Exploring Artificial Intelligence Biases in Predictive Models for Cancer Diagnosis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Search Strategy and Sources
2.3. Screening and Data Extraction
2.4. Outcomes
2.4.1. Characteristics of the Included Studies
2.4.2. AI Performance Metrics
2.4.3. AI Biases
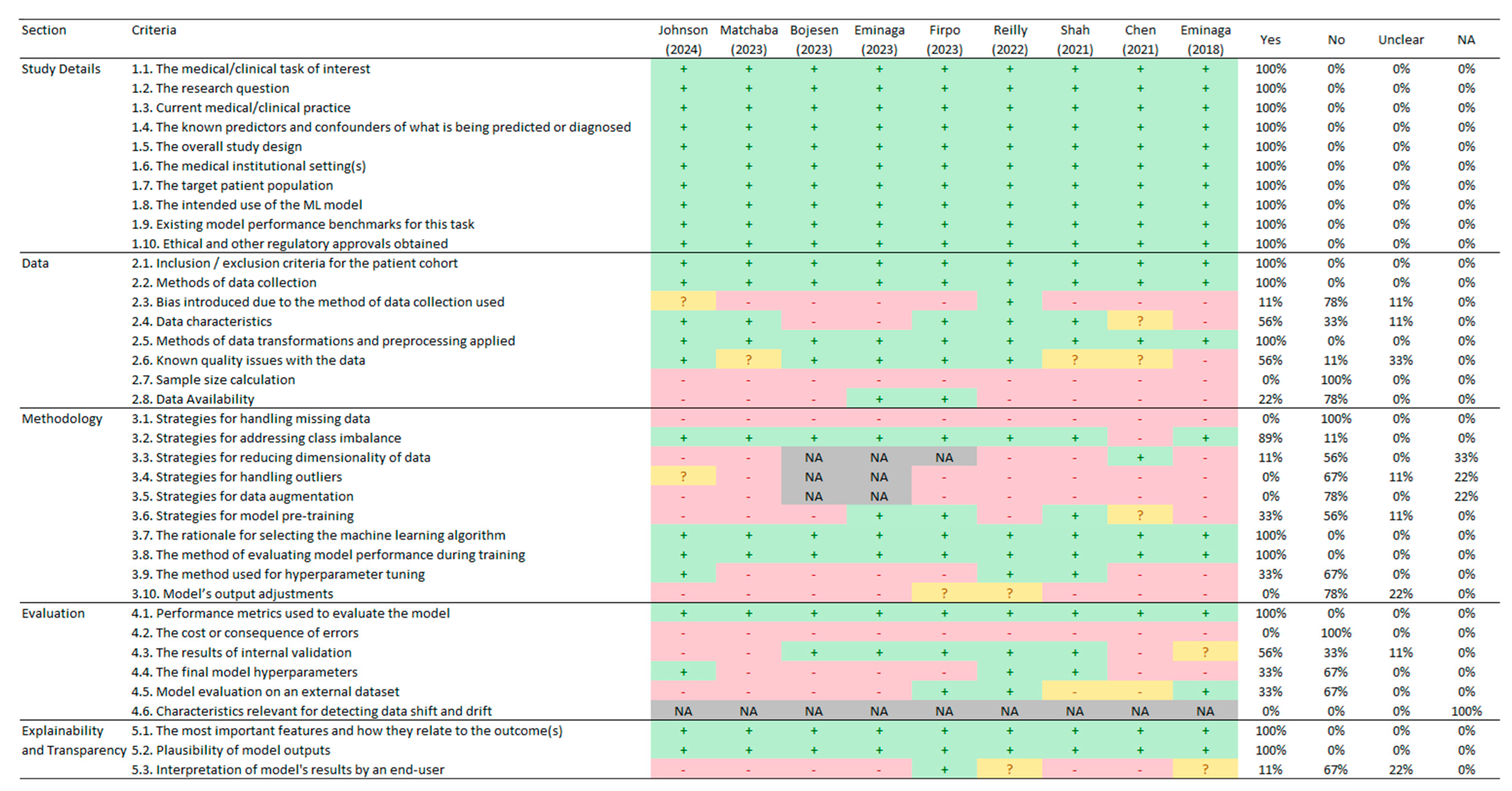
2.5. Quality Assessment
2.6. Potential Impact
2.7. Statistical Analysis
3. Results
3.1. Study Selection
3.2. Characteristics of the Included Studies
3.3. AI Performance Metrics
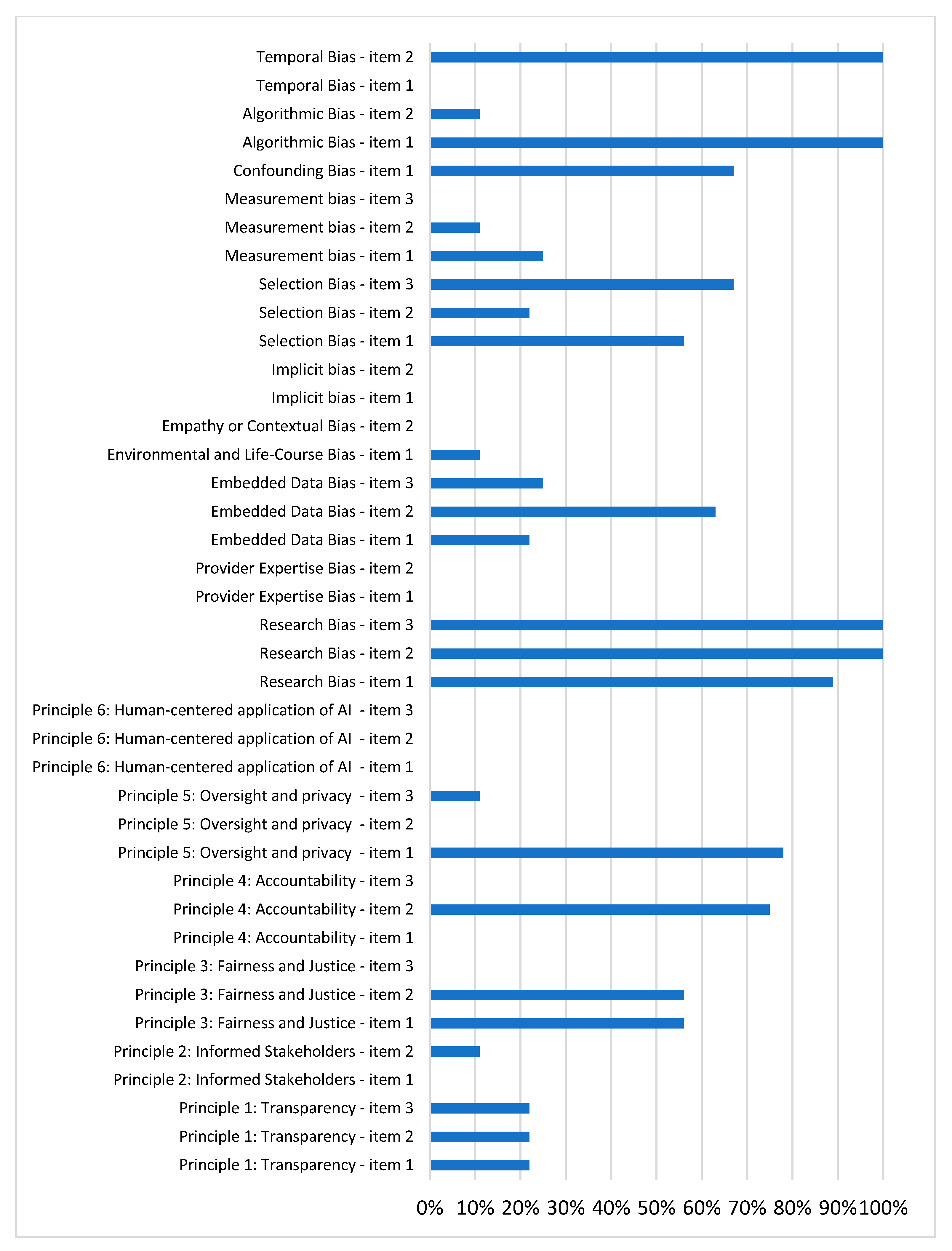
3.4. AI Biases
3.4.1. Principle 1: Transparency
3.4.2. Principle 2: Informed Stakeholders
3.4.3. Principle 3: Fairness and Justice
3.4.4. Principle 4: Accountability and Compliance with Local Regulations
3.4.5. Principle 5: Oversight and Privacy
3.4.6. Principle 6: Human-Centered AI Application
3.4.7. Research Bias
3.4.8. Provider Expertise Bias
3.4.9. Embedded Data Bias
3.4.10. Environmental and Life-Course Bias
3.4.11. Empathy or Contextual Bias
3.4.12. Implicit Bias
3.4.13. Selection Bias
3.4.14. Measurement Bias
3.4.15. Confounding Bias
3.4.16. Algorithmic Bias
3.4.17. Temporal Bias
3.4.18. Risk of Bias Using PROBAST
3.5. Quality Assessment
3.6. Potential Impact
4. Discussion
Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kanan, M.; Alharbi, H.; Alotaibi, N.; Almasuood, L.; Aljoaid, S.; Alharbi, T.; Albraik, L.; Alothman, W.; Aljohani, H.; Alzahrani, A.; et al. AI-Driven Models for Diagnosing and Predicting Outcomes in Lung Cancer: A Systematic Review and Meta-Analysis. Cancers 2024, 16, 674. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.; Li, Z.; Parwani, A.V. Artificial Intelligence’s Impact on Breast Cancer Pathology: A Literature Review. Diagn. Pathol. 2024, 19, 38. [Google Scholar] [CrossRef]
- Thong, L.T.; Chou, H.S.; Chew, H.S.J.; Lau, Y. Diagnostic Test Accuracy of Artificial Intelligence-Based Imaging for Lung Cancer Screening: A Systematic Review and Meta-Analysis. Lung Cancer 2023, 176, 4–13. [Google Scholar] [CrossRef]
- Marletta, S.; Eccher, A.; Martelli, F.M.; Santonicco, N.; Girolami, I.; Scarpa, A.; Pagni, F.; L’Imperio, V.; Pantanowitz, L.; Gobbo, S.; et al. Artificial intelligence-based algorithms for the diagnosis of prostate cancer: A systematic review. Am. J. Clin. Pathol. 2024, 161, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Koteluk, O.; Wartecki, A.; Mazurek, S.; Kołodziejczak, I.; Mackiewicz, A. How Do Machines Learn? Artificial Intelligence as a New Era in Medicine. J. Pers. Med. 2021, 11, 32. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rizzo, P.C.; Caputo, A.; Maddalena, E.; Caldonazzi, N.; Girolami, I.; Dei Tos, A.P.; Scarpa, A.; Sbaraglia, M.; Brunelli, M.; Gobbo, S.; et al. Digital pathology world tour. Digit. Health 2023, 9. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jayakumar, S.; Sounderajah, V.; Normahani, P.; Harling, L.; Markar, S.R.; Ashrafian, H.; Darzi, A. Quality Assessment Standards in Artificial Intelligence Diagnostic Accuracy Systematic Reviews: A Meta-Research Study. npj Digit. Med. 2022, 5, 11. [Google Scholar] [CrossRef]
- Dankwa-Mullan, I.; Weeraratne, D. Artificial Intelligence and Machine Learning Technologies in Cancer Care: Addressing Disparities, Bias, and Data Diversity. Cancer Discov. 2022, 12, 1423–1427. [Google Scholar] [CrossRef]
- Chen, F.; Wang, L.; Hong, J.; Jiang, J.; Zhou, L. Unmasking Bias in Artificial Intelligence: A Systematic Review of Bias Detection and Mitigation Strategies in Electronic Health Record-Based Models. J. Am. Med. Inform. Assoc. 2024, 31, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- American Society of Clinical Oncology. Artificial Intelligence Principles for Oncology Practice and Research; ASCO: Alexandria, VA, USA, 2024; Available online: https://society.asco.org/sites/new-www.asco.org/files/ASCO-AI-Principles-2024.pdf (accessed on 20 November 2024).
- World Health Organization. Ethics and Governance of Artificial Intelligence for Health: WHO Guidance; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Swann, R.; Lyratzopoulos, G.; Rubin, G.; Pickworth, E.; McPhail, S. The frequency, nature and impact of GP-assessed avoidable delays in a population-based cohort of cancer patients. Cancer Epidemiol. 2020, 64, 101617. [Google Scholar] [CrossRef] [PubMed]
- Hanna, T.P.; King, W.D.; Thibodeau, S.; Jalink, M.; Paulin, G.A.; Harvey-Jones, E.; O’Sullivan, D.E.; Booth, C.M.; Sullivan, R.; Aggarwal, A. Mortality due to cancer treatment delay: Systematic review and meta-analysis. BMJ 2020, 371, m4087. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Srivastava, S.; Koay, E.J.; Borowsky, A.D.; De Marzo, A.M.; Ghosh, S.; Wagner, P.D.; Kramer, B.S. Cancer overdiagnosis: A biological challenge and clinical dilemma. Nat. Rev. Cancer 2019, 19, 349–358. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 201. [Google Scholar] [CrossRef]
- El Emam, K.; Leung, T.I.; Malin, B.; Klement, W.; Eysenbach, G. Consolidated Reporting Guidelines for Prognostic and Diagnostic Machine Learning Models (CREMLS). J. Med. Internet Res. 2024, 26, e52508. [Google Scholar] [CrossRef] [PubMed]
- Wolff, R.F.; Moons, K.G.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S.; PROBAST Group. PROBAST: A tool to assess the risk of bias and applicability of prediction model studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) the TRIPOD statement. Circulation 2015, 131, 211–219. [Google Scholar] [CrossRef]
- Collins, G.S.; Moons, K.G.; Dhiman, P.; Riley, R.D.; Beam, A.L.; Van Calster, B.; Ghassemi, M.; Liu, X.; Reitsma, J.B.; Van Smeden, M.; et al. TRIPOD+AI statement: Updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 2024, 385, e078378. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org (accessed on 20 November 2024).
- Bojesen, A.B.; Mortensen, F.V.; Kirkegård, J. Real-Time Identification of Pancreatic Cancer Cases Using Artificial Intelligence Developed on Danish Nationwide Registry Data. JCO Clin. Cancer Inform. 2023, 7, e2300084. [Google Scholar] [CrossRef]
- Chen, Q.; Cherry, D.R.; Nalawade, V.; Qiao, E.M.; Kumar, A.; Lowy, A.M.; Simpson, D.R.; Murphy, J.D. Clinical Data Prediction Model to Identify Patients with Early-Stage Pancreatic Cancer. JCO Clin. Cancer Inform. 2021, 5, 279–287. [Google Scholar] [CrossRef]
- Eminaga, O.; Eminaga, N.; Semjonow, A.; Breil, B. Diagnostic Classification of Cystoscopic Images Using Deep Convolutional Neural Networks. JCO Clin. Cancer Inform. 2018, 2, 1–8. [Google Scholar] [CrossRef]
- Eminaga, O.; Lee, T.J.; Laurie, M.; Ge, T.J.; La, V.; Long, J.; Semjonow, A.; Bogemann, M.; Lau, H.; Shkolyar, E.; et al. Efficient Augmented Intelligence Framework for Bladder Lesion Detection. JCO Clin. Cancer Inform. 2023, 7, e2300031. [Google Scholar] [CrossRef]
- Firpo, M.A.; Boucher, K.M.; Bleicher, J.; Khanderao, G.D.; Rosati, A.; Poruk, K.E.; Kamal, S.; Marzullo, L.; De Marco, M.; Falco, A.; et al. Multianalyte Serum Biomarker Panel for Early Detection of Pancreatic Adenocarcinoma. JCO Clin. Cancer Inform. 2023, 7, e2200160. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.J.; Bhatti, E.; Toyoda, H.; He, S. Serologic Detection of Hepatocellular Carcinoma: Application of Machine Learning and Implications for Diagnostic Models. JCO Clin. Cancer Inform. 2024, 8, e2300199. [Google Scholar] [CrossRef] [PubMed]
- Matchaba, S.; Fellague-Chebra, R.; Purushottam, P.; Johns, A. Early Diagnosis of Pancreatic Cancer via Machine Learning Analysis of a National Electronic Medical Record Database. JCO Clin. Cancer Inform. 2023, 7, e2300076. [Google Scholar] [CrossRef]
- Reilly, G.; Bullock, R.G.; Greenwood, J.; Ure, D.R.; Stewart, E.; Davidoff, P.; DeGrazia, J.; Fritsche, H.; Dunton, C.J.; Bhardwaj, N.; et al. Analytical Validation of a Deep Neural Network Algorithm for the Detection of Ovarian Cancer. JCO Clin. Cancer Inform. 2022, 6, e2100192. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.P.; Selby, H.M.; Mukherjee, P.; Verma, S.; Xie, P.; Xu, Q.; Das, M.; Malik, S.; Gevaert, O.; Napel, S. Machine Learning Radiomics Model for Early Identification of Small-Cell Lung Cancer on Computed Tomography Scans. JCO Clin. Cancer Inform. 2021, 5, 746–757. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef]
- Pappas, T.C.; Roy Choudhury, M.; Chacko, B.K.; Twiggs, L.B.; Fritsche, H.; Elias, K.M.; Phan, R.T. Neural Network-Derived Multivariate Index Assay Demonstrates Effective Clinical Performance in Longitudinal Monitoring of Ovarian Cancer Risk. Gynecol. Oncol. 2024, 187, 21–29. [Google Scholar] [CrossRef]
- Reilly, G.P.; Dunton, C.J.; Bullock, R.G.; Ure, D.R.; Fritsche, H.; Ghosh, S.; Pappas, T.C.; Phan, R.T. Validation of a Deep Neural Network-Based Algorithm Supporting Clinical Management of Adnexal Mass. Front. Med. 2023, 10, 1102437. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, M.; Pappas, T.C.; Twiggs, L.B.; Caoili, E.; Fritsche, H.; Phan, R.T. Ovarian Cancer Surgical Consideration Is Markedly Improved by the Neural Network Powered-MIA3G Multivariate Index Assay. Front. Med. 2024, 11, 1374836. [Google Scholar] [CrossRef]
- Liu, H.; Mo, Z.-H.; Yang, H.; Zhang, Z.-F.; Hong, D.; Wen, L.; Lin, M.-Y.; Zheng, Y.-Y.; Zhang, Z.-W.; Xu, X.-W.; et al. Automatic Facial Recognition of Williams-Beuren Syndrome Based on Deep Convolutional Neural Networks. Front. Pediatr. 2021, 9, 648255. [Google Scholar] [CrossRef]
- Pusztai, L.; Hatzis, C.; Andre, F. Reproducibility of Research and Preclinical Validation: Problems and Solutions. Nat. Rev. Clin. Oncol. 2013, 10, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Connor, J.T. Positive Reasons for Publishing Negative Findings. Am. J. Gastroenterol. 2008, 103, 2181–2183. [Google Scholar] [CrossRef] [PubMed]
- Dilaghi, E.; Lahner, E.; Annibale, B.; Esposito, G. Systematic Review and Meta-Analysis: Artificial Intelligence for the Diagnosis of Gastric Precancerous Lesions and Helicobacter Pylori Infection. Dig. Liver Dis. 2022, 54, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Hassan, C.; Spadaccini, M.; Iannone, A.; Maselli, R.; Jovani, M.; Chandrasekar, V.T.; Antonelli, G.; Yu, H.; Areia, M.; Dinis-Ribeiro, M.; et al. Performance of Artificial Intelligence in Colonoscopy for Adenoma and Polyp Detection: A Systematic Review and Meta-Analysis. Gastrointest. Endosc. 2021, 93, 77–85.e6. [Google Scholar] [CrossRef] [PubMed]
- Krakowski, I.; Kim, J.; Cai, Z.R.; Daneshjou, R.; Lapins, J.; Eriksson, H.; Lykou, A.; Linos, E. Human-AI Interaction in Skin Cancer Diagnosis: A Systematic Review and Meta-Analysis. npj Digit. Med. 2024, 7, 78. [Google Scholar] [CrossRef]
- Jones, O.T.; Matin, R.N.; van der Schaar, M.; Prathivadi Bhayankaram, K.; Ranmuthu, C.K.I.; Islam, M.S.; Behiyat, D.; Boscott, R.; Calanzani, N.; Emery, J.; et al. Artificial Intelligence and Machine Learning Algorithms for Early Detection of Skin Cancer in Community and Primary Care Settings: A Systematic Review. Lancet Digit. Health 2022, 4, e466–e476. [Google Scholar] [CrossRef]
- Lococo, F.; Ghaly, G.; Chiappetta, M.; Flamini, S.; Evangelista, J.; Bria, E.; Stefani, A.; Vita, E.; Martino, A.; Boldrini, L.; et al. Implementation of Artificial Intelligence in Personalized Prognostic Assessment of Lung Cancer: A Narrative Review. Cancers 2024, 16, 1832. [Google Scholar] [CrossRef]
- Ng, A.Y.; Oberije, C.J.G.; Ambrózay, É.; Szabó, E.; Serfőző, O.; Karpati, E.; Fox, G.; Glocker, B.; Morris, E.A.; Forrai, G.; et al. Prospective Implementation of AI-Assisted Screen Reading to Improve Early Detection of Breast Cancer. Nat. Med. 2023, 29, 3044–3049. [Google Scholar] [CrossRef] [PubMed]
- Freedman, R.S.; Cantor, S.B.; Merriman, K.W.; Edgerton, M.E. 2013 HIPAA Changes Provide Opportunities and Challenges for Researchers: Perspectives from a Cancer Center. Clin. Cancer Res. 2016, 22, 533–539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vlahou, A.; Hallinan, D.; Apweiler, R.; Argiles, A.; Beige, J.; Benigni, A.; Bischoff, R.; Black, P.C.; Boehm, F.; Céraline, J.; et al. Data Sharing Under the General Data Protection Regulation: Time to Harmonize Law and Research Ethics? Hypertension 2021, 77, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.B.; Peng, J.; Farnell, D.; Nursey, M.; Wang, Y.; Naso, J.R.; Ren, H.; Farahani, H.; Chen, C.; Chiu, D.; et al. Synthesis of Diagnostic Quality Cancer Pathology Images by Generative Adversarial Networks. J. Pathol. 2020, 252, 178–188. [Google Scholar] [CrossRef]
- Katalinic, M.; Schenk, M.; Franke, S.; Katalinic, A.; Neumuth, T.; Dietz, A.; Stoehr, M.; Gaebel, J. Generation of a Realistic Synthetic Laryngeal Cancer Cohort for AI Applications. Cancers 2024, 16, 639. [Google Scholar] [CrossRef] [PubMed]

| Author (year) | Country | Type of Cancer | Objective | Training | Testing | Validation | Clinical Setting | Funding |
|---|---|---|---|---|---|---|---|---|
| Johnson (2024) [26] | Japan | Hepatocellular carcinoma | Diagnostic | Overall: 3473 patients (49.4% male, median age 61.0 years). With cancer: 445 (12.8%) patients (60.4% males, median age 65.0 years). Without cancer: 3028 (87.2%) patients (47.8% male, median age 60.3 years). | Not reported | Not reported | Single general hospital | Private company |
| Matchaba (2023) [27] | United States | Pancreatic cancer | Diagnostic | Overall: 15,189 participants (35% male, median age 60 years). With cancer: 8438 (56%) patients (51% male, median age 67 years). Without cancer: 6751 (44%) patients (15% male, median age 52 years). | Overall: 3798 participants (35% male, median age 60 years). With cancer: 2127 (56%) patients (51% male, median age 67 years). Without cancer: 1671 (44%) patients (15% male, median age 53 years). | Not reported | Primary care and hospital facilities nationwide | Pharmaceutical industry |
| Bojesen (2023) [21] | Denmark | Pancreatic cancer | Screening | Overall: Case-control dataset. Median age: 71 years (case and control groups). Gender not specified. | Not reported | Not reported | Primary care and hospital facilities nationwide | Foundation |
| Eminaga (2023) [24] | United States | Bladder cancer | Screening | Overall: 312 images used (number of patients not specified). Gender and age not reported. | Not reported | Overall: Videos from 68 cases with 272,799 frames. With cancer: 84,579 (31.0%) frames were labeled as regions with cancer. Without cancer: 188,220 (69.0%) frames were labeled as regions without cancer. Gender and age not reported. | Veterans Affairs Medical Centers | Private company |
| Firpo (2023) [25] | United States | Pancreatic cancer | Screening | Overall: 669 patients (60% male, median age 59 years). With cancer: 152 (19%) patients (57% male, median age 67 years). Without cancer: 517 (81%) patients (56% male, median age 57 years). | Overall: 168 participants (47% male, median age 62 years). With cancer: 30 (18%) patients (60% male, median age 69 years). Without cancer: 138 (82%) patients (44% male, median age 60 years). | Overall: 186 participants (47% male, median age 62 years). With cancer: 73 (39%) patients (47% male, median age 69 years). Without cancer: 113 (61%) patients (46% male, median age 58 years). | Single specialized hospital (cancer) | Government entity |
| Reilly (2022) [28] | United States | Ovarian Cancer | Diagnostic | Overall: 853 patients (all are women, median age 51.3 years). With cancer: 280 (33%) patients (median age not reported). Without cancer: 573 (67%) patients (median age not reported). | Overall: 214 patients (all are women, median age 50.8 years). With cancer: 56 (26%) patients (median age not reported). Without cancer: 158 (74%) patients (median age not reported). | Overall: 2000 patients (all are women, median age 47.5 years). With cancer: 98 (4.9%) patients (median age not reported). Without cancer: 1902 (95.1%) patients (median age not reported). | Unclear | Private company |
| Shah (2021) [29] | United States | Small-Cell Lung Cancer | Screening | Overall: 103 patients (98% male, median age 73 years). With cancer: 26 (25%) patients (100% male, median age 75.6 years). Without cancer: 77 (75%) patients (97% male, median age 72.1 years). | Not reported | Not reported | Veterans Affairs Medical Centers | Government entity and University |
| Chen (2021) [22] | United States | Pancreatic cancer | Screening | Overall: 56,474 patients (41% male, median age 59 years). With cancer: 3322 (5.88%) patients (50.5% male, median age 66.5 years). Without cancer: 53,152 (94.12%) patients (40.7 male, median age 59 years). | 30% of the total dataset. No information on age, sex, or by patients with and without cancer is reported. | Not reported | Single general hospital | Government entity |
| Eminaga (2018) [23] | Germany | Urothelial carcinoma | Diagnostic | Overall: 479 patients, encompassing 44 urological findings. No information on age, sex, or by patients with and without cancer is reported. | 30% of the total dataset. No information on age, sex, or by patients with and without cancer is reported. | 10% of the total dataset. No information on age, sex, or by patients with and without cancer is reported. | Single general hospital | Foundation |
| Author (year) | Stage | Models Evaluated | Sensitivity (Recall or True Positive Rate) | Specificity (True Negative Rate) | Accuracy (Probability of Correct Classification) | Precision (Positive Predictive Value) | F1 | ROC/AUC |
|---|---|---|---|---|---|---|---|---|
| Johnson (2024) [26] | Testing | RF-GALAD: Based on variables from the GALAD model (best model) RF-practical: Based on routine clinical and serological biomarkers (best model) | RF-GALAD: 90.7% RF-practical: 85.9% | RF-GALAD: 74.5% RF-practical: 86.7% | RF-GALAD: 82.6% RF-practical: 82.0% | RF-GALAD: 0.477 RF-practical: 0.467 | RF-GALAD: 0.623 RF-practical: 0.614 | RF-GALAD: 0.907 RF-practical: 0.911 |
| Matchaba (2023) [27] | Testing | SVM, RF, DT, LR, GB, EM (merges SVM, RF, DT, LR, and GB) (best model) | SVM: 41.84% RF: 95.58% DT: 72.87% LR: 93.27% GB: 84.91% EM: 85.61% | SVM: 71.39% RF: 38.42% DT: 69.71% LR: 13.22% GB: 75.88% EM: 76.18% | Not reported | Not reported | SVM: 0.5093 RF: 0.7835 DT: 0.7410 LR: 0.7135 GB: 0.8330 EM: 0.8380 | SVM: 0.53 RF: 0.80 DT: 0.71 LR: 0.61 GB: 0.88 EM: 0.89 |
| Bojesen (2023) [21] | Testing | RF (best model), BT | 23.4% (in Combined cohorts) Data reported only from the best model | Not reported | Not reported | 10.1% (in Combined cohorts) Data reported only from the best model | Not reported | 74.4% (in Combined cohorts) Data reported only from the best model |
| Eminaga (2023) [24] | Validation | ConvNeXt (best model), PlexusNet (best model), MobileNet, SwinTransformer. | Frame level: 81.4% to 88.1% Block level: 100% Does not report the value per model. | Frame level: 30.0% to 44.8% Block level: 56% to 67% Does not report the value per model. | Not reported | 32.8% to 37.0% Does not report the value per model. | 0.444 to 0.495 Does not report the value per model. | 63.9% to 74.4% Does not report the value per model. |
| Firpo (2023) [25] | Training, Testing, and Validation | GLMnet, RF, KNN, SVM, NNET, EM using stacking (merges GLMnet, KNN, NNET, RF, and SVM) (best model) | Training set: 92.8% Test set: 63.3% Validation set: 72.6% Data reported only from the best model | Training set: 99.8% Test set: 97.1% Validation set: 95.6% Data reported only from the best model | Training set: 98.2% Test set: 90.5% Validation set: 86.6% Data reported only from the best model | Not reported | Not reported | Training set: Not reported Test set: 0.944 Validation set: 0.925 Data reported only from the best model |
| Reilly (2022) [28] | Testing and Validation | MIA3G | Test Dataset Overall: 91.07% Premenopausal: 88.89% Postmenopausal: 92.11% Epithelial ovarian cancer: 93.33% Validation Dataset: Overall: 89.80% Premenopausal: 80.77% Postmenopausal: 93.06% Epithelial ovarian cancer: 94.94% | Test Dataset Overall: 87.97% Premenopausal: 95.40% Postmenopausal: 78.87% Validation Dataset: Overall: 84.02% Premenopausal: 91.86% Postmenopausal: 71.56% | Not reported | Testing Dataset Overall: 72.86% Premenopausal: 80.00% Postmenopausal: 70.00% Validation Dataset: Overall: 22.45% Premenopausal: 18.10% Postmenopausal: 24.28% | Not reported | Test Dataset: Overall: 0.938 Validation Dataset: Overall: 0.937 |
| Shah (2021) [29] | Training | LR, RF (best model), SVC, XGBoost | Not reported | Not reported | Not reported | Not reported | Not reported | Noncontrast scans: RF: 0.81 SVC: 0.77 XGBoost: 0.84 LR: 0.84. Contrast-enhanced scans: RF: 0.88 SVC: 0.87 XGBoost: 0.85 LR: 0.81 |
| Chen (2021) [22] | Merge between training and testing | XGBoost (best model) | 60% | 90% | Not reported | 0.07% to 0.23% | Not reported | 0.84 |
| Eminaga (2018) [23] | Unclear | ResNet50, VGG-19, VGG-16, InceptionV3, Xception (best model), harmonic-series concept, 90%-layer concept | Not reported | Not reported | Xception: 99.52% ResNet50: 99.48% InceptionV3: 98.73% Harmonic-series concept: 99.45% 90%-layer concept: 99.11% VGG-16: 97.42% VGG-19: 95.47% | Xception: 99.54% ResNet50: 99.48% InceptionV3: 98.86% Harmonic-series concept: 99.45% 90%-layer concept: 99.11% VGG-16: 97.82% VGG-19: 95.65% | Xception: 0.9952 ResNet50: 0.9948 InceptionV3: 0.9874 Harmonic-series concept: 0.9945 90%-layer concept: 0.9911 VGG-16: 0.9735 VGG-19: 0.9547 | Not reported |
| Bias Criterion | Items | [26] | [27] | [21] | [24] | [25] | [28] | [29] | [22] | [23] | Yes % (n) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Principle 1: Transparency | 1. Data and procedure access of the training | No | No | No | Yes | Yes | No | No | No | No | 22% (2/9) |
| 2. Data and procedure access of the testing | No | No | No | Yes | Yes | No | No | No | No | 22% (2/9) | |
| 3. Reproducibility materials access | No | No | No | Yes | Yes | No | No | No | No | 22% (2/9) | |
| Principle 2: Informed Stakeholders | 1. Professional training in AI usage | No | No | No | No | No | No | No | No | No | 0% (0/9) |
| 2. Patient’s informed consent for the use of data in AI. | No | No | No | Yes | No | No | No | No | No | 11% (1/9) | |
| Principle 3: Fairness and Justice | 1. Model fairness measures | No | No | Yes | Yes | No | Yes | Yes | Yes | No | 56% (5/9) |
| 2. Diversity of participants included is reported | Yes | Yes | No | No | Yes | Yes | Yes | No | No | 56% (5/9) | |
| 3. Compliance with specific ethical guidelines for AI models | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| Principle 4: Accountability | 1. Compliance with legal and regulatory requirements | No | No | No | No | No | No | No | No | No | 0% (0/9) |
| 2. Adherence to ethical standards | Yes | NA | Yes | Yes | Yes | Yes | Yes | No | No | 75% (6/8) | |
| 3. Statement of responsibility | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| Principle 5: Oversight and privacy | 1. Patient data privacy protection | Yes | Yes | Yes | Yes | Yes | No | Yes | No | Yes | 78% (7/9) |
| 2. Use of privacy-enhancing technologies | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| 3. Ensuring the autonomy of health professionals and patients | No | No | No | No | No | No | No | No | Yes | 11% (1/9) | |
| Principle 6: Human-centered application of AI | 1. Guaranteeing human interaction in health services | No | No | No | No | No | No | No | No | No | 0% (0/9) |
| 2. Ensuring human oversight throughout the AI lifecycle | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| 3. Clinical consent management | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| Research Bias | 1. Real-world data application | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | 89% (8/9) |
| 2. Diverse backgrounds | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 100% (9/9) | |
| 3. Funding and conflicts of interest | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 100% (9/9) | |
| Provider Expertise Bias | 1. Provider bias consideration | No | No | No | No | No | No | No | No | No | 0% (0/9) |
| 2. Consistency of clinical guidelines | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| Embedded Data Bias | 1. Data collection bias analysis | Yes | No | No | Yes | No | No | No | No | No | 22% (2/9) |
| 2. Synthetic data integration | No | Yes | No | Yes | NA | Yes | Yes | No | Yes | 63% (5/8) | |
| 3. Missing or incomplete data management | No | Yes | No | No | NA | No | No | Yes | No | 25% (2/8) | |
| Environmental and Life-Course Bias | 1. Environmental and life factors impact | Yes | No | No | No | No | No | No | No | No | 11% (1/9) |
| Empathy or Contextual Bias | 2. Knowledge of cultural or procedural factors of the data | No | No | No | No | No | No | No | No | No | 0% (0/9) |
| Implicit bias | 1. Pre-existing biases in data | No | No | No | No | No | No | No | No | No | 0% (0/9) |
| 2. Worse clinical outcomes in vulnerable groups | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| Selection Bias | 1. Population representativeness assessment | Yes | Yes | No | No | No | No | Yes | Yes | Yes | 56% (5/9) |
| 2. Participant diversity and data during the training phase | No | Yes | No | No | No | No | No | Yes | No | 22% (2/9) | |
| 3. Sampling bias assessment | Yes | Yes | Yes | Yes | Yes | Yes | No | No | No | 67% (6/9) | |
| Measurement bias | 1. Inaccuracies in data collection | No | Yes | No | No | NA | No | No | Yes | No | 25% (2/8) |
| 2. Standardization of data collection | No | No | No | Yes | No | No | No | No | No | 11% (1/9) | |
| 3. Data biases affecting AI model performance | No | No | No | No | No | No | No | No | No | 0% (0/9) | |
| Confounding Bias | 1. Confounding factors analysis | Yes | Yes | Yes | No | Yes | Yes | No | Yes | No | 67% (6/9) |
| Algorithmic Bias | 1. Performance indicators reporting | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 100% (9/9) |
| 2. Statistical assumptions check | No | No | Yes | No | No | No | No | No | No | 11% (1/9) | |
| Temporal Bias | 1. Temporal changes impact | No | No | No | No | No | No | No | No | No | 0% (0/9) |
| 2. Adjustments for temporal changes | NA | NA | NA | Yes | NA | NA | NA | NA | NA | 100% (1/1) |
| Reference | Total Citations | Mean by Year | Replicate or Use the AI Model |
|---|---|---|---|
| [26] | 3 | 3.0 | 0 |
| [27] | 2 | 1.0 | 0 |
| [21] | 1 | 0.5 | 0 |
| [24] | 0 | 0.0 | 0 |
| [25] | 3 | 1.5 | 0 |
| [28] | 10 | 3.3 | 3 |
| [29] | 7 | 1.8 | 0 |
| [22] | 31 | 7.8 | 0 |
| [23] | 71 | 10.1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smiley, A.; Reategui-Rivera, C.M.; Villarreal-Zegarra, D.; Escobar-Agreda, S.; Finkelstein, J. Exploring Artificial Intelligence Biases in Predictive Models for Cancer Diagnosis. Cancers 2025, 17, 407. https://doi.org/10.3390/cancers17030407
Smiley A, Reategui-Rivera CM, Villarreal-Zegarra D, Escobar-Agreda S, Finkelstein J. Exploring Artificial Intelligence Biases in Predictive Models for Cancer Diagnosis. Cancers. 2025; 17(3):407. https://doi.org/10.3390/cancers17030407
Chicago/Turabian StyleSmiley, Aref, C. Mahony Reategui-Rivera, David Villarreal-Zegarra, Stefan Escobar-Agreda, and Joseph Finkelstein. 2025. "Exploring Artificial Intelligence Biases in Predictive Models for Cancer Diagnosis" Cancers 17, no. 3: 407. https://doi.org/10.3390/cancers17030407
APA StyleSmiley, A., Reategui-Rivera, C. M., Villarreal-Zegarra, D., Escobar-Agreda, S., & Finkelstein, J. (2025). Exploring Artificial Intelligence Biases in Predictive Models for Cancer Diagnosis. Cancers, 17(3), 407. https://doi.org/10.3390/cancers17030407






