Artificial Intelligence-Assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
2.3. Informational Sources
2.4. Search Strategy
2.5. Data Items
3. Results
3.1. Characteristics of Sources of Evidence
3.2. Results of Individual Sources of Evidence
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro Oncol. 2022, 24, v1–v95. [Google Scholar] [CrossRef] [PubMed]
- Koshy, M.; Villano, J.L.; Dolecek, T.A.; Howard, A.; Mahmood, U.; Chmura, S.J.; Weichselbaum, R.R.; McCarthy, B.J. Improved Survival Time Trends for Glioblastoma Using the SEER 17 Population-Based Registries. J. Neurooncol. 2012, 107, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.; Rosenthal, M.A. Survival Comparison between Glioblastoma Multiforme and Other Incurable Cancers. J. Clin. Neurosci. 2010, 17, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Wirsching, H.-G.; Galanis, E.; Weller, M. Glioblastoma; Springer: Berlin/Heidelberg, Germany, 2016; pp. 381–397. [Google Scholar]
- Delgado-Martín, B.; Medina, M.Á. Advances in the Knowledge of the Molecular Biology of Glioblastoma and Its Impact in Patient Diagnosis, Stratification, and Treatment. Adv. Sci. 2020, 7, 1902971. [Google Scholar] [CrossRef]
- Del Pilar, G.P.M.; de La Fuente, M. The Role of Molecular Genetics of Glioblastoma in the Clinical Setting. In Precision Molecular Pathology of Glioblastoma; Spriger: Berlin/Heidelberg, Germany, 2021; pp. 21–33. [Google Scholar]
- Verdugo, E.; Puerto, I.; Medina, M.Á. An update on the Molecular Biology of Glioblastoma, with Clinical Implications and Progress in Its Treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef]
- D’Alessio, A.; Proietti, G.; Sica, G.; Scicchitano, B.M. Pathological and Molecular Features of Glioblastoma and Its Peritumoral Tissue. Cancers 2019, 11, 469. [Google Scholar] [CrossRef]
- Reifenberger, G.; Wirsching, H.-G.; Knobbe-Thomsen, C.B.; Weller, M. Advances in the Molecular Genetics of Gliomas—Implications for Classification and Therapy. Nat. Rev. Clin. Oncol. 2017, 14, 434–452. [Google Scholar] [CrossRef]
- Bellei, M.; Nabhan, C.; Pesce, E.A.; Conte, L.; Vose, J.M.; Foss, F.; Federico, M. The Value and Relevance of the T Cell Lymphoma Registries and International Collaborations: The Case of COMPLETE and the T-Cell Project. Curr. Hematol. Malig. Rep. 2015, 10, 448–455. [Google Scholar] [CrossRef]
- Arigliani, M.; Toraldo, D.M.; Montevecchi, F.; Conte, L.; Galasso, L.; De Rosa, F.; Lattante, C.; Ciavolino, E.; Arigliani, C.; Palumbo, A.; et al. A New Technological Advancement of the Drug-Induced Sleep Endoscopy (Dise) Procedure: The “All in One Glance” Strategy. Int. J. Environ. Res. Public Health 2020, 17, 4261. [Google Scholar] [CrossRef]
- Conte, L.; Greco, M.; Toraldo, D.M.; Arigliani, M.; Maffia, M.; De Benedetto, M. A Review of the “OMICS” for Management of patients with Obstructive Sleep Apnoea. Acta Otorhinolaryngol. Ital. 2020, 40, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Conte, L.; De Nunzio, G.; Lupo, R.; Mieli, M.; Lezzi, A.; Vitale, E.; Carriero, M.C.; Calabrò, A.; Carvello, M.; Rubbi, I.; et al. Breast Cancer Prevention: The Key Role of Population Screening, Breast Self-Examination (BSE) and Technological Tools. Survey of Italian Women. J. Cancer Educ. 2023, 38, 1728–1742. [Google Scholar] [CrossRef] [PubMed]
- Conte, L.; Lupo, R.; Lezzi, A.; Paolo, V.; Rubbi, I.; Rizzo, E.; Carvello, M.; Calabrò, A.; Botti, S.; De Matteis, E.; et al. A Nationwide Cross-Sectional Study Investigating Adherence to the Mediterranean Diet, Smoking, Alcohol and Work Habits, Hormonal dynamics between Breast Cancer Cases and Healthy Subjects. Clin. Nutr. Open Sci. 2024, 55, 1–19. [Google Scholar] [CrossRef]
- Conte, L.; Lupo, R.; Sciolti, S.; Lezzi, A.; Rubbi, I.; Botti, S.; Carvello, M.; Fanizzi, A.; Massafra, R.; Vitale, E.; et al. Exploring the Landscape of Breast Cancer Prevention among Chinese Residents in Italy: An In-Depth Analysis of Screening Adherence, Breast Self-Examination (BSE) Practices, the Role of Technological Tools, and Misconceptions Surrounding Risk Factors and Sy. Int. J. Environ. Res. Public Health 2024, 21, 308. [Google Scholar] [CrossRef] [PubMed]
- Cucci, F.; Conte, L. Artificial Intelligence-assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature; OSF: Peoria, IL, USA, 2024. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Gao, M.; Wang, X.; Han, D.; Lu, E.; Zhang, J.; Zhang, C.; Wang, L.; Yang, Q.; Jiang, Q.; Wu, J.; et al. A Six-LncRNA Signature for Immunophenotype Prediction of Glioblastoma Multiforme. Front. Genet. 2021, 11, 604655. [Google Scholar] [CrossRef]
- Han, M.-H.; Min, K.-W.; Noh, Y.-K.; Kim, J.M.; Cheong, J.H.; Ryu, J.I.; Won, Y.D.; Koh, S.-H.; Myung, J.K.; Park, J.Y.; et al. High DKK3 Expression Related to Immunosuppression Was Associated with Poor Prognosis in Glioblastoma: Machine Learning Approach. Cancer Immunol. Immunother. 2022, 71, 3013–3027. [Google Scholar] [CrossRef]
- Suman, S.; Kulshrestha, A. Characterization of Cellular Heterogeneity in Recurrent Pediatric Glioblastoma: Machine Learning-Enhanced Single-Cell RNA-Seq Unveils Regulatory Signatures. Hum. Gene 2024, 41, 201300. [Google Scholar] [CrossRef]
- Ye, L.; Tong, S.; Wang, Y.; Wang, Y.; Ma, W. Grade Scoring System Reveals Distinct Molecular Subtypes and Identifies KIF20A as a Novel Biomarker for Predicting Temozolomide Treatment Efficiency in Gliomas. J. Cancer Res. Clin. Oncol. 2023, 149, 9857–9876. [Google Scholar] [CrossRef]
- Han, S.; Zhang, Z.; Ma, W.; Gao, J.; Li, Y. Nucleotide-Binding Oligomerization Domain (NOD)-Like Receptor Subfamily C (NLRC) as a Prognostic Biomarker for Glioblastoma Multiforme Linked to Tumor Microenvironment: A Bioinformatics, Immunohistochemistry, and Machine Learning-Based Study. J. Inflamm. Res. 2023, 16, 523–537. [Google Scholar] [CrossRef]
- Tang, Y.; Qazi, M.A.; Brown, K.R.; Mikolajewicz, N.; Moffat, J.; Singh, S.K.; McNicholas, P.D. Identification of Five Important Genes to Predict Glioblastoma Subtypes. Neuro-Oncol. Adv. 2021, 3, vdab144. [Google Scholar] [CrossRef] [PubMed]
- Quddusi, D.M.; Bajcinca, N. Identification of Genomic Biomarkers and Their Pathway Crosstalks for Deciphering Mechanistic Links in Glioblastoma. IET Syst. Biol. 2023, 17, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jin, H.; Liu, H. Identification of T-cell Exhaustion-Related Gene Signature for Predicting Prognosis in Glioblastoma Multiforme. J. Cell. Mol. Med. 2023, 27, 3503–3513. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Li, M.; Li, K.; Pu, X.; Guo, Y. Immune Landscape-Based Machine-Learning–Assisted Subclassification, Prognosis, and Immunotherapy Prediction for Glioblastoma. Front. Immunol. 2022, 13, 1027631. [Google Scholar] [CrossRef]
- Yang, X.; Cai, Z.; Wang, C.; Jiang, C.; Li, J.; Chen, F.; Li, W. Integrated Multiomic Analysis Reveals Disulfidptosis Subtypes in Glioblastoma: Implications for Immunotherapy, Targeted Therapy, and Chemotherapy. Front. Immunol. 2024, 15, 1362543. [Google Scholar] [CrossRef]
- Huang, R.; Kong, Y.; Luo, Z.; Li, Q. LncRNA NDUFA6-DT: A Comprehensive Analysis of a Potential LncRNA Biomarker and Its Regulatory Mechanisms in Gliomas. Genes 2024, 15, 483. [Google Scholar] [CrossRef]
- Joyce, T.; Tasci, E.; Jagasia, S.; Shephard, J.; Chappidi, S.; Zhuge, Y.; Zhang, L.; Cooley Zgela, T.; Sproull, M.; Mackey, M.; et al. Serum CD133-Associated Proteins Identified by Machine Learning Are Connected to Neural Development, Cancer Pathways, and 12-Month Survival in Glioblastoma. Cancers 2024, 16, 2740. [Google Scholar] [CrossRef]
- Lu, C.-H.; Wei, S.-T.; Liu, J.-J.; Chang, Y.-J.; Lin, Y.-F.; Yu, C.-S.; Chang, S.L.-Y. Recognition of a Novel Gene Signature for Human Glioblastoma. Int. J. Mol. Sci. 2022, 23, 4157. [Google Scholar] [CrossRef]
- Kałuzińska, Ż.; Kołat, D.; Bednarek, A.K.; Płuciennik, E. PLEK2, RRM2, GCSH: A Novel WWOX-Dependent Biomarker Triad of Glioblastoma at the Crossroads of Cytoskeleton Reorganization and Metabolism Alterations. Cancers 2021, 13, 2955. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, N.; Wu, W.; Zhou, R.; Li, S.; Wang, Z.; Dai, Z.; Zhang, L.; Liu, Z.; Zhang, J.; et al. Machine Learning-Based Tumor-Infiltrating Immune Cell-Associated lncRNAs for Predicting Prognosis and Immunotherapy Response in Patients with Glioblastoma. Brief. Bioinform. 2022, 23, bbac386. [Google Scholar] [CrossRef]
- Zhang, W.; Dang, R.; Liu, H.; Dai, L.; Liu, H.; Adegboro, A.A.; Zhang, Y.; Li, W.; Peng, K.; Hong, J.; et al. Machine Learning-Based Investigation of Regulated Cell Death for Predicting Prognosis and Immunotherapy Response in Glioma Patients. Sci. Rep. 2024, 14, 4173. [Google Scholar] [CrossRef] [PubMed]
- Mei, N.; Lu, Y.; Yang, S.; Jiang, S.; Ruan, Z.; Wang, D.; Liu, X.; Ying, Y.; Li, X.; Yin, B. Oligodendrocyte Transcription Factor 2 as a Potential Prognostic Biomarker of Glioblastoma: Kaplan-Meier Analysis and the Development of a Binary Predictive Model Based on Visually Accessible Rembrandt Image and Magnetic Resonance Imaging Radiomic Feature. J. Comput. Assist. Tomogr. 2023, 47, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.H.B.; Leon, A.J.; Hui, W.; Lee, S.C.-E.; Batruch, I.; Faust, K.; Klekner, A.; Hutóczki, G.; Koritzinsky, M.; Richer, M.; et al. Topographic Mapping of the Glioblastoma Proteome Reveals a Triple-Axis Model of Intra-Tumoral Heterogeneity. Nat. Commun. 2022, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.; Koo, H.; Yang, Y.; Shin, S.; Zhu, Z.; Kim, D.; Cho, H.J.; Mu, Q.; Choi, S.W.; Sa, J.K.; et al. Pharmacogenomic Profiling Reveals Molecular Features of Chemotherapy Resistance in IDH Wild-Type Primary Glioblastoma. Genome Med. 2023, 15, 16. [Google Scholar] [CrossRef]
- Bahcheli, A.T.; Min, H.-K.; Bayati, M.; Zhao, H.; Fortuna, A.; Dong, W.; Dzneladze, I.; Chan, J.; Chen, X.; Guevara-Hoyer, K.; et al. Pan-Cancer Ion Transport Signature Reveals Functional Regulators of Glioblastoma Aggression. EMBO J. 2024, 43, 196–224. [Google Scholar] [CrossRef]
- Arteaga-Arteaga, H.B.; Candamil-Cortés, M.S.; Breaux, B.; Guillen-Rondon, P.; Orozco-Arias, S.; Tabares-Soto, R. Machine Learning Applications on Intratumoral Heterogeneity in Glioblastoma Using Single-Cell RNA Sequencing Data. Brief. Funct. Genom. 2023, 22, 428–441. [Google Scholar] [CrossRef]
- Molyneaux, K.; Laggner, C.; Brady-Kalnay, S.M. Artificial Intelligence-Based Computational Screening and Functional Assays Identify Candidate Small Molecule Antagonists of PTPmu-Dependent Adhesion. Int. J. Mol. Sci. 2023, 24, 4274. [Google Scholar] [CrossRef]
- Jiang, P.; Huang, S.; Fu, Z.; Sun, Z.; Lakowski, T.M.; Hu, P. Deep Graph Embedding for Prioritizing Synergistic Anticancer Drug Combinations. Comput. Struct. Biotechnol. J. 2020, 18, 427–438. [Google Scholar] [CrossRef]
- Chen, H.; Chen, Z.; Zhang, Z.; Li, Y.; Zhang, S.; Jiang, F.; Wei, J.; Ding, P.; Zhou, H.; Gu, Q.; et al. Discovery of New LXRβ Agonists as Glioblastoma Inhibitors. Eur. J. Med. Chem. 2020, 194, 112240. [Google Scholar] [CrossRef]
- Alabed, S.J.; Zihlif, M.; Taha, M. Discovery of New Potent Lysine Specific Histone Demythelase-1 Inhibitors (LSD-1) Using Structure Based and Ligand Based Molecular Modelling and Machine Learning. RSC Adv. 2022, 12, 35873–35895. [Google Scholar] [CrossRef]
- Neves, B.J.; Agnes, J.P.; do Nascimento Gomes, M.; Henriques Donza, M.R.; Gonçalves, R.M.; Delgobo, M.; Ribeiro de Souza Neto, L.; Senger, M.R.; Silva-Junior, F.P.; Ferreira, S.B.; et al. Efficient Identification of Novel Anti-Glioma Lead Compounds by Machine Learning Models. Eur. J. Med. Chem. 2020, 189, 111981. [Google Scholar] [CrossRef]
- Jang, B.-S.; Park, A.J.; Kim, I.A. Exploration of Biomedical Knowledge for Recurrent Glioblastoma Using Natural Language Processing Deep Learning Models. BMC Med. Inform. Decis. Mak. 2022, 22, 267. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Pan, P.; Lv, R.; Ma, C.; Wu, E.; Guo, R.; Zhao, Z.; Song, H.; Zhou, J.; Liu, Y.; et al. High-Throughput Glycolytic Inhibitor Discovery Targeting Glioblastoma by Graphite Dots–Assisted LDI Mass Spectrometry. Sci. Adv. 2022, 8, eabl4923. [Google Scholar] [CrossRef] [PubMed]
- Giczewska, A.; Pastuszak, K.; Houweling, M.; Abdul, K.U.; Faaij, N.; Wedekind, L.; Noske, D.; Wurdinger, T.; Supernat, A.; Westerman, B.A. Longitudinal Drug Synergy Assessment Using Convolutional Neural Network Image-Decoding of Glioblastoma Single-Spheroid Cultures. Neuro-Oncol. Adv. 2023, 5, vdad134. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, C.R.; Gutiérrez-Asorey, P.; Blanes-Rodríguez, M.; Hidalgo-Delgado, I.; de Jesús Blanco Liverio, M.; Castiñeiras Galdo, B.; Porto-Pazos, A.B.; Gestal, M.; Arrasate, S.; González-Díaz, H. Prediction of Anti-Glioblastoma Drug-Decorated Nanoparticle Delivery Systems Using Molecular Descriptors and Machine Learning. Int. J. Mol. Sci. 2021, 22, 11519. [Google Scholar] [CrossRef]
- Molyneaux, K.; Laggner, C.; Vincent, J.; Brady-Kalnay, S. Small Molecule Antagonists of PTPmu Identified by Artificial Intelligence-Based Computational Screening Block Glioma Cell Migration and Growth. PLoS ONE 2023, 18, e0288980. [Google Scholar] [CrossRef]
- Ghafoor, N.A.; Yildiz, A. Targeting MDM2–p53 Axis through Drug Repurposing for Cancer Therapy: A Multidisciplinary Approach. ACS Omega 2023, 8, 34583–34596. [Google Scholar] [CrossRef]
- Caffo, M.; Casili, G.; Caruso, G.; Barresi, V.; Campolo, M.; Paterniti, I.; Minutoli, L.; Ius, T.; Esposito, E. DKK3 Expression in Glioblastoma: Correlations with Biomolecular Markers. Int. J. Mol. Sci. 2024, 25, 4091. [Google Scholar] [CrossRef]
- Munquad, S.; Das, A.B. Uncovering the Subtype-Specific Disease Module and the Development of Drug Response Prediction Models for Glioma. Heliyon 2024, 10, e27190. [Google Scholar] [CrossRef]
- Lupo, R.; Zacchino, S.; Caldararo, C.; Calabrò, A.; Carriero, M.C.; Santoro, P.; Carvello, M.; Conte, L. The use of electronical devices and relative levels of Nomophobia within a group of Italian nurses: An Observational Study. Epidemiol. Biostat. Public Health 2022, 17, e13272-1–e13272-10. [Google Scholar] [CrossRef]
- Vitale, E.; Conte, L.; Dell’Aglio, A.; Calabrò, A.; Ilari, F.; Bardone, L.; Benedetto, A.; Caldararo, C.; Zacchino, S.; Lezzi, A.; et al. Healthcare workers perceptions in the difficult moment of the end of life and coping strategies adopted during the COVID-19 pandemic: An Italian pilot study. Acta Biomed. 2021, 92, e2021330. [Google Scholar] [CrossRef]
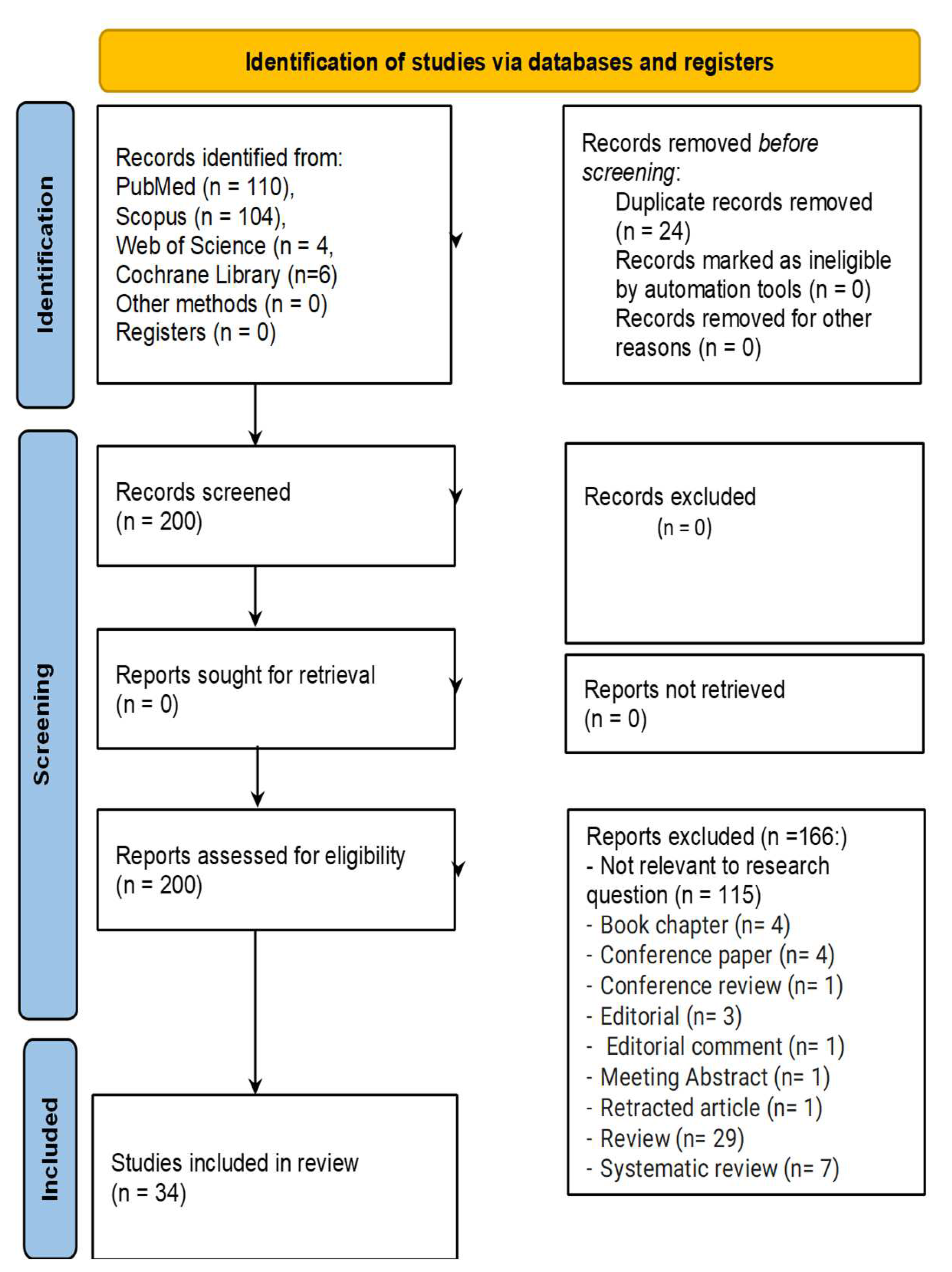

| Population | Adult and Pediatric Patients Diagnosed with GB |
|---|---|
| Concept | Studies examining the application of AI in drug discovery or biomarker identification for GB. Includes AI techniques like ML and DL applied to GB drug or biomarker discovery. |
| Context | Research conducted in clinical, translational, or in silico settings. Includes a range of healthcare environments, both retrospective and prospective studies. |
| Database | Search String |
|---|---|
| PubMed | ((Glioblastoma[Title/Abstract]) AND (Artificial Intelligence[Title/Abstract] OR deep learning[Title/Abstract] OR machine learning[Title/Abstract])) AND (Drug[Title/Abstract] OR Biomarker[Title/Abstract]) |
| The Cochrane Library | (Glioblastoma):ti,ab,kw AND (Artificial intelligence OR deep learning OR machine learning):ti,ab,kw AND (Drug OR Biomarker):ti,ab,kw |
| Scopus | (TITLE-ABS-KEY (glioblastoma) AND TITLE-ABS-KEY (artificial AND intelligence OR deep AND learning OR machine AND learning) AND TITLE-ABS-KEY (drug OR biomarker)) |
| Web of science (WOS) | ((TI = (Glioblastoma)) AND TI = (Artificial intelligence OR deep learning OR machine learning)) AND TI = (Drug OR Biomarker) |
| No. | Reference | Methods | Data Retrieval | Algorithms Used | Found Biomarkers | Summary of the Study |
|---|---|---|---|---|---|---|
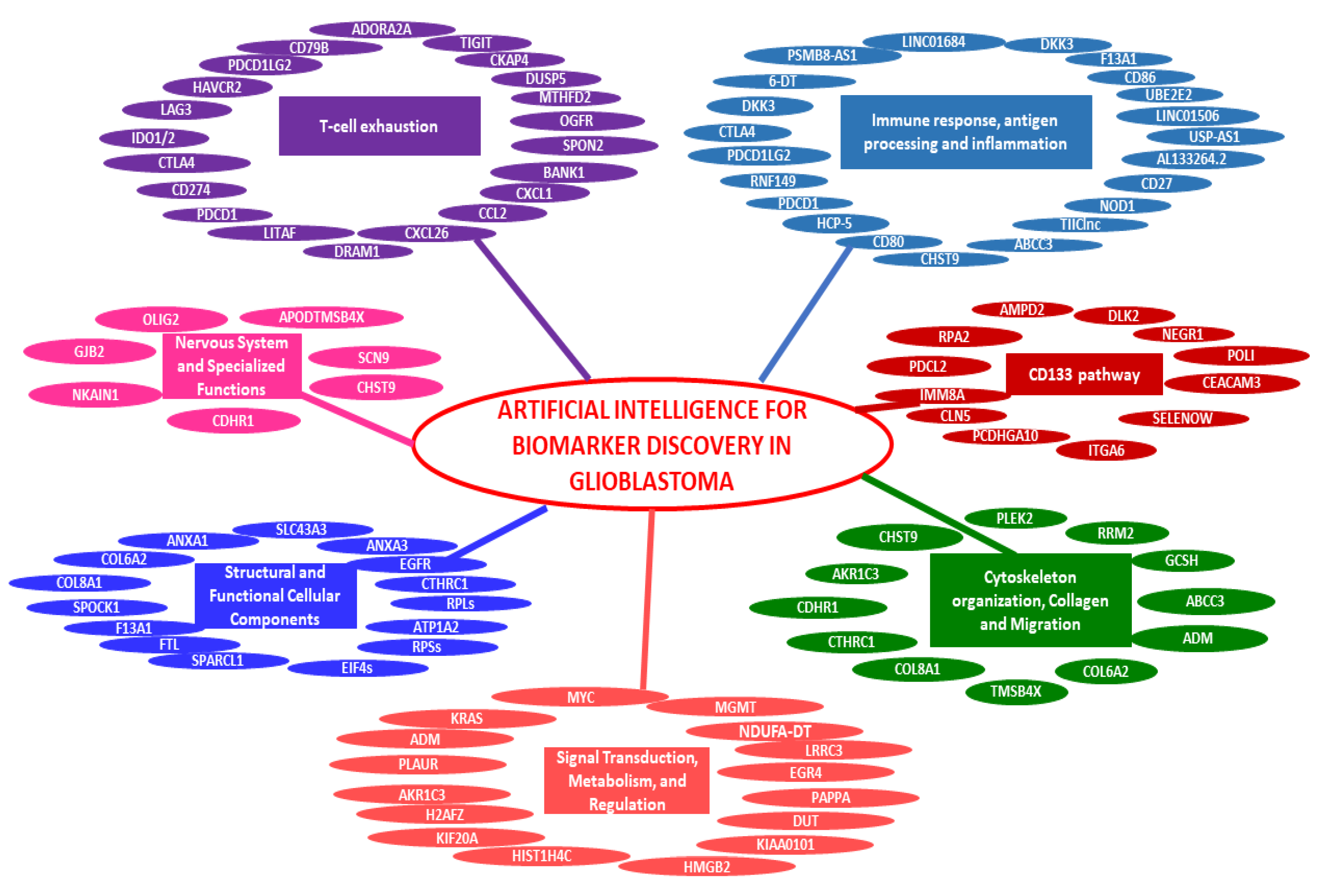
| 1 | [19] | SsGSEA, ML | TCGA | mRMR, RF | USP30-AS1, HCP5, PSMB8-AS1, AL133264.2, LINC01684, LINC01506 | Identified six lncRNA biomarkers to distinguish immune phenotypes in GB using TCGA transcriptome data, guiding immunotherapy. |
| 2 | [20] | GSEA, ML | TCGA | GBM | DKK3 | High DKK3 expression was associated with poor prognosis and immunosuppression in GB, providing a therapeutic target. |
| 3 | [21] | scRNA-Seq, ML | GEO | PCA, KNN, RF, XGBoost, DT | HMGB2, H2AFZ, HIST1H4C, KIAA0101, DUT | Used scRNA-Seq data and ML to identify gene signatures and cell types, improving understanding of recurrent pediatric GB. |
| 4 | [22] | mRNA data analysis, ML | TCGA, CGGA, GEO | UC, PCA Boruta | KIF20A | Identified KIF20A as a biomarker predicting glioma prognosis and response to TMZ treatment using a novel grade scoring system. |
| 5 | [23] | Bioinformatics, ML | TCGA | RF | NOD1 | High NOD1 expression was associated with poor prognosis and immunosuppression in GB, providing a therapeutic target. |
| 6 | [24] | ML | WSD | XGBoost | NKAIN1, UBE2E2, F13A1, RNF149, PLAUR | Developed a five-gene classifier to accurately predict GB subtypes, offering potential for diagnostic biomarker development. |
| 7 | [25] | GSEA, Liquid biopsies, ML | ArrayExpress, TEP | PCA, DT | RPLs, RPSs, ETCs, EIF4s, etc. | Numerous genomic biomarkers were identified from liquid biopsies, revealing different oncogenic pathways involved in GB. |
| 8 | [26] | Multi-omics, ML | TCGA, GEO, CGGA. | GSEA | CD79B, CKAP4, DUSP5, MTHFD2, OGFR, SPON2, BANK1, CXCL1, CCL2, CXCL26, DRAM1, LITAF | Developed a TEX signature (mRNA, miRNA, and lncRNA) to predict GB prognosis and guide immunotherapy. |
| 9 | [27] | GSEA, ML | TCGA, GlioVis, WSD | KNN, SVM-RFE, SVM, RF, XGBoost, CMap | PDCD1, CD274, CTLA4, IDO1, IDO2, LAG3, HAVCR2, PDCD1LG2, TIGIT, ADORA2A, VTCN1 , etc. | ML identified GB immune subtypes with different prognoses and immunotherapy responses. |
| 10 | [28] | SsGSEA, Transcriptomics, ML | TCGA, CGGA | LASSO | CD80, CD86, CTLA4, PDCD1, PDCD1LG2, CD27, etc. | Identified disulfidptosis subtypes in GB, highlighting immunotherapy and targeted therapy implications. |
| 11 | [29] | SsGSEA, RNA-seq, ML | UCSC Xena, TCGA, Ensembl, GEO, CGGA | GLM, RF, Boruta, GBM, XGBoost, SVM-RFE. | NDUFA6-DT | Identified lncRNA NDUFA6-DT as a prognostic biomarker in gliomas, influencing immune responses and synaptic transmission. |
| 12 | [30] | ML, proteomics | NIH | LASSO, RFECV | RPA2, AMPD2, DLK2, NEGR1, PDCL2, POLI, CEACAM3, ITGA6, PCDHGA10, SELENOW, IMM8A, CLN5, etc. | Identified serum proteins associated with CD133 using ML to predict 12-month survival in GB patients. |
| 13 | [31] | GSEA, ML | Rembrandt, G-DOC, GEO | GSEA SVM RBF | ABCC3, ADM, COL6A2, COL8A1, CTHRC1, AKR1C3, CDHR1, CHST9, etc. | Identified a 33-gene signature that may serve as a biomarker for diagnosis and treatment of GB. |
| 14 | [32] | RNA-seq, ML | TCGA, UCSC | GSEA, SVM-RFE | WWOX-dependent genes: PLEK2, RRM2, GCSH | Identified three key WWOX-dependent genes affecting cytoskeleton reorganization and metabolism in GB. |
| 15 | [33] | Transcriptomics, ML | TCGA, CGGA, GEO, Xiangya | LASSO, Boruta, XGBoost, SVM, RF, PAMR | TIIClnc signature | Developed a lncRNA signature associated with tumor-infiltrating immune cells to predict prognosis and immunotherapy response in GB |
| 16 | [34] | Multi-omics, ML | TCGA, CGGA, GEO, GLASS, GTEx, CCLE | LASSO, RSF, XGBoost, Enet, CoxBoost, Boruta | SLC43A3 | Developed a gene pair scoring system to predict prognosis and identify therapeutic targets for GB based on RCD phenotypes. |
| 17 | [35] | Kaplan–Meier, ML | NA | RFE, RF | OLIG2 | Developed a predictive model based on OLIG2 expression and MRI features for preoperative prognostic prediction in GB patients. |
| 18 | [36] | ssGSEA, Proteomics, ML | Recruited patients | XGBoost | KRAS, MYC | Used mass spectrometry and ML to map GB proteome, identifying KRAS and MYC as key drivers of tumor heterogeneity. |
| 19 | [37] | ML, pharmacogenomics | Recruited patients, TCGA | XGBoost | MGMT, EGR4, PAPPA, LRRC3, ANXA3 | Developed an ML model to predict TMZ resistance based on genomic and transcriptomic features in IDH wild-type GB. |
| 20 | [38] | ML, transcriptomics | TCGA | UC | GJB2, SCN9 | Identified ion transport genes such as GJB2 and SCN9A as regulators of GB aggression, providing new drug targets. |
| 21 | [39] | scRNA-seq, ML | GBTR | PCA, LR, XGBoost, GBM, ET, GNB DT, RF, MLP, LDA, SVM,QDA, KNN, AdaBoost | ATP1A2, SPARCL1, FTL, EGFR, SPOCK1, ANXA1, APODTMSB4X | Applied ML to classify cell groups within GB based on single-cell RNA-seq data, identifying potential biomarkers. |
| No. | Reference | Methods | Data Retrieval | Algorithms Used | Founded Drugs | Summary of the Study |
|---|---|---|---|---|---|---|
| 1 | [40] | AI-based computational screening, cell-based assays | Cell cultures | AtomNet® | Antagonists of PTPmu | This study uses the AtomNet® platform to identify small-molecule antagonists of PTPmu, targeting GB. Several compounds were tested in glioma sphere growth assays. |
| 2 | [41] | Network integration | Cell cultures | GCN, DNN, SVM, RF, EN | Anticancer drug combinations | The study applies GCN to predict synergistic anticancer drug combinations, identifying combinations for cancer cell lines, including GB. |
| 3 | [42] | ML, molecular docking, in vivo testing | ChEMBL BindingDB | SVM, NB | LXRβ agonists | LXRβ agonists were designed through ML and tested in vivo on GB xenograft models, showing promising inhibition of tumor growth. |
| 4 | [43] | Structure-based and ligand-based molecular modeling, ML | ChEMBL | RF, XGBoost | LSD-1 inhibitors | A combination of molecular modeling and ML was used to discover new LSD-1 inhibitors, tested in GB cells. |
| 5 | [44] | ML models, in vitro and in vivo testing | ChEMBL | SVM, RF, DNN | 4m, 4n | ML models were used to prioritize compounds for anti-glioma properties, leading to in vivo tests in glioma models, with two promising candidates identified that efficiently decreased malignant glioma development in mice, probably by inhibiting thioredoxin reductase activity. |
| 6 | [45] | AI-based literature mining | QA task, Clinical trials | SAPBERT, NLP | RTK inhibitors, LPA-1 antagonists | NLP was employed to explore medical corpora and clinical trials for potential drug targets for recurrent GB, identifying key drug–gene interactions. |
| 7 | [46] | High-throughput virtual screening, LDI mass spectrometry | Cell cultures, In vivo model | GLMSD | HK2 inhibitors | Discovery of glycolytic inhibitors for GB, targeting metabolic pathways. Inhibitors were tested both in vitro and in vivo models. |
| 8 | [47] | Imaging, drug interaction assays | Cell cultures | CNN | Longitudinal drug interaction assessment | This study used CNNs to assess drug synergy in 3D GB neurosphere images over time, revealing dynamic drug interactions. |
| 9 | [48] | PTML | ChEMBL | KNN, GBM, LDA, LR, DT, RF, XGBoost, GBC, BC, AdaBoost | DDNPs with anti-GB activity | PTML was used to predict drug-nanoparticle complexes targeting GB. Models predicted anti-GB drug interactions with nanoparticles. |
| 10 | [49] | AI-based AtomNet®, cell-based assays | Cell cultures, In vivo models | AtomNet® | Antagonists of PTPmu | AI-assisted screening identified small-molecule inhibitors of PTPmu that reduce glioma cell migration and tumor growth in vivo. |
| 11 | [50] | ML, molecular docking, in vitro assays | ChEMBL | RF, KNN, ET, LGB, HGB, XGBoost, DT, SGD, MLP, AdaBoost | MDM2-p53 inhibitors: cetirizine, rapatadine | This study used ML to repurpose drugs like cetirizine and rupatadine as MDM2-p53 inhibitors, showing anti-GB activity in vitro. |
| 12 | Munquad 46 | Network medicine, AI-based drug response prediction | COSMIC, UCSC Xena, TissueNet | DNN | Drug response prediction | The study developed drug response prediction models using network medicine to identify effective therapies for GB subtypes. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conte, L.; Caruso, G.; Philip, A.K.; Cucci, F.; De Nunzio, G.; Cascio, D.; Caffo, M. Artificial Intelligence-Assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature. Cancers 2025, 17, 571. https://doi.org/10.3390/cancers17040571
Conte L, Caruso G, Philip AK, Cucci F, De Nunzio G, Cascio D, Caffo M. Artificial Intelligence-Assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature. Cancers. 2025; 17(4):571. https://doi.org/10.3390/cancers17040571
Chicago/Turabian StyleConte, Luana, Gerardo Caruso, Anil K. Philip, Federico Cucci, Giorgio De Nunzio, Donato Cascio, and Maria Caffo. 2025. "Artificial Intelligence-Assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature" Cancers 17, no. 4: 571. https://doi.org/10.3390/cancers17040571
APA StyleConte, L., Caruso, G., Philip, A. K., Cucci, F., De Nunzio, G., Cascio, D., & Caffo, M. (2025). Artificial Intelligence-Assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature. Cancers, 17(4), 571. https://doi.org/10.3390/cancers17040571









