. 1–11. [Google Scholar]
- Sharma, A.; Kumar, R.; Garg, P. Deep Learning-Based Prediction Model for Diagnosing Gastrointestinal Diseases Using Endoscopy Images. Int. J. Med. Inform. 2023, 177, 105142. [Google Scholar] [CrossRef]
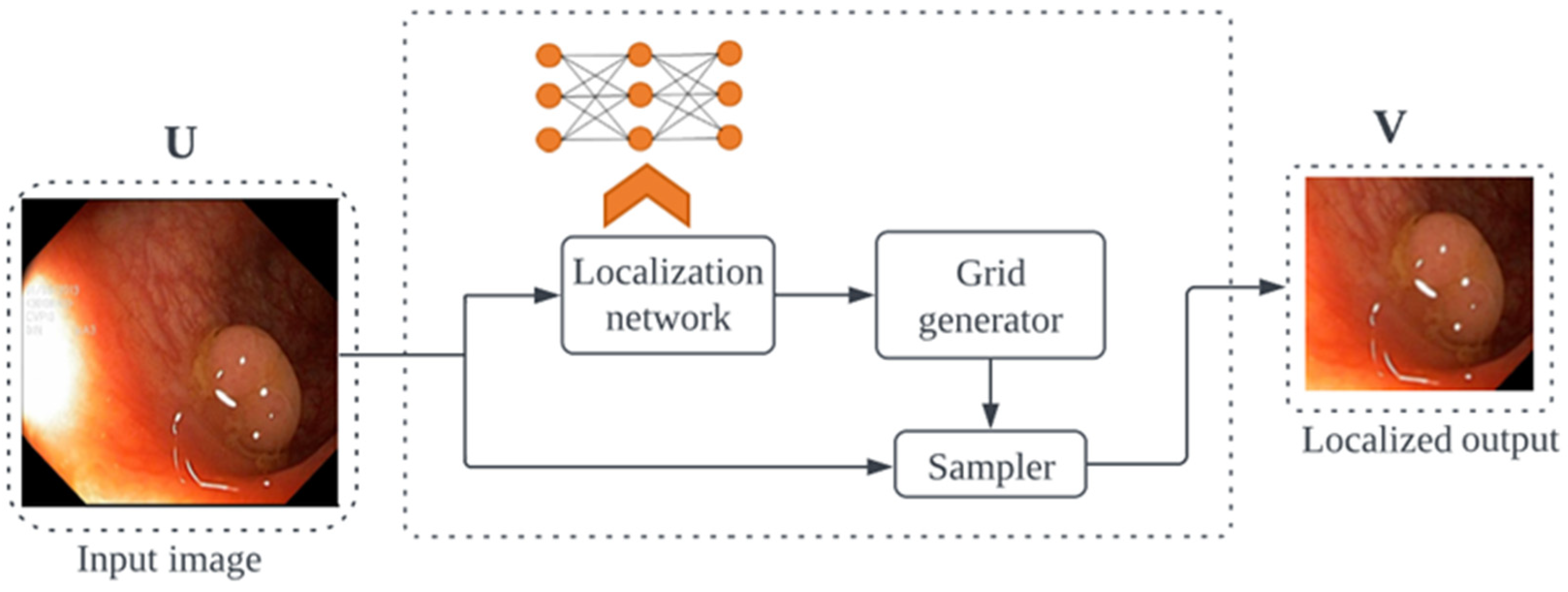
- Jaderberg, M.; Simonyan, K.; Zisserman, A.; Kavukcuoglu, K. Spatial Transformer Networks. In Proceedings of the ACM International Conference Proceeding Series; MIT Press; Cambridge, MA, USA, 2016; Volume 2, pp. 45–48. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez Aidan, N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS ’17), Long Beach, CA, USA, 4–9 December 2017; pp. 1–11. [Google Scholar]
- Alboaneen, D.; Alqarni, R.; Alqahtani, S.; Alrashidi, M.; Alhuda, R.; Alyahyan, E.; Alshammari, T. Predicting Colorectal Cancer Using Machine and Deep Learning Algorithms: Challenges and Opportunities. Big Data Cogn. Comput. 2023, 7, 74. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Jothilakshmi, S.; Suthir, S. Colorectal Cancer Detection Based on Convolutional Neural Networks (CNN) and Ranking Algorithm. Meas. Sens. 2024, 31, 100976. [Google Scholar] [CrossRef]
- Li, Y.; Shen, F.; Hu, L.; Lang, Z.; Liu, Q.; Cai, F.; Fu, L. A Stare-Down Video-Rate High-Throughput Hyperspectral Imaging System and Its Applications in Biological Sample Sensing. IEEE Sens. J. 2023, 23, 23629–23637. [Google Scholar] [CrossRef]
- Roy, S.; Menapace, W.; Oei, S.; Luijten, B.; Fini, E.; Saltori, C.; Huijben, I.; Chennakeshava, N.; Mento, F.; Sentelli, A.; et al. Deep Learning for Classification and Localization of COVID-19 Markers in Point-of-Care Lung Ultrasound. IEEE Trans. Med. Imaging 2020, 39, 2676–2687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, G.; Li, Z.; Cui, S.; Qian, D.; Yu, Y. Adaptive Context Selection for Polyp Segmentation BT—Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 253–262. [Google Scholar]
- Nguyen, T.-C.; Nguyen, T.-P.; Diep, G.-H.; Tran-Dinh, A.-H.; Nguyen, T.V.; Tran, M.-T. CCBANet: Cascading Context and Balancing Attention for Polyp Segmentation BT—Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 633–643. [Google Scholar]
- Fan, D.-P.; Ji, G.-P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. PraNet: Parallel Reverse Attention Network for Polyp Segmentation BT—Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 263–273. [Google Scholar]
- Huang, C.; Wu, H.; Lin, Y. HarDNet-MSEG: A Simple Encoder-Decoder Polyp Segmentation Neural Network That Achieves over 0.9 Mean Dice and 86 FPS. arXiv 2021, arXiv:2101.07172. [Google Scholar]
- Wei, J.; Hu, Y.; Zhang, R.; Li, Z.; Zhou, S.K.; Cui, S. Shallow Attention Network for Polyp Segmentation BT—Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 699–708. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Liu, F.; Hua, Z.; Li, J.; Fan, L. MFBGR: Multi-Scale Feature Boundary Graph Reasoning Network for Polyp Segmentation. Eng. Appl. Artif. Intell. 2023, 123, 106213. [Google Scholar] [CrossRef]
- Song, P.; Yang, Z.; Li, J.; Fan, H. DPCTN: Dual Path Context-Aware Transformer Network for Medical Image Segmentation. Eng. Appl. Artif. Intell. 2023, 124, 106634. [Google Scholar] [CrossRef]
- Khan, T.M.; Arsalan, M.; Razzak, I.; Meijering, E. Simple and Robust Depth-Wise Cascaded Network for Polyp Segmentation. Eng. Appl. Artif. Intell. 2023, 121, 106023. [Google Scholar] [CrossRef]


| Model | Accuracy (95%) | AUC (95%) | Sensitivity (95%) | Specificity (95%) | F1 (95%) | MCC (95%) | Kappa (95%) |
|---|---|---|---|---|---|---|---|
| vitb_16 | 0.9998 ± 0.001 | 0.9999 ± 0.001 | 0.9998 ± 0.001 | 0.9998 ± 0.002 | 0.9999 ± 0.001 | 0.9995 ± 0.002 | 0.9997 ± 0.001 |
| vitb_32 | 0.9864 ± 0.002 | 0.9882 ± 0.001 | 0.9860 ± 0.002 | 0.9871 ± 0.003 | 0.9868 ± 0.002 | 0.9847 ± 0.003 | 0.9842 ± 0.002 |
| vitl_32 | 0.9752 ± 0.001 | 0.9774 ± 0.002 | 0.9749 ± 0.001 | 0.9748 ± 0.002 | 0.9768 ± 0.002 | 0.9720 ± 0.002 | 0.9741 ± 0.002 |
| Model | Accuracy (95%) | AUC (95%) | Sensitivity (95%) | Specificity (95%) | F1 (95%) | MCC (95%) | Kappa (95%) |
|---|---|---|---|---|---|---|---|
| vitb_16 | 0.9845 ± 0.001 | 0.987 ± 0.001 | 0.9832 ± 0.002 | 0.9836 ± 0.003 | 0.984 ± 0.001 | 0.9817 ± 0.002 | 0.9826 ± 0.002 |
| vitb_32 | 0.9684 ± 0.001 | 0.9742 ± 0.001 | 0.9644 ± 0.02 | 0.9642 ± 0.003 | 0.966 ± 0.001 | 0.9604 ± 0.003 | 0.9622 ± 0.002 |
| vitl_32 | 0.9605 ± 0.001 | 0.969 ± 0.001 | 0.9618 ± 0.002 | 0.9616 ± 0.002 | 0.9614 ± 0.001 | 0.9598 ± 0.002 | 0.9599 ± 0.001 |
| Model | Accuracy (95%) | AUC (95%) | Sensitivity (95%) | Specificity (95%) | F1 (95%) | MCC (95%) | Kappa (95%) |
|---|---|---|---|---|---|---|---|
| ResNet50 | 0.9740 ± 0.001 | 0.9749 ± 0.001 | 0.9749 ± 0.002 | 0.9748 ± 0.002 | 0.9741 ± 0.001 | 0.9726 ± 0.003 | 0.9724 ± 0.002 |
| EfficientNetB2 | 0.9642 ± 0.005 | 0.9707 ± 0.004 | 0.9627 ± 0.003 | 0.9640 ± 0.004 | 0.9624 ± 0.006 | 0.9628 ± 0.003 | 0.9624 ± 0.003 |
| InceptionV3 | 0.9702 ± 0.001 | 0.9718 ± 0.001 | 0.9694 ± 0.003 | 0.9697 ± 0.002 | 0.9702 ± 0.001 | 0.9659 ± 0.004 | 0.9648 ± 0.004 |
| Model | mIoU | mDic | Sensitivity | Specificity | MCC |
|---|---|---|---|---|---|
| Kvasir-SEG dataset | |||||
| U-net | 0.7919 | 0.7941 | 0.7908 | 0.7916 | 0.7902 |
| ResUnet | 0.7914 | 0.8085 | 0.7819 | 0.8021 | 0.7528 |
| Unet 3+ | 0.7998 | 0.8275 | 0.8142 | 0.8156 | 0.8024 |
| DeepLabV3 | 0.7932 | 0.8154 | 0.8068 | 0.8052 | 0.8014 |
| TransUnet | 0.8058 | 0.8691 | 0.8453 | 0.8512 | 0.8439 |
| SwinUnet | 0.8206 | 0.8892 | 0.8734 | 0.8783 | 0.8642 |
| PUTS | 0.8673 | 0.9186 | 0.9095 | 0.9104 | 0.9048 |
| CVC-ClinicDB dataset | |||||
| U-net | 0.8416 | 0.8892 | 0.8793 | 0.8799 | 0.8724 |
| ResUnet | 0.8239 | 0.8792 | 0.8674 | 0.8683 | 0.8619 |
| Unet 3+ | 0.8427 | 0.8904 | 0.8817 | 0.8844 | 0.8786 |
| DeepLabV3 | 0.8397 | 0.8829 | 0.8732 | 0.8719 | 0.8694 |
| TransUnet | 0.8502 | 0.8946 | 0.8915 | 0.8924 | 0.8852 |
| SwinUnet | 0.8552 | 0.9053 | 0.8937 | 0.8915 | 0.8906 |
| PUTS | 0.9092 | 0.9484 | 0.9392 | 0.9387 | 0.9317 |
| Dataset | Paper | Year | mIoU | mDic |
|---|---|---|---|---|
| Kvasir-SEG | Zhang et al. [57] | 2020 | 0.820 | 0.884 |
| Nguyen et al. [58] | 2021 | 0.834 | 0.894 | |
| Fan et al. [59] | 2020 | 0.840 | 0.898 | |
| Huang et al. [60] | 2021 | 0.844 | 0.903 | |
| Wei et al. [61] | 2021 | 0.847 | 0.904 | |
| Xie et al. [62] | 2021 | 0.864 | 0.915 | |
| Dong et al. [48] | 2021 | 0.864 | 0.917 | |
| Li et al. [45] | 2022 | 0.876 | 0.926 | |
| Liu et al. [63] | 2023 | 0.728 | 0.803 | |
| Song et al. [64] | 2023 | 0.876 | 0.924 | |
| Khan et al. [65] | 2023 | 0.874 | 0.923 | |
| PUTS | 2023 | 0.889 | 0.935 | |
| CVC-ClinicDB | Zhang et al. [57] | 2020 | 0.828 | 0.883 |
| Nguyen et al. [58] | 2021 | 0.849 | 0.904 | |
| Fan et al. [59] | 2020 | 0.849 | 0.899 | |
| Huang et al. [60] | 2021 | 0.865 | 0.916 | |
| Wei et al. [61] | 2021 | 0.859 | 0.916 | |
| Xie et al. [62] | 2021 | 0.882 | 0.931 | |
| Dong et al. [48] | 2021 | 0.889 | 0.937 | |
| Li et al. [45] | 2022 | 0.899 | 0.942 | |
| Liu et al. [63] | 2023 | 0.893 | 0.941 | |
| Song et al. [64] | 2023 | 0.892 | 0.940 | |
| Khan et al. [65] | 2023 | 0.903 | 0.950 | |
| PUTS | 2023 | 0.909 | 0.949 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayana, G.; Barki, H.; Choe, S.-w. Pathological Insights: Enhanced Vision Transformers for the Early Detection of Colorectal Cancer. Cancers 2024, 16, 1441. https://doi.org/10.3390/cancers16071441
Ayana G, Barki H, Choe S-w. Pathological Insights: Enhanced Vision Transformers for the Early Detection of Colorectal Cancer. Cancers. 2024; 16(7):1441. https://doi.org/10.3390/cancers16071441
Chicago/Turabian StyleAyana, Gelan, Hika Barki, and Se-woon Choe. 2024. "Pathological Insights: Enhanced Vision Transformers for the Early Detection of Colorectal Cancer" Cancers 16, no. 7: 1441. https://doi.org/10.3390/cancers16071441








