Possible Role of miR-375-3p in Cervical Lymph Node Metastasis of Oral Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Cells and Cell Culture
2.3. Total RNA Extraction
2.4. miRNA Microarray
2.5. RT-qPCR
2.6. Digital PCR
2.7. Cell Growth Assay
2.8. Cell Migration Assay
2.9. Xenograft Model
2.10. Microarray and Pathway Analysis
2.11. Statistics
3. Results
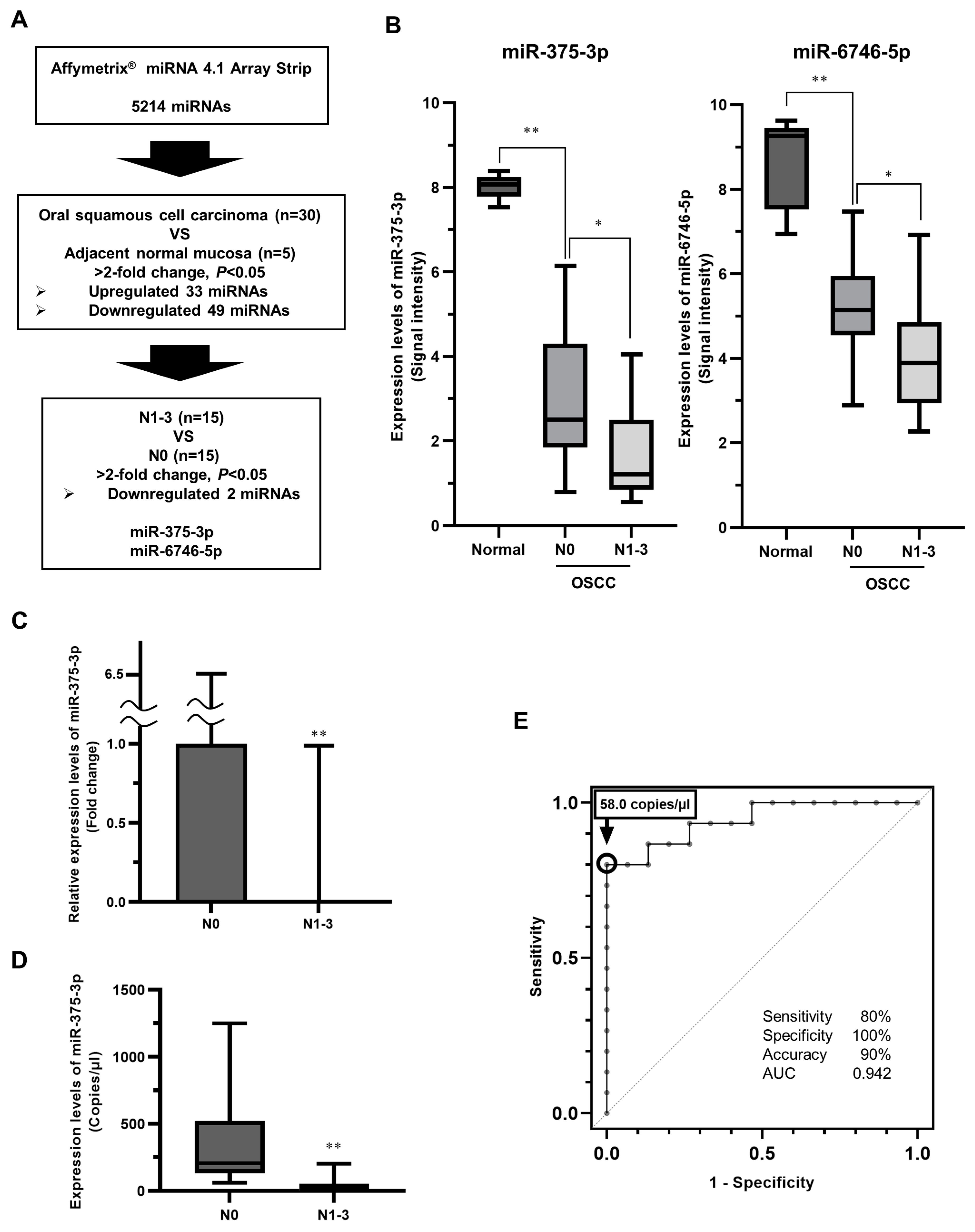
3.1. Identification of miRNAs Related to Latent Cervical LNM in Primary eOSCC Tissues
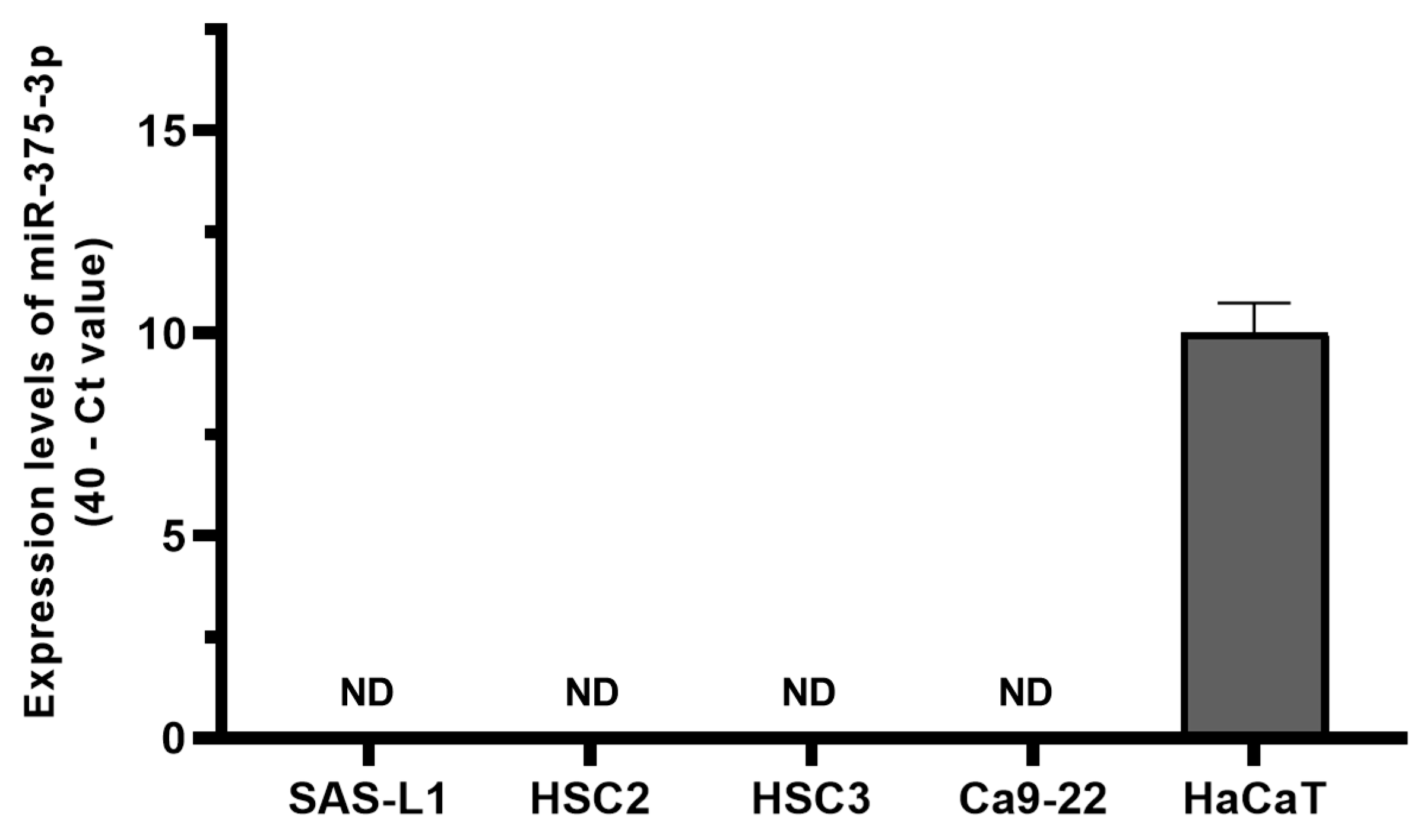
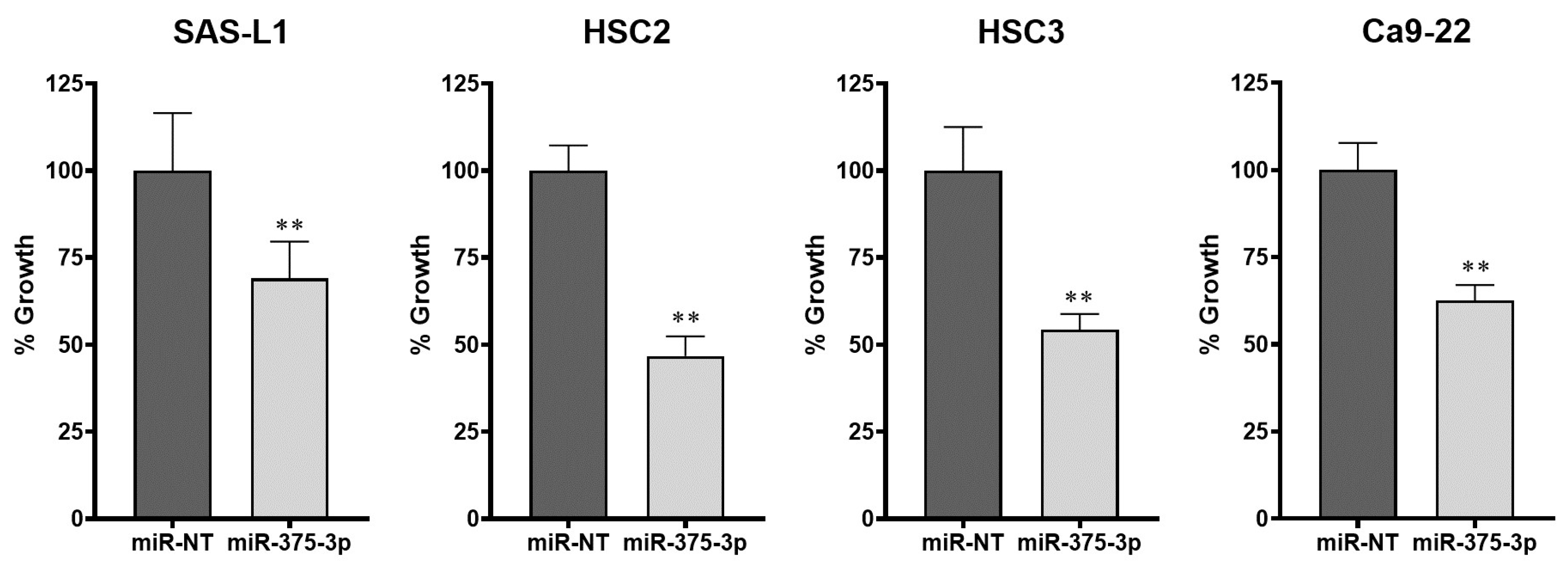
3.2. Expression and Function of miR-375-3p in Human OSCC Cells In Vitro
3.3. Effect of miR-375-3p Mimic on In Vivo Growth
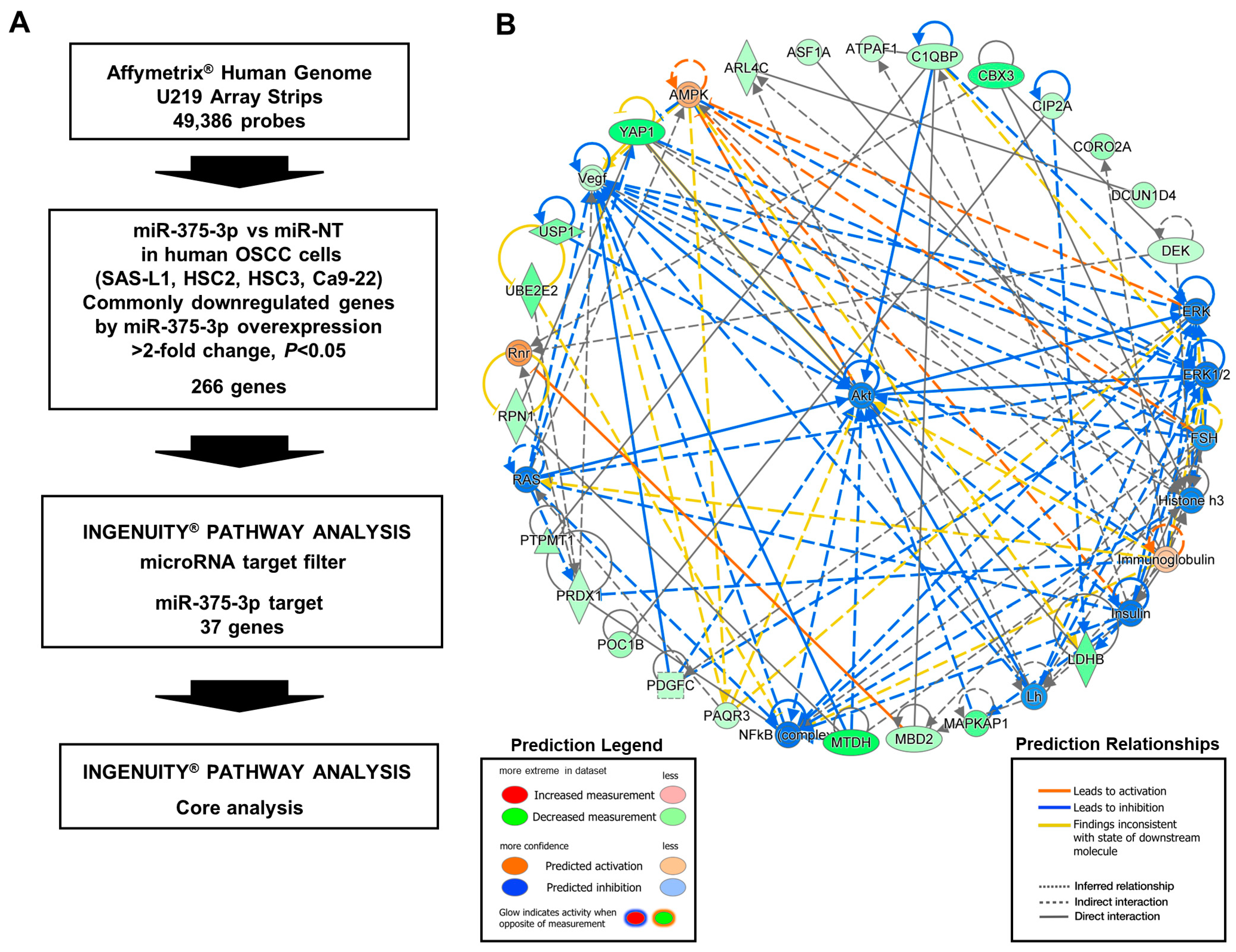
3.4. Target Gene Candidates and Pathways of miR-375-3p in Human OSCC Cells
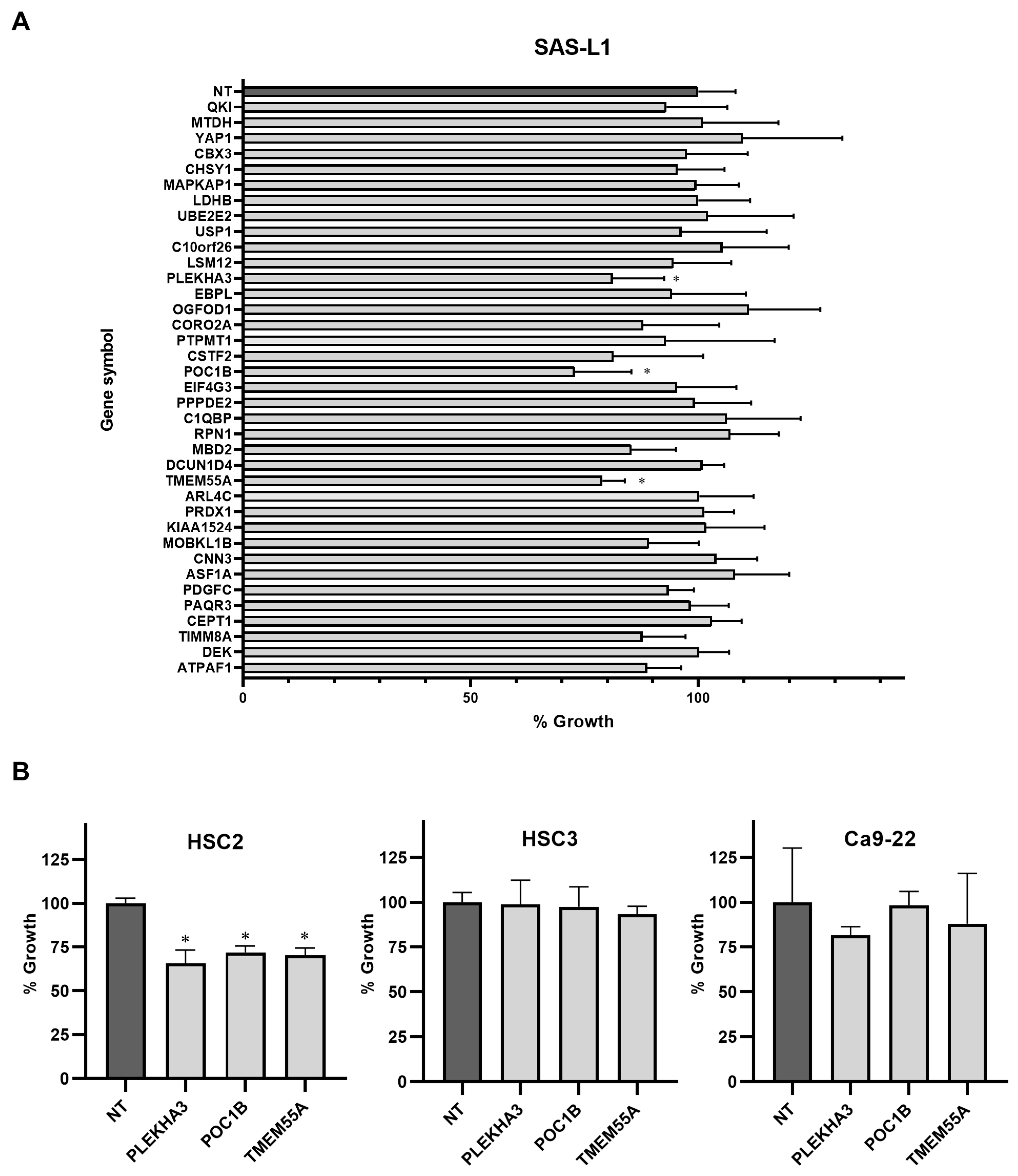
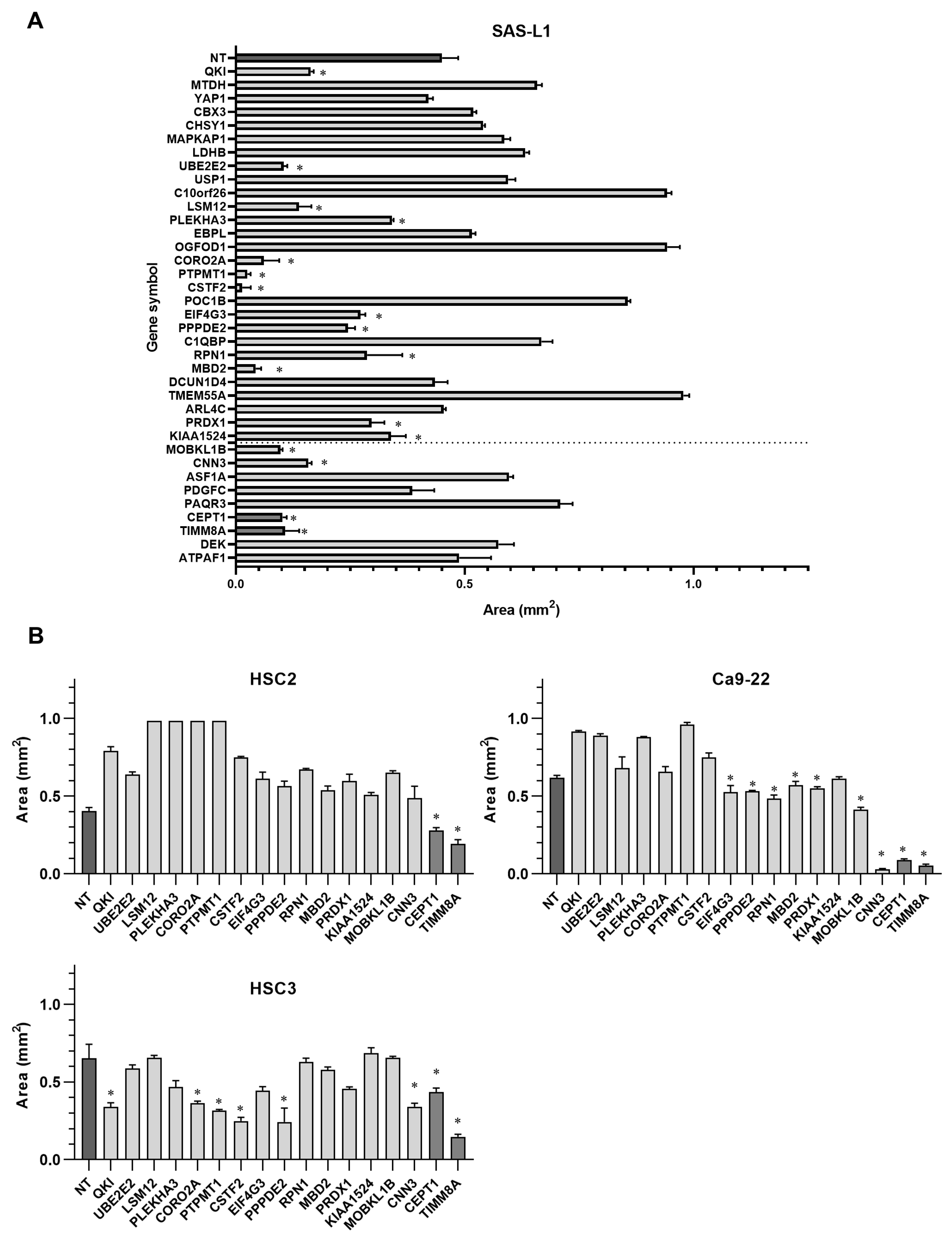
3.5. Identification of the miR-375-3p Target Gene Candidates Involved in the Growth and Migration of Human OSCC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chamoli, A.; Gosavi, A.S.; Shirwadkar, U.P.; Wangdale, K.V.; Behera, S.K.; Kurrey, N.K.; Kalia, K.; Mandoli, A. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral. Oncol. 2021, 121, 105451. [Google Scholar] [CrossRef] [PubMed]
- Almangush, A.; Mäkitie, A.A.; Triantafyllou, A.; de Bree, R.; Strojan, P.; Rinaldo, A.; Hernandez-Prera, J.C.; Suárez, C.; Kowalski, L.P.; Ferlito, A.; et al. Staging and grading of oral squamous cell carcinoma: An update. Oral. Oncol. 2020, 107, 104799. [Google Scholar] [CrossRef] [PubMed]
- Brandwein-Gensler, M.; Smith, R.V.; Wang, B.; Penner, C.; Theilken, A.; Broughel, D.; Schiff, B.; Owen, R.P.; Smith, J.; Sarta, C.; et al. Validation of the histologic risk model in a new cohort of patients with head and neck squamous cell carcinoma. Am. J. Surg. Pathol. 2010, 34, 676–688. [Google Scholar] [CrossRef]
- Ganly, I.; Patel, S.; Shah, J. Early stage squamous cell cancer of the oral tongue—Clinicopathologic features affecting outcome. Cancer 2012, 118, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, H.; Yamashita, Y.; Takeda, S.; Zhang, M.; Fukuyama, H.; Takahashi, T. Risk factors for late cervical lymph node metastases in patients with stage I or II carcinoma of the tongue. Head Neck 2002, 24, 731–736. [Google Scholar] [CrossRef] [PubMed]
- de Bree, R.; Takes, R.P.; Castelijns, J.A.; Medina, J.E.; Stoeckli, S.J.; Mancuso, A.A.; Hunt, J.L.; Rodrigo, J.P.; Triantafyllou, A.; Teymoortash, A.; et al. Advances in diagnostic modalities to detect occult lymph node metastases in head and neck squamous cell carcinoma. Head Neck 2015, 37, 1829–1839. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Shen, G. Association of neck dissection with survival for early stage N0 tongue cancer: A SEER population-based study. Medicine 2018, 97, e13633. [Google Scholar] [CrossRef] [PubMed]
- D’Cruz, A.K.; Vaish, R.; Kapre, N.; Dandekar, M.; Gupta, S.; Hawaldar, R.; Agarwal, J.P.; Pantvaidya, G.; Chaukar, D.; Deshmukh, A.; et al. Elective versus Therapeutic Neck Dissection in Node-Negative Oral Cancer. N. Engl. J. Med. 2015, 373, 521–529. [Google Scholar] [CrossRef]
- Abu-Ghanem, S.; Yehuda, M.; Carmel, N.N.; Leshno, M.; Abergel, A.; Gutfeld, O.; Fliss, D.M. Elective Neck Dissection vs Observation in Early-Stage Squamous Cell Carcinoma of the Oral Tongue With No Clinically Apparent Lymph Node Metastasis in the Neck: A Systematic Review and Meta-analysis. JAMA Otolaryngol. Head Neck Surg. 2016, 142, 857–865. [Google Scholar] [CrossRef]
- de Bree, R.; Takes, R.P.; Shah, J.P.; Hamoir, M.; Kowalski, L.P.; Robbins, K.T.; Rodrigo, J.P.; Sanabria, A.; Medina, J.E.; Rinaldo, A.; et al. Elective neck dissection in oral squamous cell carcinoma: Past, present and future. Oral. Oncol. 2019, 90, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Hanai, N.; Asakage, T.; Kiyota, N.; Homma, A.; Hayashi, R. Controversies in relation to neck management in N0 early oral tongue cancer. Jpn. J. Clin. Oncol. 2019, 49, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, A.; Fukuma, D.; Nagata, M.; Shiraishi, S.; Kawahara, K.; Matsuoka, Y.; Nakagawa, Y.; Yoshida, R.; Tanaka, T.; Yoshitake, Y.; et al. Sentinel lymph node biopsy reduces the incidence of secondary neck metastasis in patients with oral squamous cell carcinoma. Mol. Clin. Oncol. 2016, 5, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Matsuzuka, T.; Tsukahara, K.; Yoshimoto, S.; Chikamatsu, K.; Shiotani, A.; Oze, I.; Murakami, Y.; Shinozaki, T.; Enoki, Y.; Ohba, S.; et al. Predictive factors for dissection-free sentinel node micrometastases in early oral squamous cell carcinoma. Sci. Rep. 2023, 13, 6188. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Tsukahara, K.; Yoshimoto, S.; Miura, K.; Yokoyama, J.; Hirano, S.; Uemura, H.; Sugasawa, M.; Yoshizaki, T.; Homma, A.; et al. Neck dissections based on sentinel lymph node navigation versus elective neck dissections in early oral cancers: A randomized, multicenter, and noninferiority trial. J. Clin. Oncol. 2021, 39, 2025–2036. [Google Scholar] [CrossRef] [PubMed]
- Fransen, M.F.; Schoonderwoerd, M.; Knopf, P.; Camps, M.G.; Hawinkels, L.J.; Kneilling, M.; van Hall, T.; Ossendorp, F. Tumor-draining lymph nodes are pivotal in PD-1/PD-L1 checkpoint therapy. JCI Insight 2018, 3, e124507. [Google Scholar] [CrossRef] [PubMed]
- Novikov, N.M.; Zolotaryova, S.Y.; Gautreau, A.M.; Denisov, E.V. Mutational drivers of cancer cell migration and invasion. Br. J. Cancer 2021, 124, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Roepman, P.; Wessels, L.F.; Kettelarij, N.; Kemmeren, P.; Miles, A.J.; Lijnzaad, P.; Tilanus, M.G.; Koole, R.; Hordijk, G.J.; van der Vliet, P.C.; et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat. Genet. 2005, 37, 182–186. [Google Scholar] [CrossRef]
- van Hooff, S.R.; Leusink, F.K.; Roepman, P.; de Jong, R.J.B.; Speel, E.J.; van den Brekel, M.W.; van Velthuysen, M.L.; van Diest, P.J.; van Es, R.J.; Merkx, M.A.; et al. Validation of a gene expression signature for assessment of lymph node metastasis in oral squamous cell carcinoma. J. Clin. Oncol. 2012, 30, 4104–4110. [Google Scholar] [CrossRef]
- Leusink, F.K.; van Es, R.J.; de Bree, R.; de Jong, R.J.B.; van Hooff, S.R.; Holstege, F.C.; Slootweg, P.J.; Brakenhoff, R.H.; Takes, R.P. Novel diagnostic modalities for assessment of the clinically node-negative neck in oral squamous-cell carcinoma. Lancet Oncol. 2012, 13, e554–e561. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Jin, J.; Ma, T.; Zhai, H. MiR-376c-3p regulates the proliferation, invasion, migration, cell cycle and apoptosis of human oral squamous cancer cells by suppressing HOXB7. Biomed. Pharmacother. 2017, 91, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, I.; Hanazawa, T.; Kinoshita, T.; Kikkawa, N.; Koshizuka, K.; Goto, Y.; Nishikawa, R.; Chiyomaru, T.; Enokida, H.; Nakagawa, M.; et al. MicroRNA expression signature of oral squamous cell carcinoma: Functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br. J. Cancer 2015, 112, 891–900. [Google Scholar] [CrossRef]
- Hsiao, S.Y.; Weng, S.M.; Hsiao, J.R.; Wu, Y.Y.; Wu, J.E.; Tung, C.H.; Shen, W.L.; Sun, S.F.; Huang, W.T.; Lin, C.Y.; et al. MiR-455-5p suppresses PDZK1IP1 to promote the motility of oral squamous cell carcinoma and accelerate clinical cancer invasion by regulating partial epithelial-to-mesenchymal transition. J. Exp. Clin. Cancer Res. 2023, 42, 40. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Nakashiro, K.; Tokuzen, N.; Kuribayashi, N.; Goda, H.; Uchida, D. MicroRNA-361-3p is a potent therapeutic target for oral squamous cell carcinoma. Cancer Sci. 2020, 111, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Nakashiro, K.I.; Tokuzen, N.; Saika, M.; Shirai, H.; Kuribayashi, N.; Goda, H.; Uchida, D. MicroRNA-1289 Functions as a novel tumor Suppressor in Oral Squamous Cell Carcinoma. Cancers 2023, 15, 4138. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Lu, Y.; Wang, R.; Xu, X.; Liu, Q.; He, S.; Pan, H.; Liu, X.; Yuan, B.; Ding, Y.; et al. MicroRNA-375: Potential cancer suppressor and therapeutic drug. Biosci. Rep. 2021, 41, BSR20211494. [Google Scholar] [CrossRef] [PubMed]
- Gyvyte, U.; Lukosevicius, R.; Inciuraite, R.; Streleckiene, G.; Gudoityte, G.; Bekampyte, J.; Valentini, S.; Salteniene, V.; Ruzgys, P.; Satkauskas, S.; et al. The Role of miR-375-3p and miR-200b-3p in gastrointestinal stromal tumors. Int. J. Mol. Sci. 2020, 21, 5151. [Google Scholar] [CrossRef]
- Yang, X.; Nanayakkara, J.; Claypool, D.; Saghafinia, S.; Wong, J.J.M.; Xu, M.; Wang, X.; Nicol, C.J.B.; Michael, I.P.; Hafner, M.; et al. A miR-375/YAP axis regulates neuroendocrine differentiation and tumorigenesis in lung carcinoid cells. Sci. Rep. 2021, 11, 10455. [Google Scholar] [CrossRef]
- Hu, A.; Huang, J.J.; Xu, W.H.; Jin, X.J.; Li, J.P.; Tang, Y.J.; Huang, X.F.; Cui, H.J.; Sun, G.B.; Li, R.L.; et al. MiR-21/miR-375 ratio is an independent prognostic factor in patients with laryngeal squamous cell carcinoma. Am. J. Cancer Res. 2015, 5, 1775–1785. [Google Scholar]
- Osako, Y.; Seki, N.; Kita, Y.; Yonemori, K.; Koshizuka, K.; Kurozumi, A.; Omoto, I.; Sasaki, K.; Uchikado, Y.; Kurahara, H.; et al. Regulation of MMP13 by anti-tumor microRNA-375 markedly inhibits cancer cell migration and invasion in esophageal squamous cell carcinoma. Int. J. Oncol. 2016, 49, 2255–2264. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Wang, J.; Dong, Y.H. The inhibitory effect of miR-375 targeting sp1 in colorectal cancer cell proliferation. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.T.; Li, B.X.; Yuan, Y.J.; Tan, M.; Tan, J.F.; Dai, W.G.; Feng, W.D.; Zuo, J.D. Deregulation of microRNA-375 inhibits proliferation and migration in gastric cancer in association with autophagy-mediated AKT/mTOR signaling pathways. Technol. Cancer Res. Treat. 2018, 17, 1533033818806499. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Gao, Y.; Sui, G.; Jiao, D.; Sun, L.; Fu, Q.; Jin, C. miR-375-3p/YWHAZ/β-catenin axis regulates migration, invasion, EMT in gastric cancer cells. Clin. Exp. Pharmacol. Physiol. 2019, 46, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Yuan, P.; Wang, D.; Jin, H.; Chen, H. Expression and prognostic significance of miR-375 and miR-221 in liver cancer. Oncol. Lett. 2017, 14, 2305–2309. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jin, J.; Wang, L.; Hu, Y.; Liang, D.; Yang, H.; Liu, Y.; Shan, B. Evaluation of miR-21 and miR-375 as prognostic biomarkers in oesophageal cancer in high-risk areas in China. Clin. Exp. Metastasis 2017, 34, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, Y.; Hou, D.; Shi, Q.; Yang, S.; Li, Q. MicroRNA-375 Inhibits Growth and Enhances Radiosensitivity in Oral Squamous Cell Carcinoma by Targeting Insulin Like Growth Factor 1 Receptor. Cell Physiol. Biochem. 2017, 42, 2105–2117. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Yang, J.; Li, S.; Shan, X.; Liu, X.; Hua, H.; Zhao, C.; Feng, Z.; Cai, Z.; Zhang, L.; et al. Potential involvement of miR-375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget 2015, 6, 40172–40185. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.F.; Chang, K.W.; Lin, S.C.; Hung, W.W.; Ji, S.H.; Wu, H.L.; Liu, C.J. Aberrant miR-10b, miR-372, and miR-375 expression in the cytobrushed samples from oral potentially malignant disorders. J. Dent. Sci. 2022, 17, 688–695. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, X.; Song, B.; Qiu, X.; Zhao, J. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. 2017, 6, 1686–1697. [Google Scholar] [CrossRef]
- Cao, Z.H.; Cheng, J.L.; Zhang, Y.; Bo, C.X.; Li, Y.L. MicroRNA-375 inhibits oral squamous cell carcinoma cell migration and invasion by targeting platelet-derived growth factor-A. Mol. Med. Rep. 2017, 15, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.F.; Lin, L.H.; Chang, K.W.; Cheng, H.W.; Liu, C.J. Exploiting salivary miR-375 as a clinical biomarker of oral potentially malignant disorder. J. Dent. Sci. 2022, 17, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, S.; Xu, F.; Fu, J.; Sun, J.; Gan, X.; Yang, C.; Mao, Z. ncRNAs-mediated high expression of TIMM8A correlates with poor prognosis and act as an oncogene in breast cancer. Cancer Cell Int. 2022, 22, 177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, L.; Wu, Y.; Bing, P.; Zhou, J.; Yu, W. Upregulation of TIMM8A is correlated with prognosis and immune regulation in BC. Front. Oncol. 2022, 12, 922178. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saika, M.; Nakashiro, K.-i.; Tokuzen, N.; Shirai, H.; Uchida, D. Possible Role of miR-375-3p in Cervical Lymph Node Metastasis of Oral Squamous Cell Carcinoma. Cancers 2024, 16, 1492. https://doi.org/10.3390/cancers16081492
Saika M, Nakashiro K-i, Tokuzen N, Shirai H, Uchida D. Possible Role of miR-375-3p in Cervical Lymph Node Metastasis of Oral Squamous Cell Carcinoma. Cancers. 2024; 16(8):1492. https://doi.org/10.3390/cancers16081492
Chicago/Turabian StyleSaika, Masato, Koh-ichi Nakashiro, Norihiko Tokuzen, Hiroyuki Shirai, and Daisuke Uchida. 2024. "Possible Role of miR-375-3p in Cervical Lymph Node Metastasis of Oral Squamous Cell Carcinoma" Cancers 16, no. 8: 1492. https://doi.org/10.3390/cancers16081492
APA StyleSaika, M., Nakashiro, K. -i., Tokuzen, N., Shirai, H., & Uchida, D. (2024). Possible Role of miR-375-3p in Cervical Lymph Node Metastasis of Oral Squamous Cell Carcinoma. Cancers, 16(8), 1492. https://doi.org/10.3390/cancers16081492




