Novel Recurrent Cytogenetic Abnormalities Predict Overall Survival in Tetraploid/Near-Tetraploid Myelodysplastic Syndrome and Acute Myeloid Leukemia
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection
2.2. Hematopathology and Conventional Cytogenetic Analyses
2.3. CytoGPS
2.4. Statistical Analyses
3. Results
3.1. Clinical Findings
3.2. Recurrent Cytogenetic Abnormalities in T/NT MDS/AML Identified by CytoGPS
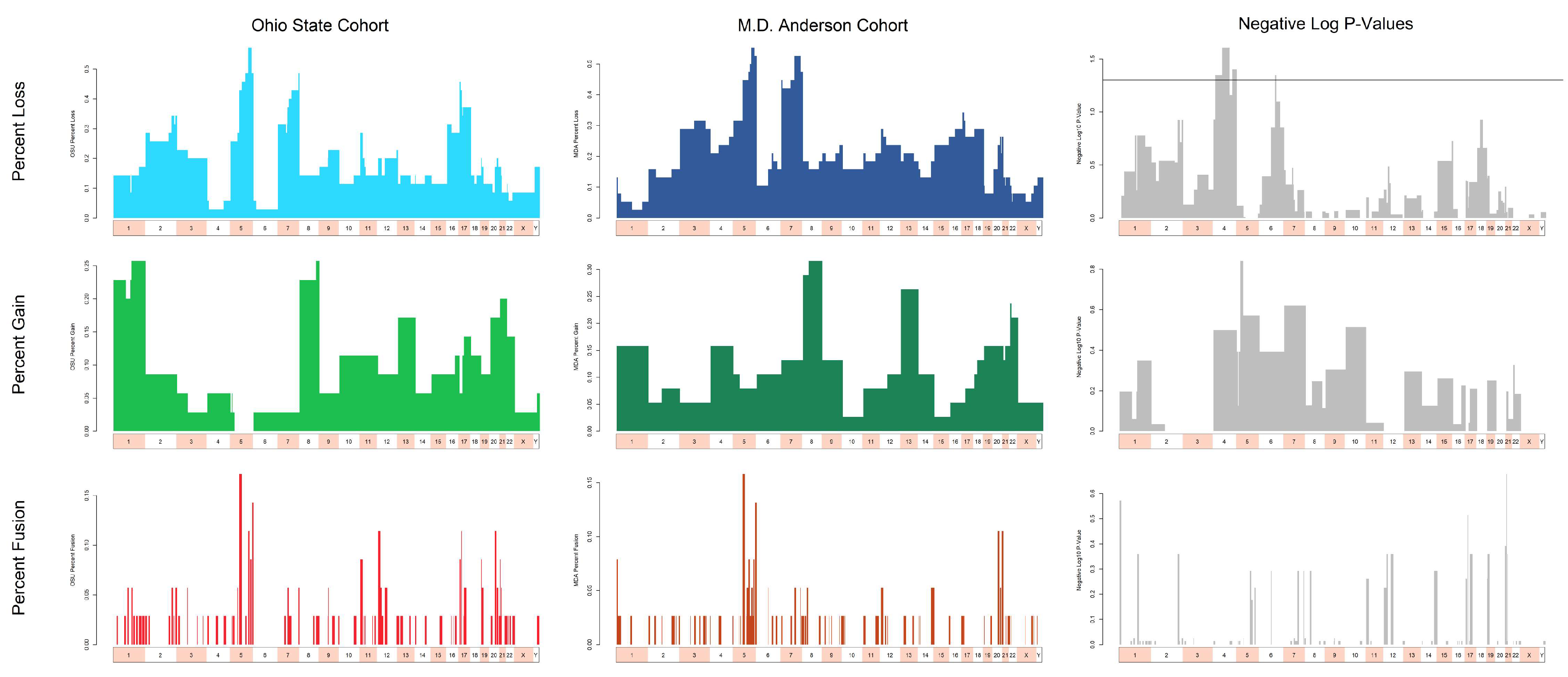
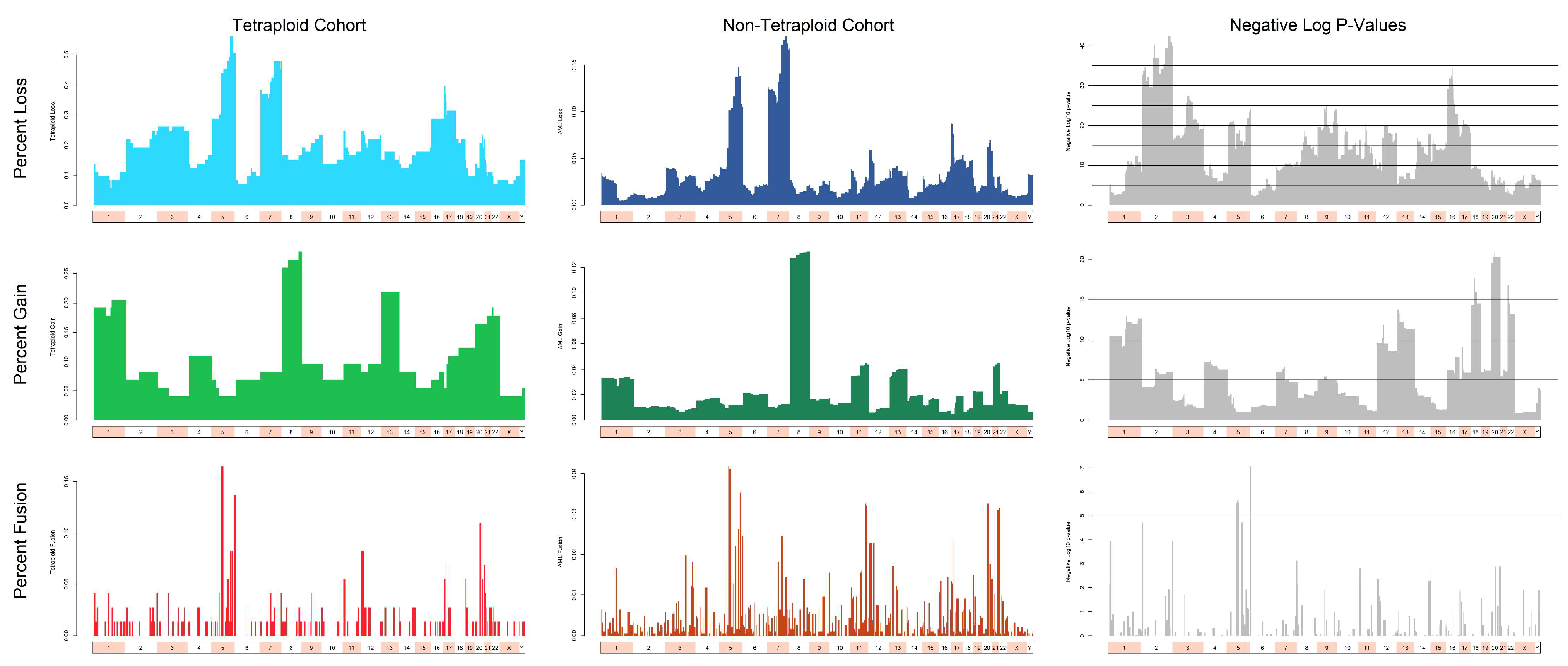
3.3. Cytogenetic Abnormalities Over- and Under-Represented in T/NT Compared to Non-T/NT Karyotypes
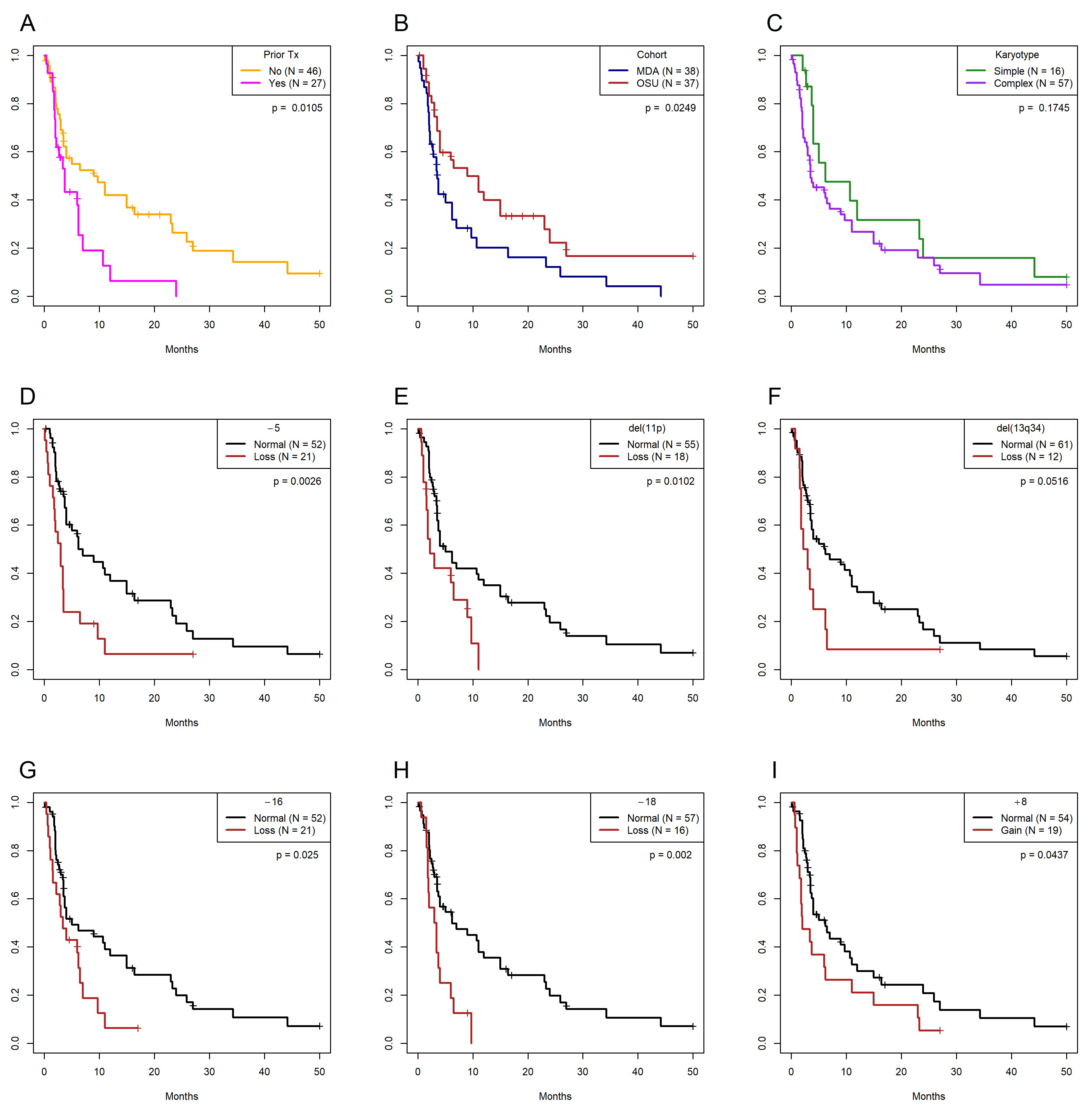
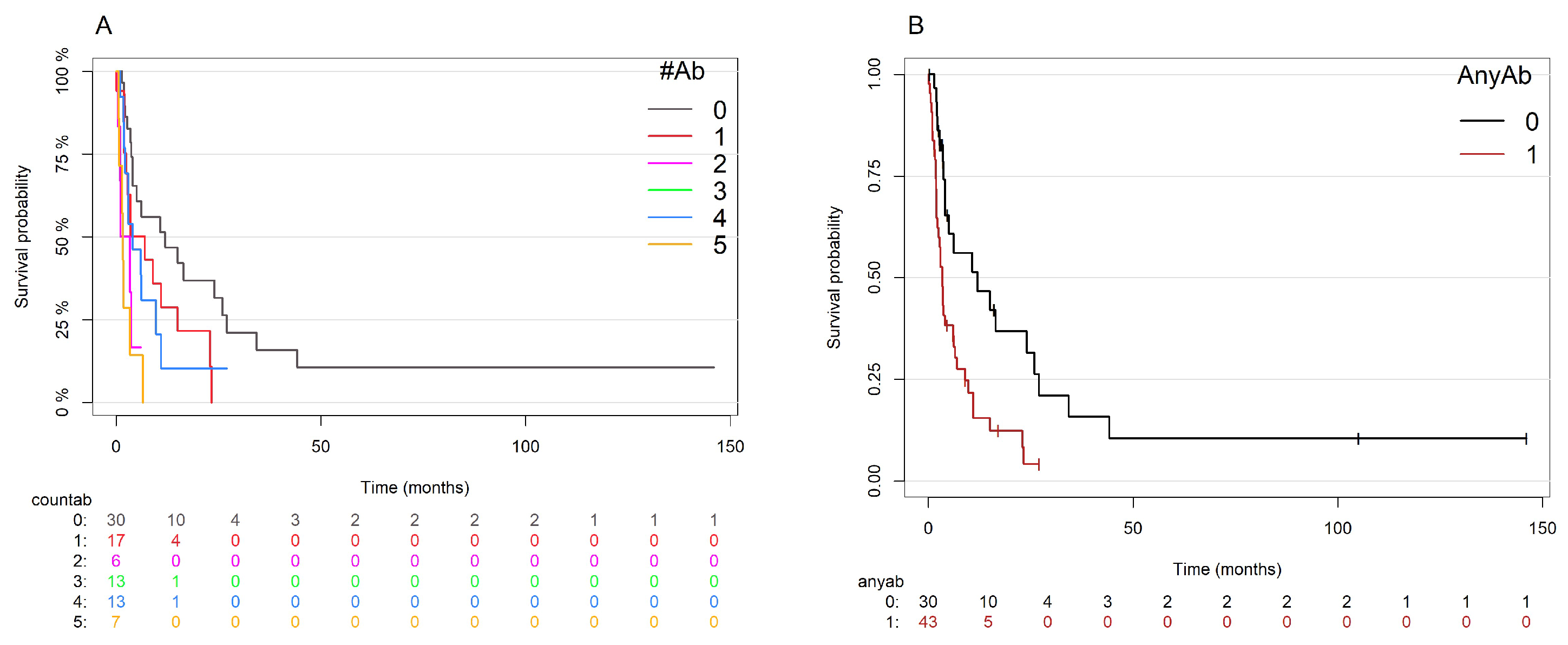
3.4. Univariate and Multivariate Models of Overall Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Inokuchi, K.; Yamaguchi, H.; Mizuki, T.; Tanosaki, S.; Shimada, T.; Dan, K. Near-Triploidy and near-Tetraploidy in Hematological Malignancies and Mutation of the P53 Gene. Clin. Lab. Haematol. 2004, 26, 25–30. [Google Scholar] [CrossRef]
- Huang, L.; Wang, S.A.; DiNardo, C.; Li, S.; Hu, S.; Xu, J.; Zhou, W.; Goswami, M.; Medeiros, L.J.; Tang, G. Tetraploidy/near-Tetraploidy Acute Myeloid Leukemia. Leuk. Res. 2017, 53, 20–27. [Google Scholar] [CrossRef]
- McGowan-Jordan, J.; Simons, A.; Schmid, M. ISCN 2016: An International System for Human Cytogenomic Nomenclature (2016); Karger Medical and Scientific Publishers: Basel, Switzerland, 2016; ISBN 978-3-318-05857-4. [Google Scholar]
- Xie, J.; Nachabe, A.; Hathaway, L.J.; Farah, B.; Berbari, B.; Li, Y.; Brown, T.C.; Schmid, J.L.; Socola, F.; Saba, N.S.; et al. The Prognostic Implications of Tetraploidy/near-Tetraploidy in Acute Myeloid Leukemia: A Case Series and Systematic Review of the Literature. Leuk. Lymphoma 2021, 62, 203–210. [Google Scholar] [CrossRef]
- Yang, R.; Jiang, M.; Zhao, J.; Chen, H.; Gong, J.; You, Y.; Song, L.; Li, Z.; Li, Q. Identification of Chromosomal Abnormalities and Genomic Features in Near-Triploidy/Tetraploidy-Acute Leukemia by Fluorescence in Situ Hybridization. Cancer Manag. Res. 2019, 11, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.V.; Sait, S.N.J.; Matsui, S.-I.; Block, A.W.; Barcos, M.; Slack, J.L.; Wetzler, M.; Baer, M.R. Massive Hyperdiploidy and Tetraploidy in Acute Myelocytic Leukemia and Myelodysplastic Syndrome. Cancer Genet. Cytogenet. 2004, 148, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Shankar, N.; Bao, L.; Haag, M.; Carstens, B.; Pang, C.S. Near-Tetraploidy/Tetraploidy Acute Myeloid Leukemia with Clinical, Pathologic and Molecular Characteristics. Leuk. Lymphoma 2024, 65, 1820–1829. [Google Scholar] [CrossRef]
- Tanaka, K.; Goto, H.; Nishimura, Y.; Kasahara, K.; Mizoguchi, A.; Inagaki, M. Tetraploidy in Cancer and Its Possible Link to Aging. Cancer Sci. 2018, 109, 2632–2640. [Google Scholar] [CrossRef]
- Attarbaschi, A.; Mann, G.; König, M.; Steiner, M.; Dworzak, M.N.; Gadner, H.; Haas, O.A.; Austrian Berlin-Frankfurt-Münster Cooperative Study Group. Near-Tetraploidy in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia Is a Highly Specific Feature of ETV6/RUNX1-Positive Leukemic Cases. Genes Chromosomes Cancer 2006, 45, 608–611. [Google Scholar] [CrossRef]
- Miller, C.R.; Ruppert, A.S.; Heerema, N.A.; Maddocks, K.J.; Labanowska, J.; Breidenbach, H.; Lozanski, G.; Zhao, W.; Gordon, A.L.; Jones, J.A.; et al. Near-Tetraploidy Is Associated with Richter Transformation in Chronic Lymphocytic Leukemia Patients Receiving Ibrutinib. Blood Adv. 2017, 1, 1584–1588. [Google Scholar] [CrossRef]
- Abrams, Z.B.; Zhang, L.; Abruzzo, L.V.; Heerema, N.A.; Li, S.; Dillon, T.; Rodriguez, R.; Coombes, K.R.; Payne, P.R.O. CytoGPS: A Web-Enabled Karyotype Analysis Tool for Cytogenetics. Bioinformatics 2019, 35, 5365–5366. [Google Scholar] [CrossRef] [PubMed]
- Abrams, Z.B.; Li, S.; Zhang, L.; Coombes, C.E.; Payne, P.R.O.; Heerema, N.A.; Abruzzo, L.V.; Coombes, K.R. CytoGPS: A Large-Scale Karyotype Analysis of CML Data. Cancer Genet. 2020, 248–249, 34–38. [Google Scholar] [CrossRef]
- Clark, R.E.; Apperley, J.F.; Copland, M.; Cicconi, S. Additional Chromosomal Abnormalities at Chronic Myeloid Leukemia Diagnosis Predict an Increased Risk of Progression. Blood Adv. 2021, 5, 1102–1109. [Google Scholar] [CrossRef]
- Mrózek, K. Cytogenetic, Molecular Genetic, and Clinical Characteristics of Acute Myeloid Leukemia with a Complex Karyotype. Semin. Oncol. 2008, 35, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Parr, T. The Definitive ANTLR4 Reference; The Pragmatic Programmers: Raleigh, NC, USA, 2013. [Google Scholar]
- Abrams, Z.B.; Tally, D.G.; Abruzzo, L.V.; Coombes, K.R. RCytoGPS: An R Package for Reading and Visualizing Cytogenetics Data. Bioinformatics 2021, 37, 4589–4590. [Google Scholar] [CrossRef] [PubMed]
- Newcombe, R.G. Interval Estimation for the Difference between Independent Proportions: Comparison of Eleven Methods. Stat. Med. 1998, 17, 873–890. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Pang, C.S.; Pettenati, M.J.; Pardee, T.S. Clinicopathological Analysis of Near-Tetraploidy/Tetraploidy Acute Myeloid Leukaemia. J. Clin. Pathol. 2015, 68, 236–240. [Google Scholar] [CrossRef]
- Abe, R.; Raza, A.; Preisler, H.D.; Tebbi, C.K.; Sandberg, A.A. Chromosomes and Causation of Human Cancer and Leukemia. LIV. Near-Tetraploidy in Acute Leukemia. Cancer Genet. Cytogenet. 1985, 14, 45–59. [Google Scholar] [CrossRef]
- Testa, J.R.; Oguma, N.; Pollak, A.; Wiernik, P.H. Near-Tetraploid Clones in Acute Leukemia. Blood 1983, 61, 71–78. [Google Scholar] [CrossRef]
- Xiao, Z.; Liu, S.; Liu, X.; Yu, M.; Hao, Y. Tetraploidy or Near-Tetraploidy Clones with Double 8;21 Translocation: A Non-Random Additional Anomaly of Acute Myeloid Leukemia with t(8;21)(Q22;Q22). Haematologica 2005, 90, 413–414. [Google Scholar] [PubMed]
- Xue, Y.; Pan, Y.; Liu, Z.; Li, J.; Guo, Y.; Xie, X. Tetraploid or Near-Tetraploid Clones Characterized by Two 8;21 Translocations and Other Chromosomal Abnormalities in Two Patients with Acute Myeloblastic Leukemia. Cancer Genet. Cytogenet. 1996, 92, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; He, J.; Wang, Y.; Guo, Y.; Xie, X.; He, Y.; Chai, Y.; Ruan, Z. Secondary Near-Pentaploidy and/or near-Tetraploidy Characterized by the Duplication of 8;21 Translocation in the M2 Subtype of Acute Myeloid Leukemia. Int. J. Hematol. 2000, 71, 359–365. [Google Scholar]
- Yamamoto, K.; Nagata, K.; Kida, A.; Tsurukubo, Y.; Hamaguchi, H. CD7+ Near-Tetraploid Acute Myeloblastic Leukemia M2 with Double t(8;21)(Q22;Q22) Translocations and Aml1/ETO Rearrangements Detected by Fluorescence in Situ Hybridization Analysis. Int. J. Hematol. 2001, 74, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Hemsing, A.L.; Hovland, R.; Tsykunova, G.; Reikvam, H. Trisomy 8 in Acute Myeloid Leukemia. Expert Rev. Hematol. 2019, 12, 947–958. [Google Scholar] [CrossRef]
- Berger, R.; Le Coniat, M.; Derré, J.; Flexor, M.A.; Hillion, J. Abnormalities of Chromosome 18 in Myelodysplastic Syndromes and Secondary Leukemia. Cancer Genet. Cytogenet. 1992, 63, 97–99. [Google Scholar] [CrossRef]
- Mitev, L.; Grachylova, L.; Asenova, A. Del(18)(P11). Available online: https://atlasgeneticsoncology.org/haematological/1314/del(18)(p11) (accessed on 19 June 2023).
- Rogers, H.J.; Hsi, E.D.; Tang, G.; Wang, S.A.; Bueso-Ramos, C.E.; Lubin, D.; Morrissette, J.J.D.; Bagg, A.; Cherukuri, D.P.; George, T.I.; et al. Most Myeloid Neoplasms with Deletion of Chromosome 16q Are Distinct from Acute Myeloid Leukemia with Inv(16)(P13.1q22): A Bone Marrow Pathology Group Multicenter Study. Am. J. Clin. Pathol. 2017, 147, 411–419. [Google Scholar] [CrossRef]
- Yamamoto, K.; Nagata, K.; Kida, A.; Hamaguchi, H. Deletion of 16q11 Is a Recurrent Cytogenetic Aberration in Acute Myeloblastic Leukemia during Disease Progression. Cancer Genet. Cytogenet. 2001, 131, 65–68. [Google Scholar] [CrossRef]
- Xiao, Y.; Geng, Z.; Deng, T.; Wang, D.; Jiang, L. Tumor Necrosis Factor Receptor Type 1-Associated Death Domain Protein Is a Potential Prognostic Biomarker in Acute Myeloid Leukemia. Am. J. Med. Sci. 2019, 357, 111–115. [Google Scholar] [CrossRef]
- van Zutven, L.J.C.M.; Onen, E.; Velthuizen, S.C.J.M.; van Drunen, E.; von Bergh, A.R.M.; van den Heuvel-Eibrink, M.M.; Veronese, A.; Mecucci, C.; Negrini, M.; de Greef, G.E.; et al. Identification of NUP98 Abnormalities in Acute Leukemia: JARID1A (12p13) as a New Partner Gene. Genes Chromosomes Cancer 2006, 45, 437–446. [Google Scholar] [CrossRef]
| Case | Age * (Yrs)/Gender | Interval † (Months) | Diagnosis | Prior Treatment | Treatment | T/NT Size § (%) | T/NT Persist. ‡ | OS (Months) | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 63/M | 0 | MDS-EB1 | No | Chemo | 21 | No | 2.5 | DOD |
| 2 | 65/M | 0 | AML-MRC | No | Chemo | 95 | No | 3 | DOD |
| 3 | 89/M | 0 | Histiocytic sarcoma | No | No | 20 | NA | 1 | DOD |
| 4 | 89/F | 0 | AML-MRC | No | Chemo | 75 | NA | 1.5 | DOD |
| 5 | 84/M | 0 | AML-MRC | No | Chemo | 85 | Yes | 15 | DOD |
| 6 | 78/F | 0 | AML-MRC | No | Chemo | 74 | Yes | 2 | DOD |
| 7 | 67/M | 0 | MDS-EB1/ evolving AML | No | Chemo | 25 | Yes | 11 | DOD |
| 8 | 85/M | 0 | AML-MRC | No | Chemo | 35 | NA | 27 | DOD |
| 9 | 69/M | 48 | AML, NOS | Yes | Chemo | 75 | Yes | 24 | DOD |
| 10 | 79/M | 2.5 | AML, NOS | Yes | Chemo | 10 | No | 3 | LFU—hospice |
| 11 | 87/M | 0 | AML-MRC | No | No | 90 | NA | 0.25 | LFU—hospice |
| 12 | 58/M | 0 | AML-MRC | No | Chemo | 85 | NA | 3.5 | DOD |
| 13 | 70/F | 0 | Myeloid sarcoma/AUL | No | Chemo | 90 | No | 4.5 | LFU—hospice |
| 14 | 57/M | 0 | AML-MRC | No | Chemo | 100 | No | 15 | DOD |
| 15 | 72/M | 84 | AML, NOS | Yes | Chemo | 25 | Yes | 2 | DOD |
| 16 | 61/M | 0 | AML-MRC | NA | Chemo | 30 | No | 3.5 | DOD |
| 17 | 65/M | NA | AML-MRC | NA | Chemo | 90 | No | 4 | DOD |
| 18 | 52/M | 0 | MDS-EB2 | No | Chemo | 95 | NA | 4 | DOD |
| 19 | 58/M | 0 | AML-MRC | No | Chemo | 15 | Yes | 11 | DOD |
| 20 | 66/M | 0 | AML-MRC | No | Chemo | 25 | Yes | 1 | DOD |
| 21 | 75/F | 8 | AML t(8;21) | Yes | Chemo | 100 | Yes | 6 | LFU |
| 22 | 24/M | 0 | AML-MRC | No | Chemo, SCT | 85 | No | 146 | ANED |
| 23 | 71/M | 0 | AML, NOS | No | Chemo | 55 | Yes | 23 | DOD |
| 24 | 69/F | 0 | AML-MRC | No | Chemo, SCT | 100 | No | 6.5 | DOD |
| 25 | 29/F | 0 | AUL | No | Chemo, SCT | 13 | No | 105 | ANED |
| 26 | 62/F | 21 | AML-MRC | Yes | Chemo | 85 | Yes | 1.5 | DOD |
| 27 | 77/M | 0 | MDS-MLD | No | No | 35 | NA | 4 | DOD |
| 28 | 61/M | 0 | AML-MRC | No | Chemo, SCT | 43 | No | 9 | DOD |
| 29 | 69/F | 0 | AML-MRC | No | Chemo | 10 | Yes | 16 | LFU |
| 30 | 69/F | NA | AML-MRC | Yes | Chemo | 20 | Yes | 1.5 | LFU |
| 31 | 78/M | 4 | AML-MRC | Yes | Chemo | 70 | Yes | 6 | DOD |
| 32 | 69/M | 0 | MDS-EB2 | No | Chemo | 35 | No | 27 | ANED |
| 33 | 38/F | 0 | AUL | No | Chemo, SCT | 22 | No | 19 | ANED |
| 34 | 46/M | 0 | AML, NOS | No | Chemo, SCT | 22 | No | 21 | ANED |
| 35 | 82/F | 0 | AML-MRC | No | Chemo | 60 | Yes | 3 | DOD |
| 36 | 61/F | 20 | AML inv(3) | Yes | Chemo, SCT | 5 | Yes | 12 | DOD |
| 37 | 74/F | 0 | AML-MRC | No | Chemo | 10 | Yes | 17 | AWD |
| Case | Complexity | Karyotype |
|---|---|---|
| 1 | C | 44~49,XY,-2,-5,del(7)(q22),-19,+21,add(21)(p11.2)x2,+2-5mar[cp9]/ 47~49,sl,add(12)(q24.3)[cp3]/87~113,slx2[cp4]/46,XY[3] |
| 2 | C | 86~92<4n>,XXYY,del(2)(p23p25)x2,-5,-5,-9,-9,add(11)(p15)x2,-13,-17,-17,+22, +mar1x2,+mar2x2,+mar3x2,dmin[cp19]/46,XY[1].ish dmin(CMYC+) |
| 3 | C | 72~98<4n>,XYY,-X,+1,i(1)(q10),+8,+8,-11,+12,+12,+16[4,three w/nonclonal abnormalities]/46,XY[16,one is 4n]/nonclonal[1] |
| 4 | C | 46,XX,del(5)(q13q33),del(20)(q11.2q13.1)[2]/77~89,idemx2,del(1)(q21),-2,-3, del(3)(p25),+8,add(10)(q24),hsr(11)(q23),hsr(11)(q23),-11,add(13)(q34),-16,-17,-17, -17,del(18)(q21),+19,+19,add(19)(p13),der(19)t(17;19)(q21;p13)x2,+20,+20,-del(20), -del(20),+mar[cp18]/46,XX[4]/nonclonal[1] .ish hsr(11)(amp MLL),hsr(11)(amp MLL) |
| 5 | C | 83~88<4n>,XXYY,i(1)(q10),-2,-3,-7,-9,-17,i(17)(q10),+18[cp18]/46,XY[2]/4n[1] |
| 6 | C | 85~90<4n>,XXXX,-8,-9,-9,-12,-17,i(17)(q10),+mar1x2,+mar2,+dmin[cp10]/ 81~89,idem,-2,-5,-5,-7[cp7]/46,XX[6] |
| 7 | C | 45~50,XY,-5,der(7)t(5;7)(p14;p12)add(7)(q11.2),der(13;14)(q10;q10)c,+21,+mar1, +mar2,+mar3,+mar4,+mar5[cp8]/44,sl,add(4)(p16),add(12)(p12),add(19)(p13), -mar1,-mar4,-mar5[cp3]/85~87,slx2,+1,add(19)(p13)x2,-mar1,-mar1,-mar2,-mar3, -mar4,-mar5,+mar6[cp4]/45,XY,der(13;14)(q10;q10)c[1]. ish der(7)t(5;7)(D5S23:D5S721+,D7Z1+,D7S486-) |
| 8 | C | 46,XY,t(9;22)(q34;q11.2)[11]/89-93,idemx2,+4,add(4)(q31)x2[7]/nonclonal Ph+[2] |
| 9 | N | 94<4n>,XXYY,+13,+13[cp15]/46,XY[3]/nonclonal[2] |
| 10 | N | 46,XY,del(6)(p23)[17]/90-92,sl,x2[2]/46,sl,add(19)(p13.3)[1] |
| 11 | C | 90<4n>,XXYY,add(2)(q31),-7,-7[4]/90,sl,del(5)(q31q35)[cp14]/46,XY[2] |
| 12 | C | 97,XXYY<4n>,+1,t(1;8)(q32;q24),der(1)t(1;8),-3,+4,del(5)(q31q35),del(7)(q10), +10,+12,add(12)(q13)x3,+21,+22[17]/nonclonal w/clonal abnormalities[3] |
| 13 | C | 47,XX,+10[2]/94-99<4n>,XXXX,+X,+X(4),-2,-7(6),-9,+10,+10,+10,-16,+20(9), +der(?)t(?;1)(?;q25)x2,+mar1[cp18] |
| 14 | C | 90<4n>,XX,-Y,-Y,del(5)(q14)[15]/93,idem,+8,+11,+13[5] |
| 15 | C | 90<4n>,XX,-Y,-Y,add(7)(q36)x2[cp5]/46,XY[13]/nonclonal[2] |
| 16 | C | 45~47,XX,del(3)(p13),del(5)(q11.2q35),add(17)(p13),-20,+mar1[cp5]/ 48,idem,-del(3)(p13),+add(20)(q11.2),+22,+mar2[cp8]/76~97,idemx2,-del(3)(p13)x2, -5,+add(20)(q11.2)x2,+22,+mar2x2[cp4]/120~134,idemx3[cp2]/46,XX[1] |
| 17 | N | 92<4n>,XXYY,der(5)t(1;5)(q21;q31)[18]/46,XY[2] |
| 18 | C | 71~83<4n>,XXYY,add(2)(q37),del(2)(q33q35),-7,-13,-13,-13,add(13)(p12),-14, add(14)(p13),-15,-16,-17,-18,-20,+21,idic(21)(p13),psu dic(21;13)(p11.2;p13),-22, +mar1,+mar2,+mar3,+mar4,+mar5[cp5]/74~83,idem,-add(13)[cp14]/46,XY[1] |
| 19 | C | 46~48,X,-Y,dup(1)(q23q42),del(5)(q13q33),-7,del(7)(q11.2),+8, der(11)t(Y;11)(q11.2;p13),+20,add(20)(q11.2)x2,+mar1[cp3]/ 42,sl,add(3)(p25),-8,-12,-17,-add(20),-mar1[7]/42,sl,+1,-dup(1),-3,+7,-del(7),-8, del(12)(p11.2p13),-16,-mar1,+mar2[cp7]/88-90,sdl2x2,+mar3[cp3] |
| 20 | C | 46,XY,-1,-5,+8,+11,add(11)(p11.2)x2,-17,der(17)t(1;17)(p22;p11.2),+mar[12]/ 77~79,idemx2,-2,-3,-8,-8,-add(11),-add(11),-12,-14,-16,-mar[cp5]/46,XY[3] |
| 21 | C | 73~87<4n>,XXXX,-7,-7,t(8;21)(q22;q22)x2,-11,-14,-16,-20[cp20] |
| 22 | C | 44,X,-Y,add(5)(q11.2),-9,-12,der(17;18)(q10;q10),+mar1,+mar2[2]/ 85,idemx2,-2,-3,-7[17]/nonclonal[1] |
| 23 | C | 99~110<4n>,XXYY,+2,+3,+8,+10,+12,+13,+13,+13,+16,+18,+19,+20,+21,+22,+22,+mar1x2[cp8]/81~82<3n>,XY,-X,+Y,+2,+9,+10,+13,+13,+14,+14,+15,+15,+19,+20,+21,+22[cp3]/ 46,XY[9] |
| 24 | C | 82~88<4n>XXXX,-1,t(3;11)(p13;q21),-5,del(5)(q13q31),dic(9;21)(q12;p13),-13,-13,-16, -17,-17,dic(17;21)(p13;p12),-18,-18,-18,+21,add(22)(p13),+der(?)t(?;1)(?;p13), +der(?)t(?;1)(?:q21),+mar1,+mar2[cp8]/82-88,sl,-der(11)t(3;11)[cp16] .ish t(3;11)(MLL+,MLL-) |
| 25 | N | 94<4n>,XXXX,+13,+13[cp3]/46,XX[19]/4n[1] |
| 26 | C | 92~95<4n>,XXXX,+2,+2,del(2)(q33q37)x3,+13,+13,-21[6]/90~95,sl,+2,del(2)[3]/90-95,sdl1,-21[5]/86~94,sl,-21[3]/46,XX,t(1;12)(p36.1;p13)[2]/46,XX[1] |
| 27 | N | 92<4n>,XXYY[7]/46,XY[13] |
| 28 | C | 45,X,-Y,del(5)(q13q33),add(14)(q24),-17,-19,+mar1,+mar2[cp9]/ 90,slx2,add(11)(p15)x2[cp10]/46,XY[4] |
| 29 | C | 46,XX,add(7)(q36),-10,+mar[17]/92,slx2[2]/46,XX[1] |
| 30 | C | 46,XX,del(5)(q13q35),add(7)(q11.2),-11,add(17)(p11.2),+20,del(20)(q11.2q13.3), dic(20;22)(q11.2;p11.2),+der(?)t(?;11)(?;q12),dmin[6,two w/nonclonal abnormalities]/45,sl,der(16)t(1;16)(p13;p11.2),-del(20)[6,two w/nonclonal abnormalities]/89,sdl1x2,+16,-der(16)t(1;16)x2[2]/ 90,sdl2,+1[2]/47,sl,+ider(?)(11qter->11q12::?::11q12->11qter)[2,one is 4n]/46,XX[2] |
| 31 | C | 43,X,-Y,del(5)(q22q35),add(7)(q21),+8,add(9)(q13),-10,add(11)(p15), der(12)del(12)(p12p13)add(12)(q13),+15,add(15)(q22),-16,-17,-18,-19,add(20)(p12), +mar1[2,one w/nonclonal abnormalities]/ 82~85,slx2,idic(1)(q44),-del(7)x2,+17,+18[cp4]/75,sdl1,+1,+add(1)(q21),-dic(1;1)[cp3]/ 72~79,sdl2,+add(1)(q25),-add(1)(q21),-15[cp4]/69~75,sdl1,-15,-15,-21,-mar1[cp3]/ 46,XY[4] |
| 32 | C | 43~51,XY,+1,+2,-3,-5,+6,-7,+8,del(9)(q13q22),add(12)(p13),+13,+14,+15,+15,-17,add(17)(p11.2),+18,der(20)(20pter->20q13.1::?::5p15.2->5p15.2::?),+21,+21, der(21)add(21)(p13)del(21)(q22),der(21)t(9;21)(q13;p13)x2,+22,+mar[cp4]/ 42~55,sl,add(3)(p13),+9,del(9)(q13q22),+11,-14,-22[cp5]/84,sdl1x2,-X,-1,-1,-2,-2, -del(3),add(4)(q21),-8,-9,-del(9),-10,-13,-13,-13,-add(17),-19, -der(21)add(21)del(21)[cp3]/51~81,sl,der(1)t(1;9)(p13;q13),-2,+8,del(9)(q13q22), -15,+19,+21,+der(21)add(21)del(21),+der(21)t(9;21)[cp4,2 are 4n]/ 82~84,slx2,-X,-2,-2,-4,+8,-12,-13,-22,-22[cp2]/46,XY[2] .ish add(12)(ETV6-),der(20)(D5S23:D5S721+,D20S108+),der(21)t(9;21)(RUNX1+) |
| 33 | Inc | 91<4n>,XXXX,-17,inc[cp5]/46,XX[15]/nonclonal[3] |
| 34 | Inc | 88~91<4n>,XXYY,inc[cp5]/46,XY[18] |
| 35 | C | 47,XX,+10[1]/85,slx2,-5,-6,-7,-8,-10,-15,-16,-17,-18[7]/85,sdl1,-2,+r[5]/ 46,sl,dic(7;16)(p13;q12.1)[5]/46,XX[2] |
| 36 | N | 90<4n>,XXXX,inv(3)(q21.3q26.2)x2[1]//46,XY[19]/nonclonal[1] |
| 37 | C | 45,XX,der(5)t(5;16)(q13;q22),del(13)(q12q22),-16,add(17)(p13),der(17)del(17)(q11.2)del(17)(q11.2),add(22)(q13),+psu dic(?;17)(?::17q21->17q21::17p11.2->17q11.2::17q21->17q21::?)[18,one w/nonclonal abnormalities]/92,slx2[cp2].ish der(2)cryp ins(2;17)(TP53+),der(5)t(5;16)(CBFB+),add(17)(TP53+),psu dic(?;17)(TP53-,RARA+,D17Z1+,RARA+) |
| Factor | Number of Cases | Deaths | Median Survival (Range) | Coefficient | Hazard Ratio | Log-Rank Test | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Age in years (continuous) | NA | NA | NA | NA | 0.019 | 1.019 | 4.59 | 0.032 |
| Age in years (categorical) | <60 | 23 | 15 | 4 (3.4–∞) | NA | NA | NA | NA |
| ≥60 | 52 | 42 | 5 (3.4–10.7 | 10,433 | 1.542 | 1.954 | 0.162 | |
| Gender | Female | 25 | 16 | 3.7 (3.4–∞) | NA | NA | NA | NA |
| Male | 50 | 41 | 6 (3.5–11) | 0.031 | 1.032 | 0.011 | 0.916 | |
| Prior Rx | No | 46 | 34 | 9 (3.5–23) | NA | NA | NA | NA |
| Yes | 27 | 21 | 3.7 (2–7) | 0.743 | 2.102 | 6.541 | 0.011 | |
| Karyotype * | Simple | 16 | 12 | 6.2 (4–∞) | NA | NA | NA | NA |
| Complex | 57 | 45 | 3.5 (3–9) | 0.443 | 1.1557 | 1.843 | 0.175 | |
| Cohort | MDACC | 38 | 32 | 3.5 (2.2–7) | NA | NA | NA | NA |
| OSU | 37 | 25 | (4–24) | −0.598 | 0.55 | 5.032 | 0.025 | |
| Interval † | NA | NA | NA | NA | 0.004 | 1.004 | 0.654 | 0.419 |
| T/NT Clone Size | NA | NA | NA | NA | 0.005 | 1.005 | 1.007 | 0.316 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avenarius, M.R.; Abrams, Z.B.; Guo, L.; Blachly, J.S.; Miller, C.R.; Heerema, N.A.; Tang, G.; Coombes, K.R.; Abruzzo, L.V. Novel Recurrent Cytogenetic Abnormalities Predict Overall Survival in Tetraploid/Near-Tetraploid Myelodysplastic Syndrome and Acute Myeloid Leukemia. Cancers 2025, 17, 1277. https://doi.org/10.3390/cancers17081277
Avenarius MR, Abrams ZB, Guo L, Blachly JS, Miller CR, Heerema NA, Tang G, Coombes KR, Abruzzo LV. Novel Recurrent Cytogenetic Abnormalities Predict Overall Survival in Tetraploid/Near-Tetraploid Myelodysplastic Syndrome and Acute Myeloid Leukemia. Cancers. 2025; 17(8):1277. https://doi.org/10.3390/cancers17081277
Chicago/Turabian StyleAvenarius, Matthew R., Zachary B. Abrams, Ling Guo, James S. Blachly, Cecelia R. Miller, Nyla A. Heerema, Guilin Tang, Kevin R. Coombes, and Lynne V. Abruzzo. 2025. "Novel Recurrent Cytogenetic Abnormalities Predict Overall Survival in Tetraploid/Near-Tetraploid Myelodysplastic Syndrome and Acute Myeloid Leukemia" Cancers 17, no. 8: 1277. https://doi.org/10.3390/cancers17081277
APA StyleAvenarius, M. R., Abrams, Z. B., Guo, L., Blachly, J. S., Miller, C. R., Heerema, N. A., Tang, G., Coombes, K. R., & Abruzzo, L. V. (2025). Novel Recurrent Cytogenetic Abnormalities Predict Overall Survival in Tetraploid/Near-Tetraploid Myelodysplastic Syndrome and Acute Myeloid Leukemia. Cancers, 17(8), 1277. https://doi.org/10.3390/cancers17081277






