Molecular Mechanisms in the Transformation from Indolent to Aggressive B Cell Malignancies
Simple Summary
Abstract
1. Introduction
1.1. Chronic Lymphocytic Leukemia
1.2. Follicular Lymphoma
1.3. Marginal Zone Lymphomas
1.4. Lymphoplasmacytic Lymphoma
2. Molecular Mechanisms in CLL Transformation to RT
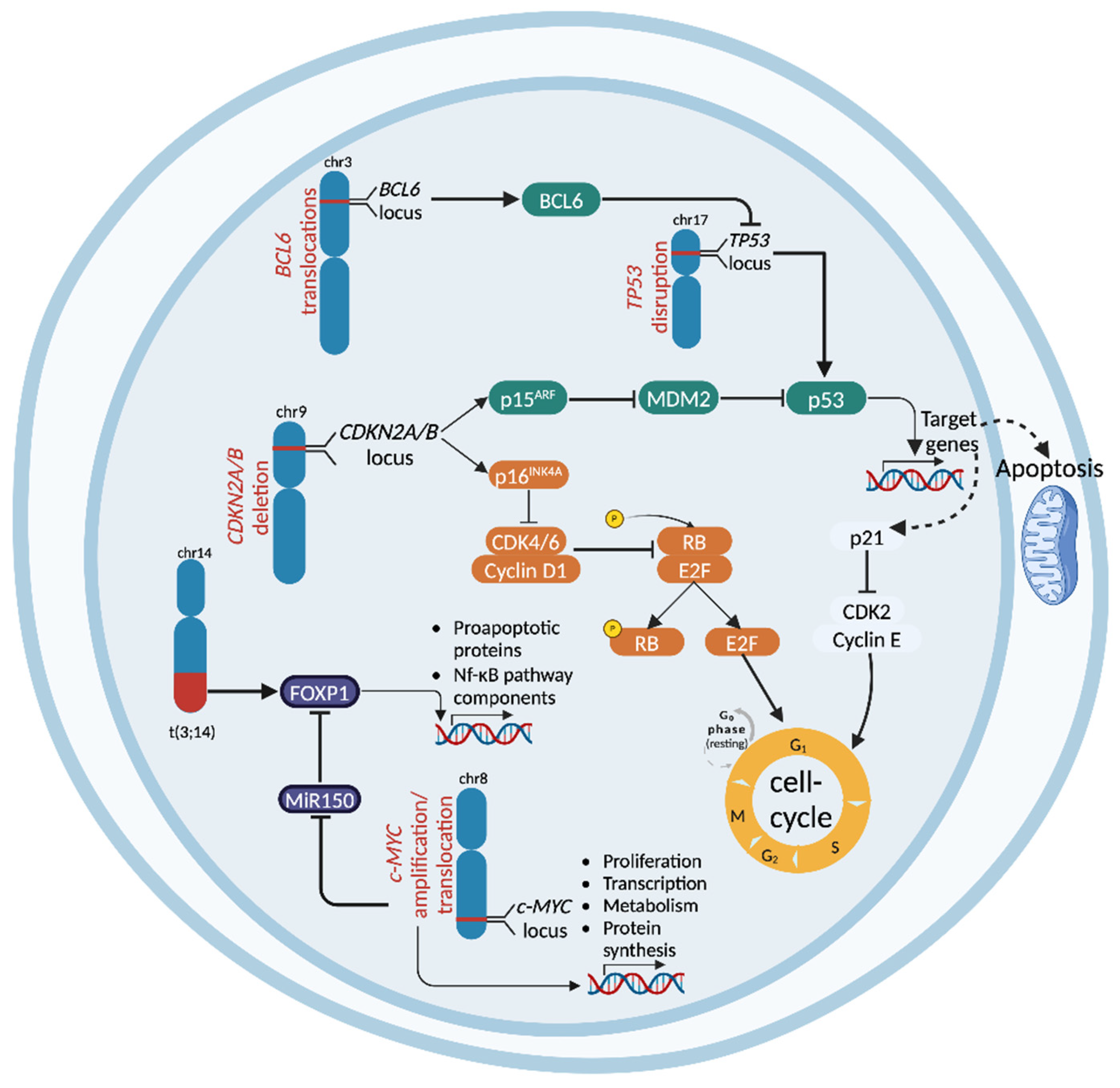
2.1. TP53 Disruption
2.2. CDKN2A Deletion
2.3. NOTCH1 Mutational Activation
2.4. c-MYC Abnormalities
2.5. BCR Pathway Dysregulation
2.6. Dysregulation of Immune Checkpoints
2.7. Novel Molecular Insights in DLBCL-Type RT
3. Molecular Mechanisms in the Transformation of FL
3.1. CDKN2A/B Deletion
3.2. c-MYC Abnormalities
3.3. TP53 Disruption
3.4. BCL-6 Genetic Lesions
3.5. NF-κB Pathway Dysregulation
3.6. tFL Cell of Origin
3.7. Microenvironmental Alterations
4. Molecular Mechanisms in the Transformation of MZLs
4.1. CDKN2A/B Deletion in SMZL
4.2. NF-κB Pathway Dysregulation in SMZL
4.3. Copy Number Alterations in SMZL
4.4. Epigenetic Alterations
4.5. FOXP1 Aberrations in MALT Lymphomas
4.6. Mechanisms of HT in NMZL
5. Molecular Mechanisms in the Transformation of LPL
CXCR4 Mutations
6. Molecular Crossroads: The Shared Pathways Fueling Lymphoma Transformation
| Disease | Gene Lesion | Biological Effect | Frequency Before HT | Frequency at HT | Reference |
|---|---|---|---|---|---|
| CLL | TP53 | DNA damage response, cell-cycle regulation | 10–15% | 60–80% | [46,47] |
| CDKN2A | Cell-cycle regulation | 7% | 30% | [54,172] | |
| NOTCH1 | NF-κB signaling | 8% | 31% | [61] | |
| c-MYC | Proliferation and survival | - | 30–35% | [54] | |
| MGA | c-MYC inhibition | 3% | 6% | [70] | |
| SETD2 | Epigenetic regulation of gene expression | 3% | 30% | [173,174] | |
| FL | CDKN2A | Cell-cycle regulation | 5% | 45% | [30,31,103,105,106] |
| c-MYC | Proliferation and survival | 5–10% | 40% | [30,31,107] | |
| TP53 | DNA damage response, cell-cycle regulation | 5% | 25% | [30,31,107,109,110] | |
| BCL6 | B cell differentiation | 10% | 25% | [119] | |
| c-REL | NF-κB signaling | - | 11% | [30] | |
| TNFAIP3 | NF-κB signaling | 5–10% | 15% | [30,121,122] | |
| MYD88 | NF-κB/BCR signaling | 5% | 11% | [31,121] | |
| CD58 | Microenviroment dysregulation | - | 5 | [31,121] | |
| B2M | NF-κB signaling | - | 12 | [31,121] | |
| TNFRSF14 | Microenviroment dysregulation | 15–20% | 30–40% | [30,121,132,133,134] | |
| MZLs | CDKN2A | Cell-cycle regulation | 5% | 40% | [140,146,147,148,149] |
| TNFAIP3 | NF-κB signaling | 15% | 60% | [145] | |
| KLF2 | NF-κB signaling and B cell differentiation | 20–25% | 35% | [139,145,147,154,155] | |
| KMT2D | Epigenetic regulation of gene expression | 10–15% | 45% | [157,158] | |
| LPL | CXCR4 | Proliferation and survival | 30% | 55% | [41,165] |
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mouhssine, S.; Gaidano, G. Richter Syndrome: From Molecular Pathogenesis to Druggable Targets. Cancers 2022, 14, 4644. [Google Scholar] [CrossRef]
- Cerhan, J.R. Epidemiology of Follicular Lymphoma. Hematol. Oncol. Clin. N. Am. 2020, 34, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Conconi, A.; Franceschetti, S.; Aprile von Hohenstaufen, K.; Margiotta-Casaluci, G.; Stathis, A.; Moccia, A.A.; Bertoni, F.; Ramponi, A.; Mazzucchelli, L.; Cavalli, F.; et al. Histologic transformation in marginal zone lymphomas. Ann. Oncol. 2015, 26, 2329–2335. [Google Scholar] [CrossRef]
- Castillo, J.J.; Gustine, J.; Meid, K.; Dubeau, T.; Hunter, Z.R.; Treon, S.P. Histological transformation to diffuse large B-cell lymphoma in patients with Waldenström macroglobulinemia. Am. J. Hematol. 2016, 91, 1032–1035. [Google Scholar] [CrossRef] [PubMed]
- Parry, E.M.; Roulland, S.; Okosun, J. DLBCL arising from indolent lymphomas: How are they different? Semin. Hematol. 2023, 60, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Gall, E.A.; Mallory, T.B. Malignant Lymphoma: A Clinico-Pathologic Survey of 618 Cases. Am. J. Pathol. 1942, 18, 381–429. [Google Scholar]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; International Agency for Research on Cancer Lyon: Lyon, France, 2017; Volume 2. [Google Scholar]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Surveillance Epidemiology and End Results Program (SEER). Cancer Stat Facts: Leukemia—Chronic Lymphocytic Leukemia (CLL). Available online: https://seer.cancer.gov/statfacts/html/clyl.html (accessed on 21 December 2024).
- Mouhssine, S.; Maher, N.; Matti, B.F.; Alwan, A.F.; Gaidano, G. Targeting BTK in B Cell Malignancies: From Mode of Action to Resistance Mechanisms. Int. J. Mol. Sci. 2024, 25, 3234. [Google Scholar] [CrossRef]
- Maher, N.; Mouhssine, S.; Matti, B.F.; Alwan, A.F.; Gaidano, G. Treatment Refractoriness in Chronic Lymphocytic Leukemia: Old and New Molecular Biomarkers. Int. J. Mol. Sci. 2023, 24, 10374. [Google Scholar] [CrossRef]
- Shadman, M. Diagnosis and Treatment of Chronic Lymphocytic Leukemia: A Review. JAMA 2023, 329, 918–932. [Google Scholar] [CrossRef]
- Mouhssine, S.; Maher, N.; Kogila, S.; Cerchione, C.; Martinelli, G.; Gaidano, G. Current Therapeutic Sequencing in Chronic Lymphocytic Leukemia. Hematol. Rep. 2024, 16, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Eichhorst, B.; Ghia, P.; Niemann, C.U.; Kater, A.P.; Gregor, M.; Hallek, M.; Jerkeman, M.; Buske, C. ESMO Clinical Practice Guideline interim update on new targeted therapies in the first line and at relapse of chronic lymphocytic leukaemia. Ann. Oncol. 2024, 35, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Spina, V.; Gaidano, G. Biology and treatment of Richter syndrome. Blood 2018, 131, 2761–2772. [Google Scholar] [CrossRef] [PubMed]
- Hampel, P.J.; Rabe, K.G.; Wang, Y.; Hwang, S.R.; Kenderian, S.S.; Muchtar, E.; Leis, J.F.; Koehler, A.B.; Tsang, M.; Hilal, T.; et al. Incidence of Richter transformation of chronic lymphocytic leukemia/small lymphocytic lymphoma in the targeted therapy era. Leukemia 2024, 39, 503–507. [Google Scholar] [CrossRef]
- Nadeu, F.; Royo, R.; Massoni-Badosa, R.; Playa-Albinyana, H.; Garcia-Torre, B.; Duran-Ferrer, M.; Dawson, K.J.; Kulis, M.; Diaz-Navarro, A.; Villamor, N.; et al. Detection of early seeding of Richter transformation in chronic lymphocytic leukemia. Nat. Med. 2022, 28, 1662–1671. [Google Scholar] [CrossRef]
- Al-Sawaf, O.; Robrecht, S.; Bahlo, J.; Fink, A.M.; Cramer, P.; Tresckow, J.V.; Lange, E.; Kiehl, M.; Dreyling, M.; Ritgen, M.; et al. Richter transformation in chronic lymphocytic leukemia (CLL)—A pooled analysis of German CLL Study Group (GCLLSG) front line treatment trials. Leukemia 2021, 35, 169–176. [Google Scholar] [CrossRef]
- Mao, Z.; Quintanilla-Martinez, L.; Raffeld, M.; Richter, M.; Krugmann, J.; Burek, C.; Hartmann, E.; Rudiger, T.; Jaffe, E.S.; Müller-Hermelink, H.K.; et al. IgVH mutational status and clonality analysis of Richter’s transformation: Diffuse large B-cell lymphoma and Hodgkin lymphoma in association with B-cell chronic lymphocytic leukemia (B-CLL) represent 2 different pathways of disease evolution. Am. J. Surg. Pathol. 2007, 31, 1605–1614. [Google Scholar] [CrossRef]
- El Hussein, S.; Medeiros, L.J.; Lyapichev, K.A.; Fang, H.; Jelloul, F.Z.; Fiskus, W.; Chen, J.; Wei, P.; Schlette, E.; Xu, J.; et al. Immunophenotypic and genomic landscape of Richter transformation diffuse large B-cell lymphoma. Pathology 2023, 55, 514–524. [Google Scholar] [CrossRef]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Müller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef]
- Wang, Y.; Tschautscher, M.A.; Rabe, K.G.; Call, T.G.; Leis, J.F.; Kenderian, S.S.; Kay, N.E.; Muchtar, E.; Van Dyke, D.L.; Koehler, A.B.; et al. Clinical characteristics and outcomes of Richter transformation: Experience of 204 patients from a single center. Haematologica 2020, 105, 765–773. [Google Scholar] [CrossRef]
- Favini, C.; Talotta, D.; Almasri, M.; Andorno, A.; Rasi, S.; Adhinaveni, R.; Kogila, S.; Awikeh, B.; Schipani, M.; Boggione, P.; et al. Clonally unrelated Richter syndrome are truly de novo diffuse large B-cell lymphomas with a mutational profile reminiscent of clonally related Richter syndrome. Br. J. Haematol. 2022, 198, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.; Eyre, T.A.; Cheah, C.Y. Emerging Therapies for the Management of Richter Transformation. J. Clin. Oncol. 2023, 41, 395–409. [Google Scholar] [CrossRef]
- Weiss, L.M.; Warnke, R.A.; Sklar, J.; Cleary, M.L. Molecular analysis of the t(14;18) chromosomal translocation in malignant lymphomas. N. Engl. J. Med. 1987, 317, 1185–1189. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, E. Follicular lymphoma: 2023 update on diagnosis and management. Am. J. Hematol. 2022, 97, 1638–1651. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Roulland, S.; Gloghini, A.; Younes, A.; von Keudell, G.; López-Guillermo, A.; Fitzgibbon, J. Follicular lymphoma. Nat. Rev. Dis. Primers 2019, 5, 83. [Google Scholar] [CrossRef]
- Fischer, T.; Zing, N.P.C.; Chiattone, C.S.; Federico, M.; Luminari, S. Transformed follicular lymphoma. Ann. Hematol. 2018, 97, 17–29. [Google Scholar] [CrossRef]
- Alcoceba, M.; García-Álvarez, M.; Medina, A.; Maldonado, R.; González-Calle, V.; Chillón, M.C.; Sarasquete, M.E.; González, M.; García-Sanz, R.; Jiménez, C. MYD88 Mutations: Transforming the Landscape of IgM Monoclonal Gammopathies. Int. J. Mol. Sci. 2022, 23, 5570. [Google Scholar] [CrossRef]
- Okosun, J.; Bödör, C.; Wang, J.; Araf, S.; Yang, C.Y.; Pan, C.; Boller, S.; Cittaro, D.; Bozek, M.; Iqbal, S.; et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014, 46, 176–181. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Khiabanian, H.; Fangazio, M.; Vasishtha, M.; Messina, M.; Holmes, A.B.; Ouillette, P.; Trifonov, V.; Rossi, D.; Tabbò, F.; et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014, 6, 130–140. [Google Scholar] [CrossRef]
- Link, B.K.; Maurer, M.J.; Nowakowski, G.S.; Ansell, S.M.; Macon, W.R.; Syrbu, S.I.; Slager, S.L.; Thompson, C.A.; Inwards, D.J.; Johnston, P.B.; et al. Rates and outcomes of follicular lymphoma transformation in the immunochemotherapy era: A report from the University of Iowa/MayoClinic Specialized Program of Research Excellence Molecular Epidemiology Resource. J. Clin. Oncol. 2013, 31, 3272–3278. [Google Scholar] [CrossRef]
- Zucca, E.; Rossi, D.; Bertoni, F. Marginal zone lymphomas. Hematol. Oncol. 2023, 41 (Suppl. S1), 88–91. [Google Scholar] [CrossRef] [PubMed]
- Cerhan, J.R.; Habermann, T.M. Epidemiology of Marginal Zone Lymphoma. Ann. Lymphoma 2021, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Cheah, C.Y.; Seymour, J.F. Marginal zone lymphoma: 2023 update on diagnosis and management. Am. J. Hematol. 2023, 98, 1645–1657. [Google Scholar] [CrossRef] [PubMed]
- Cheah, C.Y.; Zucca, E.; Rossi, D.; Habermann, T.M. Marginal zone lymphoma: Present status and future perspectives. Haematologica 2022, 107, 35–43. [Google Scholar] [CrossRef]
- Varettoni, M.; Boveri, E.; Zibellini, S.; Tedeschi, A.; Candido, C.; Ferretti, V.V.; Rizzo, E.; Doni, E.; Merli, M.; Farina, L.; et al. Clinical and molecular characteristics of lymphoplasmacytic lymphoma not associated with an IgM monoclonal protein: A multicentric study of the Rete Ematologica Lombarda (REL) network. Am. J. Hematol. 2019, 94, 1193–1199. [Google Scholar] [CrossRef]
- Minderman, M.; Lantermans, H.; van der Zwaan, C.; Hoogendijk, A.J.; van den Biggelaar, M.; Kersten, M.J.; Spaargaren, M.; Pals, S.T. The oncogenic human B-cell lymphoma MYD88 L265P mutation genocopies activation by phosphorylation at the Toll/interleukin-1 receptor (TIR) domain. Blood Cancer J. 2023, 13, 125. [Google Scholar] [CrossRef]
- McMaster, M.L. The epidemiology of Waldenström macroglobulinemia. Semin. Hematol. 2023, 60, 65–72. [Google Scholar] [CrossRef]
- Kaiser, L.M.; Hunter, Z.R.; Treon, S.P.; Buske, C. CXCR4 in Waldenström’s Macroglobulinema: Chances and challenges. Leukemia 2021, 35, 333–345. [Google Scholar] [CrossRef]
- Berendsen, M.R.; van Bladel, D.A.G.; Hesius, E.; Berganza Irusquieta, C.; Rijntjes, J.; van Spriel, A.B.; van der Spek, E.; Pruijt, J.F.M.; Kroeze, L.I.; Hebeda, K.M.; et al. Clonal Relationship and Mutation Analysis in Lymphoplasmacytic Lymphoma/Waldenström Macroglobulinemia Associated With Diffuse Large B-cell Lymphoma. HemaSphere 2023, 7, e976. [Google Scholar] [CrossRef]
- Condoluci, A.; Rossi, D. Biology and Treatment of Richter Transformation. Front. Oncol. 2022, 12, 829983. [Google Scholar] [CrossRef]
- Kaźmierczak, M.; Kroll-Balcerzak, R.; Balcerzak, A.; Czechowska, E.; Gil, L.; Sawiński, K.; Szczepaniak, A.; Komarnicki, M. Hodgkin lymphoma transformation of chronic lymphocytic leukemia: Cases report and discussion. Med. Oncol. 2014, 31, 800. [Google Scholar] [CrossRef] [PubMed]
- Tsimberidou, A.M.; Keating, M.J. Richter syndrome: Biology, incidence, and therapeutic strategies. Cancer 2005, 103, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.; Gaidano, G.; Mouhssine, S. Immunological Aspects of Richter Syndrome: From Immune Dysfunction to Immunotherapy. Cancers 2023, 15, 1015. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Spina, V.; Deambrogi, C.; Rasi, S.; Laurenti, L.; Stamatopoulos, K.; Arcaini, L.; Lucioni, M.; Rocque, G.B.; Xu-Monette, Z.Y.; et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood 2011, 117, 3391–3401. [Google Scholar] [CrossRef]
- Bertossi, C.; Robrecht, S.; Ligtvoet, R.; Schneider, C.; Yosifov, D.; Zenz, T.; Jebaray, B.; Riecke, A.; Goede, V.; Fürstenau, M.; et al. S101—The Landscape of TP53 Mutations and Their Prognostic Impact in Chronic Lymphocytic Leukemia—EHA2024 Hybrid Congress. HemaSphere 2024, 8, e104. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 as a target for cancer treatment. Eur. J. Cancer 2017, 83, 258–265. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, J.; Long, Y.; Maimaitijiang, A.; Su, Z.; Li, W.; Li, J. Unraveling the Guardian: p53’s Multifaceted Role in the DNA Damage Response and Tumor Treatment Strategies. Int. J. Mol. Sci. 2024, 25, 12928. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef]
- Gatz, S.A.; Wiesmüller, L. p53 in recombination and repair. Cell Death Differ. 2006, 13, 1003–1016. [Google Scholar] [CrossRef]
- Chipuk, J.E.; Kuwana, T.; Bouchier-Hayes, L.; Droin, N.M.; Newmeyer, D.D.; Schuler, M.; Green, D.R. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 2004, 303, 1010–1014. [Google Scholar] [CrossRef]
- Wang, J.; Thomas, H.R.; Li, Z.; Yeo, N.C.F.; Scott, H.E.; Dang, N.; Hossain, M.I.; Andrabi, S.A.; Parant, J.M. Puma, noxa, p53, and p63 differentially mediate stress pathway induced apoptosis. Cell Death Dis. 2021, 12, 659. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, G.; Khiabanian, H.; Holmes, A.B.; Wang, J.; Messina, M.; Mullighan, C.G.; Pasqualucci, L.; Rabadan, R.; Dalla-Favera, R. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J. Exp. Med. 2013, 210, 2273–2288. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Ink4-Arf locus in cancer and aging. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001, 2, 731–737. [Google Scholar] [CrossRef]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef]
- Pellarin, I.; Dall’Acqua, A.; Favero, A.; Segatto, I.; Rossi, V.; Crestan, N.; Karimbayli, J.; Belletti, B.; Baldassarre, G. Cyclin-dependent protein kinases and cell cycle regulation in biology and disease. Signal Transduct. Target. Ther. 2025, 10, 11. [Google Scholar] [CrossRef]
- Hernández Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Chakraborty, S.; Martines, C.; Porro, F.; Fortunati, I.; Bonato, A.; Dimishkovska, M.; Piazza, S.; Yadav, B.S.; Innocenti, I.; Fazio, R.; et al. B-cell receptor signaling and genetic lesions in TP53 and CDKN2A/CDKN2B cooperate in Richter transformation. Blood 2021, 138, 1053–1066. [Google Scholar] [CrossRef]
- Fabbri, G.; Rasi, S.; Rossi, D.; Trifonov, V.; Khiabanian, H.; Ma, J.; Grunn, A.; Fangazio, M.; Capello, D.; Monti, S.; et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of NOTCH1 mutational activation. J. Exp. Med. 2011, 208, 1389–1401. [Google Scholar] [CrossRef]
- Andersson, E.R.; Sandberg, R.; Lendahl, U. Notch signaling: Simplicity in design, versatility in function. Development 2011, 138, 3593–3612. [Google Scholar] [CrossRef]
- Rosati, E.; Baldoni, S.; De Falco, F.; Del Papa, B.; Dorillo, E.; Rompietti, C.; Albi, E.; Falzetti, F.; Di Ianni, M.; Sportoletti, P. NOTCH1 Aberrations in Chronic Lymphocytic Leukemia. Front. Oncol. 2018, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- Di Ianni, M.; Baldoni, S.; Rosati, E.; Ciurnelli, R.; Cavalli, L.; Martelli, M.F.; Marconi, P.; Screpanti, I.; Falzetti, F. A new genetic lesion in B-CLL: A NOTCH1 PEST domain mutation. Br. J. Haematol. 2009, 146, 689–691. [Google Scholar] [CrossRef] [PubMed]
- Arruga, F.; Gizdic, B.; Bologna, C.; Cignetto, S.; Buonincontri, R.; Serra, S.; Vaisitti, T.; Gizzi, K.; Vitale, N.; Garaffo, G.; et al. Mutations in NOTCH1 PEST domain orchestrate CCL19-driven homing of chronic lymphocytic leukemia cells by modulating the tumor suppressor gene DUSP22. Leukemia 2017, 31, 1882–1893. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Rasi, S.; Spina, V.; Fangazio, M.; Monti, S.; Greco, M.; Ciardullo, C.; Famà, R.; Cresta, S.; Bruscaggin, A.; et al. Different impact of NOTCH1 and SF3B1 mutations on the risk of chronic lymphocytic leukemia transformation to Richter syndrome. Br. J. Haematol. 2012, 158, 426–429. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Palomero, T.; Lim, W.K.; Odom, D.T.; Sulis, M.L.; Real, P.J.; Margolin, A.; Barnes, K.C.; O’Neil, J.; Neuberg, D.; Weng, A.P.; et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 2006, 103, 18261–18266. [Google Scholar] [CrossRef]
- De Paoli, L.; Cerri, M.; Monti, S.; Rasi, S.; Spina, V.; Bruscaggin, A.; Greco, M.; Ciardullo, C.; Famà, R.; Cresta, S.; et al. MGA, a suppressor of MYC, is recurrently inactivated in high risk chronic lymphocytic leukemia. Leuk. Lymphoma 2013, 54, 1087–1090. [Google Scholar] [CrossRef]
- Martines, C.; Chakraborty, S.; Vujovikj, M.; Gobessi, S.; Vaisitti, T.; Deaglio, S.; Laurenti, L.; Dimovski, A.J.; Efremov, D.G. Macrophage- and BCR- but not TLR-derived signals support the growth of CLL and Richter Syndrome murine models in vivo. Blood 2022, 140, 2335–2347. [Google Scholar] [CrossRef]
- Herishanu, Y.; Pérez-Galán, P.; Liu, D.; Biancotto, A.; Pittaluga, S.; Vire, B.; Gibellini, F.; Njuguna, N.; Lee, E.; Stennett, L.; et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-κB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood 2011, 117, 563–574. [Google Scholar] [CrossRef]
- Ringshausen, I.; Schneller, F.; Bogner, C.; Hipp, S.; Duyster, J.; Peschel, C.; Decker, T. Constitutively activated phosphatidylinositol-3 kinase (PI-3K) is involved in the defect of apoptosis in B-CLL: Association with protein kinase Cdelta. Blood 2002, 100, 3741–3748. [Google Scholar] [CrossRef] [PubMed]
- Kohlhaas, V.; Blakemore, S.J.; Al-Maarri, M.; Nickel, N.; Pal, M.; Roth, A.; Hövelmeyer, N.; Schäfer, S.C.; Knittel, G.; Lohneis, P.; et al. Active Akt signaling triggers CLL toward Richter transformation via overactivation of Notch1. Blood 2021, 137, 646–660. [Google Scholar] [CrossRef] [PubMed]
- Dühren-von Minden, M.; Übelhart, R.; Schneider, D.; Wossning, T.; Bach, M.P.; Buchner, M.; Hofmann, D.; Surova, E.; Follo, M.; Köhler, F.; et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 2012, 489, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Chiorazzi, N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 2013, 34, 592–601. [Google Scholar] [CrossRef]
- Stevenson, F.K.; Forconi, F.; Kipps, T.J. Exploring the pathways to chronic lymphocytic leukemia. Blood 2021, 138, 827–835. [Google Scholar] [CrossRef]
- Stamatopoulos, K.; Belessi, C.; Moreno, C.; Boudjograh, M.; Guida, G.; Smilevska, T.; Belhoul, L.; Stella, S.; Stavroyianni, N.; Crespo, M.; et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood 2007, 109, 259–270. [Google Scholar] [CrossRef]
- Gerousi, M.; Laidou, S.; Gemenetzi, K.; Stamatopoulos, K.; Chatzidimitriou, A. Distinctive Signaling Profiles With Distinct Biological and Clinical Implications in Aggressive CLL Subsets With Stereotyped B-Cell Receptor Immunoglobulin. Front. Oncol. 2021, 11, 771454. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Cerri, M.; Rasi, S.; Deambrogi, C.; De Paoli, L.; Laurenti, L.; Maffei, R.; Forconi, F.; Bertoni, F.; et al. Stereotyped B-cell receptor is an independent risk factor of chronic lymphocytic leukemia transformation to Richter syndrome. Clin. Cancer Res. 2009, 15, 4415–4422. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Bomben, R.; Rasi, S.; Dal-Bo, M.; Bruscaggin, A.; Rossi, F.M.; Monti, S.; Degan, M.; Ciardullo, C.; et al. Association between molecular lesions and specific B-cell receptor subsets in chronic lymphocytic leukemia. Blood 2013, 121, 4902–4905. [Google Scholar] [CrossRef]
- Gounari, M.; Ntoufa, S.; Apollonio, B.; Papakonstantinou, N.; Ponzoni, M.; Chu, C.C.; Rossi, D.; Gaidano, G.; Chiorazzi, N.; Stamatopoulos, K.; et al. Excessive antigen reactivity may underlie the clinical aggressiveness of chronic lymphocytic leukemia stereotyped subset #8. Blood 2015, 125, 3580–3587. [Google Scholar] [CrossRef]
- Nabki, J.; Al Deeban, B.; Sium, A.M.; Cosentino, C.; Almasri, M.; Awikeh, B.; Maher, N.; Bellia, M.; Dondolin, R.; Mouhssine, S.; et al. Immunoglobulin light chain mutational status refines IGHV prognostic value in identifying chronic lymphocytic leukemia patients with early treatment requirement. Leukemia 2024, 39, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Salmaninejad, A.; Valilou, S.F.; Shabgah, A.G.; Aslani, S.; Alimardani, M.; Pasdar, A.; Sahebkar, A. PD-1/PD-L1 pathway: Basic biology and role in cancer immunotherapy. J. Cell Physiol. 2019, 234, 16824–16837. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sinha, S.; Wellik, L.E.; Secreto, C.R.; Rech, K.L.; Call, T.G.; Parikh, S.A.; Kenderian, S.S.; Muchtar, E.; Hayman, S.R.; et al. Distinct immune signatures in chronic lymphocytic leukemia and Richter syndrome. Blood Cancer J. 2021, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Behdad, A.; Griffin, B.; Chen, Y.-H.; Ma, S.; Kelemen, K.; Lu, X.; Chen, Q.C. PD-1 is highly expressed by neoplastic B-cells in Richter transformation. Br. J. Haematol. 2019, 185, 370–373. [Google Scholar] [CrossRef]
- Li, X.; Du, H.; Zhan, S.; Liu, W.; Wang, Z.; Lan, J.; PuYang, L.; Wan, Y.; Qu, Q.; Wang, S.; et al. The interaction between the soluble programmed death ligand-1 (sPD-L1) and PD-1(+) regulator B cells mediates immunosuppression in triple-negative breast cancer. Front. Immunol. 2022, 13, 830606. [Google Scholar] [CrossRef]
- Xiao, X.; Lao, X.M.; Chen, M.M.; Liu, R.X.; Wei, Y.; Ouyang, F.Z.; Chen, D.P.; Zhao, X.Y.; Zhao, Q.; Li, X.F.; et al. PD-1hi Identifies a Novel Regulatory B-cell Population in Human Hepatoma That Promotes Disease Progression. Cancer Discov. 2016, 6, 546–559. [Google Scholar] [CrossRef]
- Wang, X.; Wang, G.; Wang, Z.; Liu, B.; Han, N.; Li, J.; Lu, C.; Liu, X.; Zhang, Q.; Yang, Q.; et al. PD-1-expressing B cells suppress CD4+ and CD8+ T cells via PD-1/PD-L1-dependent pathway. Mol. Immunol. 2019, 109, 20–26. [Google Scholar] [CrossRef]
- Augé, H.; Notarantonio, A.-B.; Morizot, R.; Quinquenel, A.; Fornecker, L.-M.; Hergalant, S.; Feugier, P.; Broséus, J. Microenvironment Remodeling and Subsequent Clinical Implications in Diffuse Large B-Cell Histologic Variant of Richter Syndrome. Front. Immunol. 2020, 11, 594841. [Google Scholar] [CrossRef]
- Triebel, F.; Jitsukawa, S.; Baixeras, E.; Roman-Roman, S.; Genevee, C.; Viegas-Pequignot, E.; Hercend, T. LAG-3, a novel lymphocyte activation gene closely related to CD4. Journal of Experimental Medicine 1990, 171, 1393–1405. [Google Scholar] [CrossRef]
- Hannier, S.; Triebel, F. The MHC class II ligand lymphocyte activation gene-3 is co-distributed with CD8 and CD3–TCR molecules after their engagement by mAb or peptide–MHC class I complexes. Int. Immunol. 1999, 11, 1745–1752. [Google Scholar] [CrossRef]
- Ruffo, E.; Wu, R.C.; Bruno, T.C.; Workman, C.J.; Vignali, D.A.A. Lymphocyte-activation gene 3 (LAG3): The next immune checkpoint receptor. Semin. Immunol. 2019, 42, 101305. [Google Scholar] [CrossRef] [PubMed]
- Hemon, P.; Jean-Louis, F.; Ramgolam, K.; Brignone, C.; Viguier, M.; Bachelez, H.; Triebel, F.; Charron, D.; Aoudjit, F.; Al-Daccak, R.; et al. MHC Class II Engagement by Its Ligand LAG-3 (CD223) Contributes to Melanoma Resistance to Apoptosis. J. Immunol. 2011, 186, 5173–5183. [Google Scholar] [CrossRef] [PubMed]
- Harjunpää, H.; Guillerey, C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 2020, 200, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Qi, G.; Miller, J.S.; Zheng, S.G. CD226: An Emerging Role in Immunologic Diseases. Front. Cell Dev. Biol. 2020, 8, 564. [Google Scholar] [CrossRef]
- Arruga, F.; Iannello, A.; Ioannou, N.; Todesco, A.M.; Coscia, M.; Moia, R.; Gaidano, G.; Allan, J.N.; Furman, R.; Vaisitti, T.; et al. The Tigit/CD226/CD155 Immunomodulatory Axis Is Deregulated in CLL and Contributes to B-Cell Anergy. Blood 2021, 138, 3718. [Google Scholar] [CrossRef]
- Arruga, F.; Gyau, B.B.; Iannello, A.; Vitale, N.; Vaisitti, T.; Deaglio, S. Immune Response Dysfunction in Chronic Lymphocytic Leukemia: Dissecting Molecular Mechanisms and Microenvironmental Conditions. Int. J. Mol. Sci. 2020, 21, 1825. [Google Scholar] [CrossRef]
- Tedeschi, A.; Frustaci, A.M.; Condoluci, A.; Coscia, M.; Chiarle, R.; Zinzani, P.L.; Motta, M.; Gaidano, G.; Quaresmini, G.; Scarfò, L.; et al. Atezolizumab, venetoclax, and obinutuzumab combination in Richter transformation diffuse large B-cell lymphoma (MOLTO): A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2024, 25, 1298–1309. [Google Scholar] [CrossRef]
- Al-Sawaf, O.; Ligtvoet, R.; Robrecht, S.; Stumpf, J.; Fink, A.M.; Tausch, E.; Schneider, C.; Boettcher, S.; Mikusko, M.; Ritgen, M.; et al. Tislelizumab plus zanubrutinib for Richter transformation: The phase 2 RT1 trial. Nat. Med. 2024, 30, 240–248. [Google Scholar] [CrossRef]
- Parry, E.M.; Ten Hacken, E.; Wu, C.J. Richter syndrome: Novel insights into the biology of transformation. Blood 2023, 142, 11–22. [Google Scholar] [CrossRef]
- Parry, E.M.; Leshchiner, I.; Guièze, R.; Johnson, C.; Tausch, E.; Parikh, S.A.; Lemvigh, C.; Broséus, J.; Hergalant, S.; Messer, C.; et al. Evolutionary history of transformation from chronic lymphocytic leukemia to Richter syndrome. Nat. Med. 2023, 29, 158–169. [Google Scholar] [CrossRef]
- Rossi, D. XIII. Molecular pathogenesis of transformed lymphomas. Hematol. Oncol. 2015, 33 (Suppl. S1), 70–74. [Google Scholar] [CrossRef] [PubMed]
- Alcoceba, M.; García-Álvarez, M.; Okosun, J.; Ferrero, S.; Ladetto, M.; Fitzgibbon, J.; García-Sanz, R. Genetics of Transformed Follicular Lymphoma. Hemato 2022, 3, 615–633. [Google Scholar] [CrossRef]
- Alhejaily, A.; Day, A.G.; Feilotter, H.E.; Baetz, T.; Lebrun, D.P. Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin. Cancer Res. 2014, 20, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- Elenitoba-Johnson, K.S.; Gascoyne, R.D.; Lim, M.S.; Chhanabai, M.; Jaffe, E.S.; Raffeld, M. Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood 1998, 91, 4677–4685. [Google Scholar] [CrossRef]
- Kridel, R.; Chan, F.C.; Mottok, A.; Boyle, M.; Farinha, P.; Tan, K.; Meissner, B.; Bashashati, A.; McPherson, A.; Roth, A.; et al. Histological Transformation and Progression in Follicular Lymphoma: A Clonal Evolution Study. PLoS Med. 2016, 13, e1002197. [Google Scholar] [CrossRef]
- Musilova, K.; Devan, J.; Cerna, K.; Seda, V.; Pavlasova, G.; Sharma, S.; Oppelt, J.; Pytlik, R.; Prochazka, V.; Prouzova, Z.; et al. miR-150 downregulation contributes to the high-grade transformation of follicular lymphoma by upregulating FOXP1 levels. Blood 2018, 132, 2389–2400. [Google Scholar] [CrossRef]
- O’Shea, D.; O’Riain, C.; Taylor, C.; Waters, R.; Carlotti, E.; Macdougall, F.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. The presence of TP53 mutation at diagnosis of follicular lymphoma identifies a high-risk group of patients with shortened time to disease progression and poorer overall survival. Blood 2008, 112, 3126–3129. [Google Scholar] [CrossRef]
- Lo Coco, F.; Gaidano, G.; Louie, D.C.; Offit, K.; Chaganti, R.S.; Dalla-Favera, R. p53 mutations are associated with histologic transformation of follicular lymphoma. Blood 1993, 82, 2289–2295. [Google Scholar] [CrossRef]
- Davies, A.J.; Lee, A.M.; Taylor, C.; Clear, A.J.; Goff, L.K.; Iqbal, S.; Cuthbert-Heavens, D.; Calaminici, M.; Norton, A.J.; Lister, T.A.; et al. A limited role for TP53 mutation in the transformation of follicular lymphoma to diffuse large B-cell lymphoma. Leukemia 2005, 19, 1459–1465. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Green, M.R. Epigenetic Programing of B-Cell Lymphoma by BCL6 and Its Genetic Deregulation. Front. Cell Dev. Biol. 2019, 7, 272. [Google Scholar] [CrossRef]
- Phan, R.T.; Dalla-Favera, R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 2004, 432, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Ranuncolo, S.M.; Polo, J.M.; Dierov, J.; Singer, M.; Kuo, T.; Greally, J.; Green, R.; Carroll, M.; Melnick, A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat. Immunol. 2007, 8, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Phan, R.T.; Saito, M.; Basso, K.; Niu, H.; Dalla-Favera, R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 2005, 6, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.H.; Lista, F.; Lo Coco, F.; Knowles, D.M.; Offit, K.; Chaganti, R.S.; Dalla-Favera, R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 1993, 262, 747–750. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.J.; Kim, S.H. Diagnostic Approach for Double-Hit and Triple-Hit Lymphoma Based on Immunophenotypic and Cytogenetic Characteristics of Bone Marrow Specimens. Ann. Lab. Med. 2020, 40, 361–369. [Google Scholar] [CrossRef]
- Kridel, R.; Mottok, A.; Farinha, P.; Ben-Neriah, S.; Ennishi, D.; Zheng, Y.; Chavez, E.A.; Shulha, H.P.; Tan, K.; Chan, F.C.; et al. Cell of origin of transformed follicular lymphoma. Blood 2015, 126, 2118–2127. [Google Scholar] [CrossRef]
- Akasaka, T.; Lossos, I.S.; Levy, R. BCL6 gene translocation in follicular lymphoma: A harbinger of eventual transformation to diffuse aggressive lymphoma. Blood 2003, 102, 1443–1448. [Google Scholar] [CrossRef]
- Iqbal, J.; Greiner, T.C.; Patel, K.; Dave, B.J.; Smith, L.; Ji, J.; Wright, G.; Sanger, W.G.; Pickering, D.L.; Jain, S.; et al. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia 2007, 21, 2332–2343. [Google Scholar] [CrossRef]
- Bouska, A.; McKeithan, T.W.; Deffenbacher, K.E.; Lachel, C.; Wright, G.W.; Iqbal, J.; Smith, L.M.; Zhang, W.; Kucuk, C.; Rinaldi, A.; et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood 2014, 123, 1681–1690. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Burns, K.; Martinon, F.; Esslinger, C.; Pahl, H.; Schneider, P.; Bodmer, J.L.; Di Marco, F.; French, L.; Tschopp, J. MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 1998, 273, 12203–12209. [Google Scholar] [CrossRef] [PubMed]
- Dunne, A.; Ejdeback, M.; Ludidi, P.L.; O’Neill, L.A.; Gay, N.J. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J. Biol. Chem. 2003, 278, 41443–41451. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Commane, M.; Ninomiya-Tsuji, J.; Matsumoto, K.; Li, X. IRAK-mediated translocation of TRAF6 and TAB2 in the interleukin-1-induced activation of NFkappa B. J. Biol. Chem. 2001, 276, 41661–41667. [Google Scholar] [CrossRef] [PubMed]
- Deguine, J.; Barton, G.M. MyD88: A central player in innate immune signaling. F1000Prime Rep. 2014, 6, 97. [Google Scholar] [CrossRef]
- Bouska, A.; Zhang, W.; Gong, Q.; Iqbal, J.; Scuto, A.; Vose, J.; Ludvigsen, M.; Fu, K.; Weisenburger, D.D.; Greiner, T.C.; et al. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia 2017, 31, 83–91. [Google Scholar] [CrossRef]
- Kwiecinska, A.; Ichimura, K.; Berglund, M.; Dinets, A.; Sulaiman, L.; Collins, V.P.; Larsson, C.; Porwit, A.; Lagercrantz, S.B. Amplification of 2p as a genomic marker for transformation in lymphoma. Genes Chromosomes Cancer 2014, 53, 750–768. [Google Scholar] [CrossRef]
- Davies, A.J.; Rosenwald, A.; Wright, G.; Lee, A.; Last, K.W.; Weisenburger, D.D.; Chan, W.C.; Delabie, J.; Braziel, R.M.; Campo, E.; et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma proceeds by distinct oncogenic mechanisms. Br. J. Haematol. 2007, 136, 286–293. [Google Scholar] [CrossRef]
- Cai, G.; Freeman, G.J. The CD160, BTLA, LIGHT/HVEM pathway: A bidirectional switch regulating T-cell activation. Immunol. Rev. 2009, 229, 244–258. [Google Scholar] [CrossRef]
- Murphy, K.M.; Nelson, C.A.; Sedý, J.R. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat. Rev. Immunol. 2006, 6, 671–681. [Google Scholar] [CrossRef]
- O’Shea, D.; O’Riain, C.; Gupta, M.; Waters, R.; Yang, Y.; Wrench, D.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. Regions of acquired uniparental disomy at diagnosis of follicular lymphoma are associated with both overall survival and risk of transformation. Blood 2009, 113, 2298–2301. [Google Scholar] [CrossRef]
- García-Álvarez, M.; Alonso-Álvarez, S.; Prieto-Conde, I.; Jiménez, C.; Sarasquete, M.E.; Chillón, M.C.; Medina, A.; Balanzategui, A.; Maldonado, R.; Antón, A.; et al. Genetic complexity impacts the clinical outcome of follicular lymphoma patients. Blood Cancer J. 2021, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Shah, S.P.; Steidl, C.; Johnson, N.; Relander, T.; Telenius, A.; Lai, B.; Murphy, K.P.; Lam, W.; Al-Tourah, A.J.; et al. Genome-wide profiling of follicular lymphoma by array comparative genomic hybridization reveals prognostically significant DNA copy number imbalances. Blood 2009, 113, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Kotsiou, E.; Okosun, J.; Besley, C.; Iqbal, S.; Matthews, J.; Fitzgibbon, J.; Gribben, J.G.; Davies, J.K. TNFRSF14 aberrations in follicular lymphoma increase clinically significant allogeneic T-cell responses. Blood 2016, 128, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Boice, M.; Salloum, D.; Mourcin, F.; Sanghvi, V.; Amin, R.; Oricchio, E.; Jiang, M.; Mottok, A.; Denis-Lagache, N.; Ciriello, G.; et al. Loss of the HVEM Tumor Suppressor in Lymphoma and Restoration by Modified CAR-T Cells. Cell 2016, 167, 405–418.e413. [Google Scholar] [CrossRef]
- Carreras, J.; Lopez-Guillermo, A.; Kikuti, Y.Y.; Itoh, J.; Masashi, M.; Ikoma, H.; Tomita, S.; Hiraiwa, S.; Hamoudi, R.; Rosenwald, A.; et al. High TNFRSF14 and low BTLA are associated with poor prognosis in Follicular Lymphoma and in Diffuse Large B-cell Lymphoma transformation. J. Clin. Exp. Hematop. 2019, 59, 1–16. [Google Scholar] [CrossRef]
- Casulo, C.; Friedberg, J. Transformation of marginal zone lymphoma (and association with other lymphomas). Best. Pr. Res. Clin. Haematol. 2017, 30, 131–138. [Google Scholar] [CrossRef]
- Parry, M.; Rose-Zerilli, M.J.; Ljungström, V.; Gibson, J.; Wang, J.; Walewska, R.; Parker, H.; Parker, A.; Davis, Z.; Gardiner, A.; et al. Genetics and Prognostication in Splenic Marginal Zone Lymphoma: Revelations from Deep Sequencing. Clin. Cancer Res. 2015, 21, 4174–4183. [Google Scholar] [CrossRef]
- Rossi, D.; Trifonov, V.; Fangazio, M.; Bruscaggin, A.; Rasi, S.; Spina, V.; Monti, S.; Vaisitti, T.; Arruga, F.; Famà, R.; et al. The coding genome of splenic marginal zone lymphoma: Activation of NOTCH2 and other pathways regulating marginal zone development. J. Exp. Med. 2012, 209, 1537–1551. [Google Scholar] [CrossRef]
- Kiel, M.J.; Velusamy, T.; Betz, B.L.; Zhao, L.; Weigelin, H.G.; Chiang, M.Y.; Huebner-Chan, D.R.; Bailey, N.G.; Yang, D.T.; Bhagat, G.; et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J. Exp. Med. 2012, 209, 1553–1565. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006, 7, 678–689. [Google Scholar] [CrossRef]
- Sanchez-Irizarry, C.; Carpenter, A.C.; Weng, A.P.; Pear, W.S.; Aster, J.C.; Blacklow, S.C. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell Biol. 2004, 24, 9265–9273. [Google Scholar] [CrossRef] [PubMed]
- Trøen, G.; Wlodarska, I.; Warsame, A.; Hernández Llodrà, S.; De Wolf-Peeters, C.; Delabie, J. NOTCH2 mutations in marginal zone lymphoma. Haematologica 2008, 93, 1107–1109. [Google Scholar] [CrossRef] [PubMed]
- Grau, M.; López, C.; Navarro, A.; Frigola, G.; Nadeu, F.; Clot, G.; Bastidas-Mora, G.; Alcoceba, M.; Baptista, M.J.; Blanes, M.; et al. Unraveling the genetics of transformed splenic marginal zone lymphoma. Blood Adv. 2023, 7, 3695–3709. [Google Scholar] [CrossRef] [PubMed]
- Salido, M.; Baró, C.; Oscier, D.; Stamatopoulos, K.; Dierlamm, J.; Matutes, E.; Traverse-Glehen, A.; Berger, F.; Felman, P.; Thieblemont, C.; et al. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: A multicenter study of the Splenic B-Cell Lymphoma Group. Blood 2010, 116, 1479–1488. [Google Scholar] [CrossRef]
- Rinaldi, A.; Mian, M.; Chigrinova, E.; Arcaini, L.; Bhagat, G.; Novak, U.; Rancoita, P.M.; De Campos, C.P.; Forconi, F.; Gascoyne, R.D.; et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood 2011, 117, 1595–1604. [Google Scholar] [CrossRef]
- Martínez, N.; Almaraz, C.; Vaqué, J.P.; Varela, I.; Derdak, S.; Beltran, S.; Mollejo, M.; Campos-Martin, Y.; Agueda, L.; Rinaldi, A.; et al. Whole-exome sequencing in splenic marginal zone lymphoma reveals mutations in genes involved in marginal zone differentiation. Leukemia 2014, 28, 1334–1340. [Google Scholar] [CrossRef]
- Fresquet, V.; Robles, E.F.; Parker, A.; Martinez-Useros, J.; Mena, M.; Malumbres, R.; Agirre, X.; Catarino, S.; Arteta, D.; Osaba, L.; et al. High-throughput sequencing analysis of the chromosome 7q32 deletion reveals IRF5 as a potential tumour suppressor in splenic marginal-zone lymphoma. Br. J. Haematol. 2012, 158, 712–726. [Google Scholar] [CrossRef]
- Pujari, R.; Hunte, R.; Khan, W.N.; Shembade, N. A20-mediated negative regulation of canonical NF-κB signaling pathway. Immunol. Res. 2013, 57, 166–171. [Google Scholar] [CrossRef]
- Jaramillo Oquendo, C.; Parker, H.; Oscier, D.; Ennis, S.; Gibson, J.; Strefford, J.C. Systematic Review of Somatic Mutations in Splenic Marginal Zone Lymphoma. Sci. Rep. 2019, 9, 10444. [Google Scholar] [CrossRef]
- Bonfiglio, F.; Bruscaggin, A.; Guidetti, F.; Terzi di Bergamo, L.; Faderl, M.; Spina, V.; Condoluci, A.; Bonomini, L.; Forestieri, G.; Koch, R.; et al. Genetic and phenotypic attributes of splenic marginal zone lymphoma. Blood 2022, 139, 732–747. [Google Scholar] [CrossRef]
- Wittner, J.; Schuh, W. Krüppel-like factor 2: A central regulator of B cell differentiation and plasma cell homing. Front. Immunol. 2023, 14, 1172641. [Google Scholar] [CrossRef]
- Spina, V.; Khiabanian, H.; Messina, M.; Monti, S.; Cascione, L.; Bruscaggin, A.; Spaccarotella, E.; Holmes, A.B.; Arcaini, L.; Lucioni, M.; et al. The genetics of nodal marginal zone lymphoma. Blood 2016, 128, 1362–1373. [Google Scholar] [CrossRef] [PubMed]
- Clipson, A.; Wang, M.; de Leval, L.; Ashton-Key, M.; Wotherspoon, A.; Vassiliou, G.; Bolli, N.; Grove, C.; Moody, S.; Escudero-Ibarz, L.; et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia 2015, 29, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Arribas, A.J.; Rinaldi, A.; Mensah, A.A.; Kwee, I.; Cascione, L.; Robles, E.F.; Martinez-Climent, J.A.; Oscier, D.; Arcaini, L.; Baldini, L.; et al. DNA methylation profiling identifies two splenic marginal zone lymphoma subgroups with different clinical and genetic features. Blood 2015, 125, 1922–1931. [Google Scholar] [CrossRef]
- Arribas, A.J.; Bertoni, F. Methylation patterns in marginal zone lymphoma. Best. Pr. Res. Clin. Haematol. 2017, 30, 24–31. [Google Scholar] [CrossRef]
- Sagaert, X.; de Paepe, P.; Libbrecht, L.; Vanhentenrijk, V.; Verhoef, G.; Thomas, J.; Wlodarska, I.; De Wolf-Peeters, C. Forkhead box protein P1 expression in mucosa-associated lymphoid tissue lymphomas predicts poor prognosis and transformation to diffuse large B-cell lymphoma. J. Clin. Oncol. 2006, 24, 2490–2497. [Google Scholar] [CrossRef]
- Streubel, B.; Vinatzer, U.; Lamprecht, A.; Raderer, M.; Chott, A. T(3;14)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia 2005, 19, 652–658. [Google Scholar] [CrossRef]
- Haralambieva, E.; Adam, P.; Ventura, R.; Katzenberger, T.; Kalla, J.; Höller, S.; Hartmann, M.; Rosenwald, A.; Greiner, A.; Muller-Hermelink, H.K.; et al. Genetic rearrangement of FOXP1 is predominantly detected in a subset of diffuse large B-cell lymphomas with extranodal presentation. Leukemia 2006, 20, 1300–1303. [Google Scholar] [CrossRef]
- Barrans, S.L.; Fenton, J.A.; Banham, A.; Owen, R.G.; Jack, A.S. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood 2004, 104, 2933–2935. [Google Scholar] [CrossRef]
- Qian, L.; Soderquist, C.; Schrank-Hacker, A.; Strauser, H.; Dupoux, V.; Tang, C.N.; Smith, J.R.; Sun, A.; Majumdar, S.; Nguyen, T.; et al. Deletion 20q12 is associated with histological transformation of nodal marginal zone lymphoma to diffuse large B-cell lymphoma. Am. J. Hematol. 2020, 95, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Treon, S.P.; Gustine, J.; Xu, L.; Manning, R.J.; Tsakmaklis, N.; Demos, M.; Meid, K.; Guerrera, M.L.; Munshi, M.; Chan, G.; et al. MYD88 wild-type Waldenstrom Macroglobulinaemia: Differential diagnosis, risk of histological transformation, and overall survival. Br. J. Haematol. 2018, 180, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Hunter, Z.R.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Manning, R.J.; Tripsas, C.; Patterson, C.J.; Sheehy, P.; et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood 2014, 123, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Varettoni, M.; Zibellini, S.; Defrancesco, I.; Ferretti, V.V.; Rizzo, E.; Malcovati, L.; Gallì, A.; Della Porta, M.G.; Boveri, E.; Arcaini, L.; et al. Pattern of somatic mutations in patients with Waldenström macroglobulinemia or IgM monoclonal gammopathy of undetermined significance. Haematologica 2017, 102, 2077–2085. [Google Scholar] [CrossRef]
- Rossi, D.; Zlotnik, A. The biology of chemokines and their receptors. Annu. Rev. Immunol. 2000, 18, 217–242. [Google Scholar] [CrossRef]
- Cao, Y.; Hunter, Z.R.; Liu, X.; Xu, L.; Yang, G.; Chen, J.; Patterson, C.J.; Tsakmaklis, N.; Kanan, S.; Rodig, S.; et al. The WHIM-like CXCR4(S338X) somatic mutation activates AKT and ERK, and promotes resistance to ibrutinib and other agents used in the treatment of Waldenstrom’s Macroglobulinemia. Leukemia 2015, 29, 169–176. [Google Scholar] [CrossRef]
- Wang, H.; Guo, M.; Wei, H.; Chen, Y. Targeting p53 pathways: Mechanisms, structures and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 92. [Google Scholar] [CrossRef]
- Weber, L.I.; Hartl, M. Strategies to target the cancer driver MYC in tumor cells. Front. Oncol. 2023, 13, 1142111. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Teierle, S.M.; Huang, Y.; Kittai, A.S.; Bhat, S.A.; Grever, M.R.; Rogers, K.A.; Zhao, W.; Jones, D.; Byrd, J.C.; Avenarius, M.R.; et al. FISH Analysis of CDKN2A/B Deletion in Chronic Lymphocytic Leukemia. Blood 2022, 140, 7016–7017. [Google Scholar] [CrossRef]
- Klintman, J.; Appleby, N.; Stamatopoulos, B.; Ridout, K.; Eyre, T.A.; Robbe, P.; Pascua, L.L.; Knight, S.J.L.; Dreau, H.; Cabes, M.; et al. Genomic and transcriptomic correlates of Richter transformation in chronic lymphocytic leukemia. Blood 2021, 137, 2800–2816. [Google Scholar] [CrossRef]
- Parker, H.; Rose-Zerilli, M.J.J.; Larrayoz, M.; Clifford, R.; Edelmann, J.; Blakemore, S.; Gibson, J.; Wang, J.; Ljungström, V.; Wojdacz, T.K.; et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia 2016, 30, 2179–2186. [Google Scholar] [CrossRef]
- Mouhssine, S.; Maher, N.; Gaidano, G. A STEP ahead for CAR-T cell therapy of large B cell lymphoma: Understanding the molecular determinants of resistance. Transl. Cancer Res. 2023, 12, 2970–2975. [Google Scholar] [CrossRef]
- Sworder, B.J.; Kurtz, D.M.; Alig, S.K.; Frank, M.J.; Shukla, N.; Garofalo, A.; Macaulay, C.W.; Shahrokh Esfahani, M.; Olsen, M.N.; Hamilton, J.; et al. Determinants of resistance to engineered T cell therapies targeting CD19 in large B cell lymphomas. Cancer Cell 2023, 41, 210–225.e215. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maher, N.; Mouhssine, S.; Matti, B.F.; Alwan, A.F.; Gaidano, G. Molecular Mechanisms in the Transformation from Indolent to Aggressive B Cell Malignancies. Cancers 2025, 17, 907. https://doi.org/10.3390/cancers17050907
Maher N, Mouhssine S, Matti BF, Alwan AF, Gaidano G. Molecular Mechanisms in the Transformation from Indolent to Aggressive B Cell Malignancies. Cancers. 2025; 17(5):907. https://doi.org/10.3390/cancers17050907
Chicago/Turabian StyleMaher, Nawar, Samir Mouhssine, Bassam Francis Matti, Alaa Fadhil Alwan, and Gianluca Gaidano. 2025. "Molecular Mechanisms in the Transformation from Indolent to Aggressive B Cell Malignancies" Cancers 17, no. 5: 907. https://doi.org/10.3390/cancers17050907
APA StyleMaher, N., Mouhssine, S., Matti, B. F., Alwan, A. F., & Gaidano, G. (2025). Molecular Mechanisms in the Transformation from Indolent to Aggressive B Cell Malignancies. Cancers, 17(5), 907. https://doi.org/10.3390/cancers17050907







