Identification of Molecular Subtypes of Clear-Cell Renal Cell Carcinoma in Patient-Derived Xenografts Using Multi-Omics
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of ccRCC PDXs
2.2. Publicly Available Datasets Used in This Study
2.3. Proteomic Profiling of ccRCC PDX Tissues by Liquid Chromatography–Mass Spectrometry (LC-MS)
2.4. ccRCC PDX Proteomic Data Processing
2.5. ccRCC PDX Metabolic Data
2.6. Comparison Methods
2.7. GSEA
2.8. Statistical Analysis
3. Results
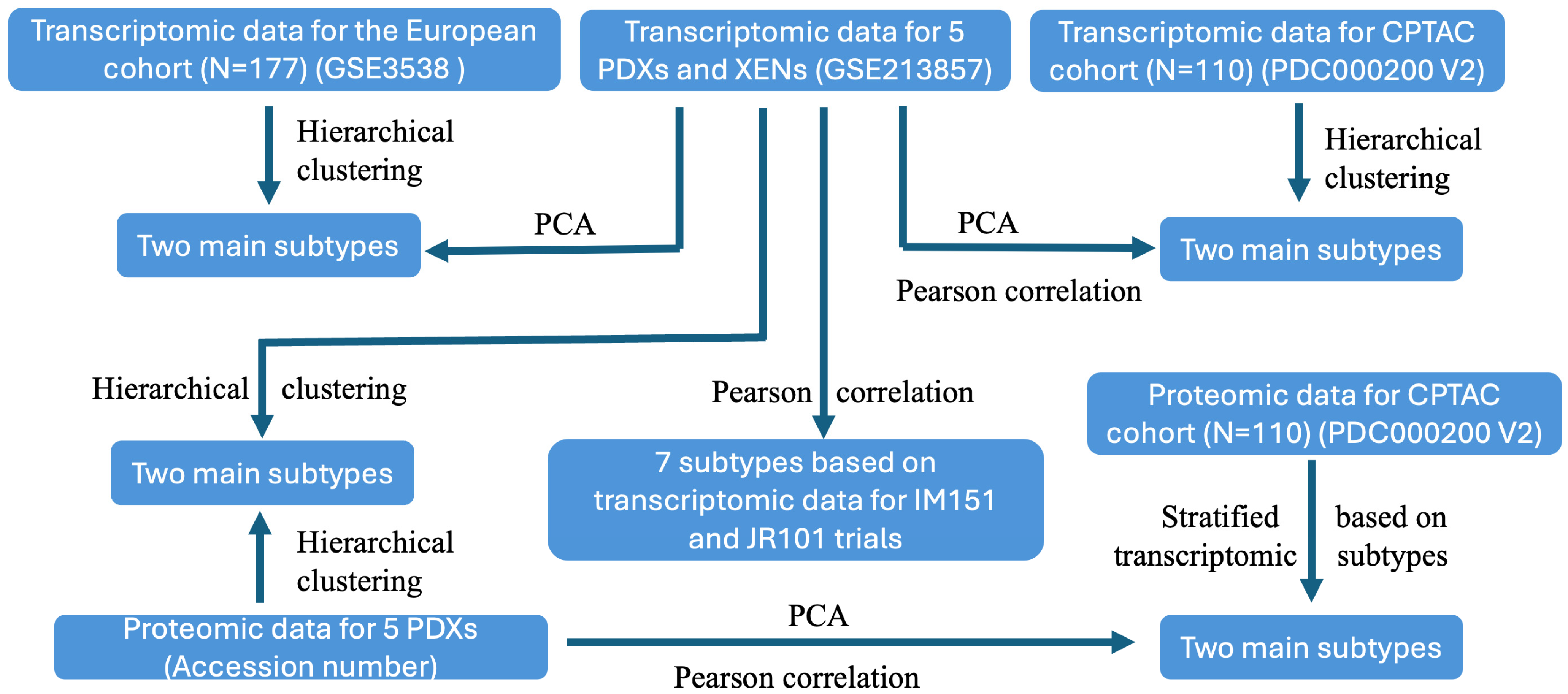
3.1. Study Design
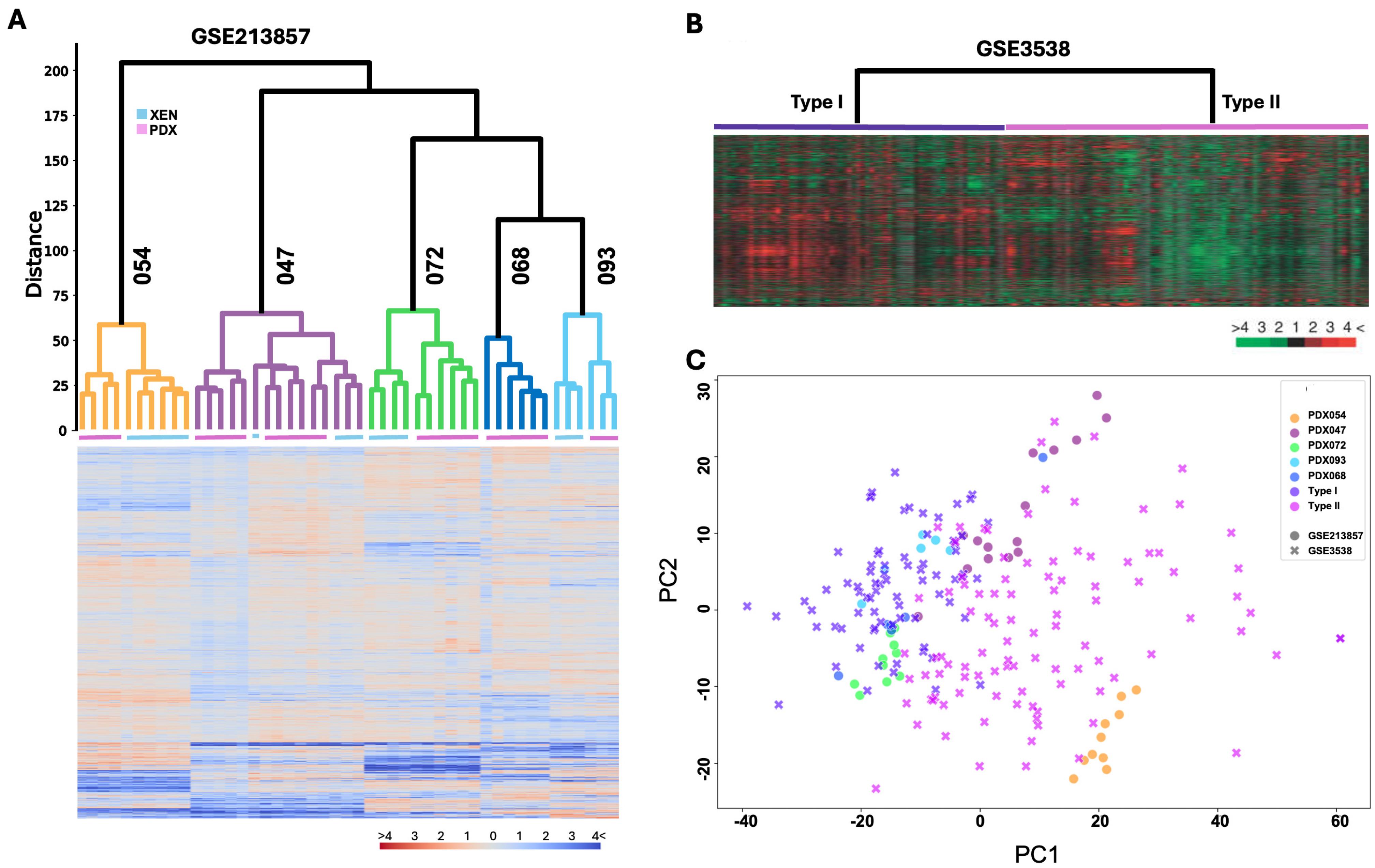
3.2. Transcriptomic Programs Are Retained in ccRCC PDXs with Different Passages and Between PDX and Their Respective XEN Counterparts
3.3. Transcriptomic Programs in ccRCC PDXs Resemble Molecular Subtypes of ccRCC Patients
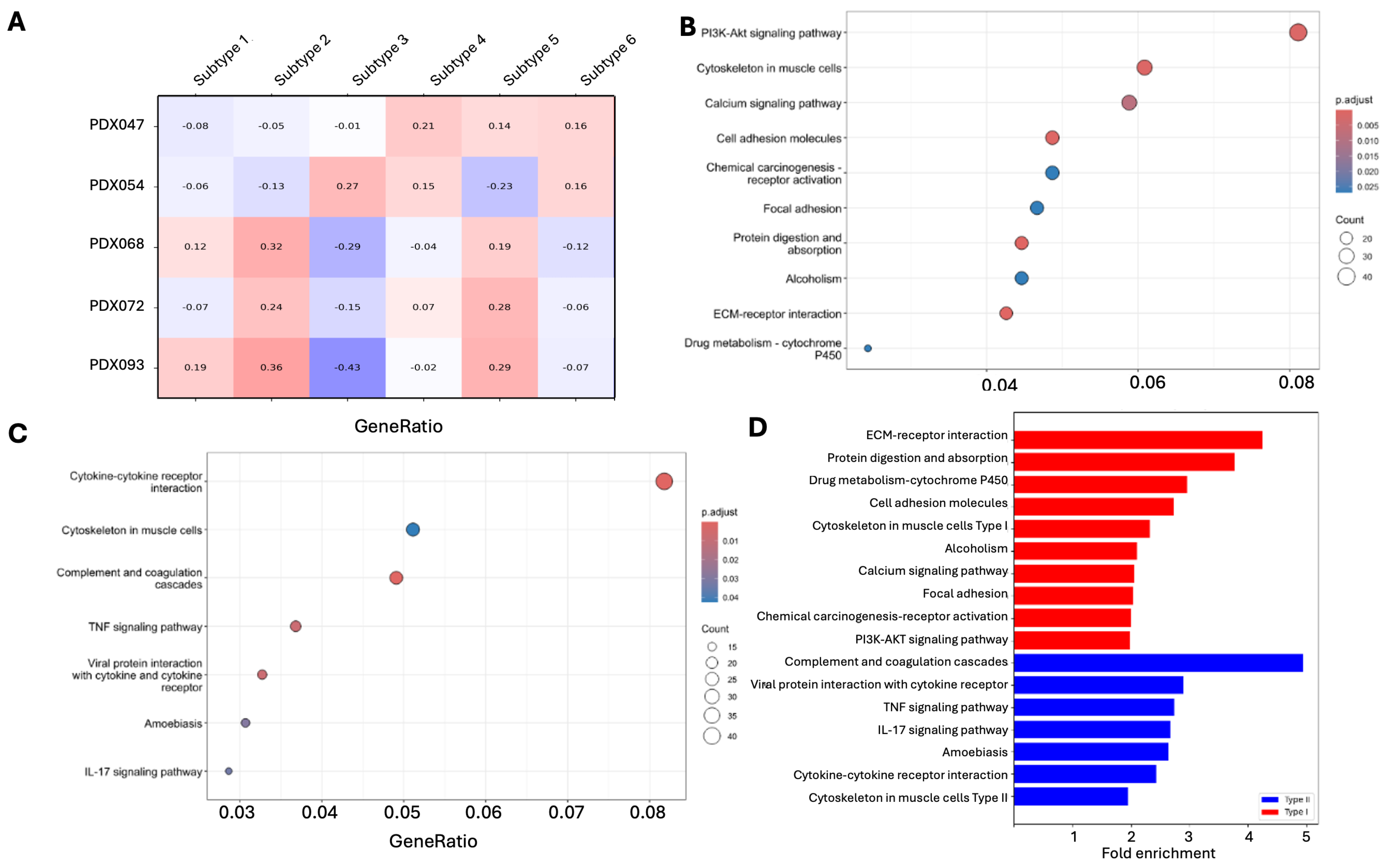
3.4. Different Pathways Are Enriched in PDX Subtypes
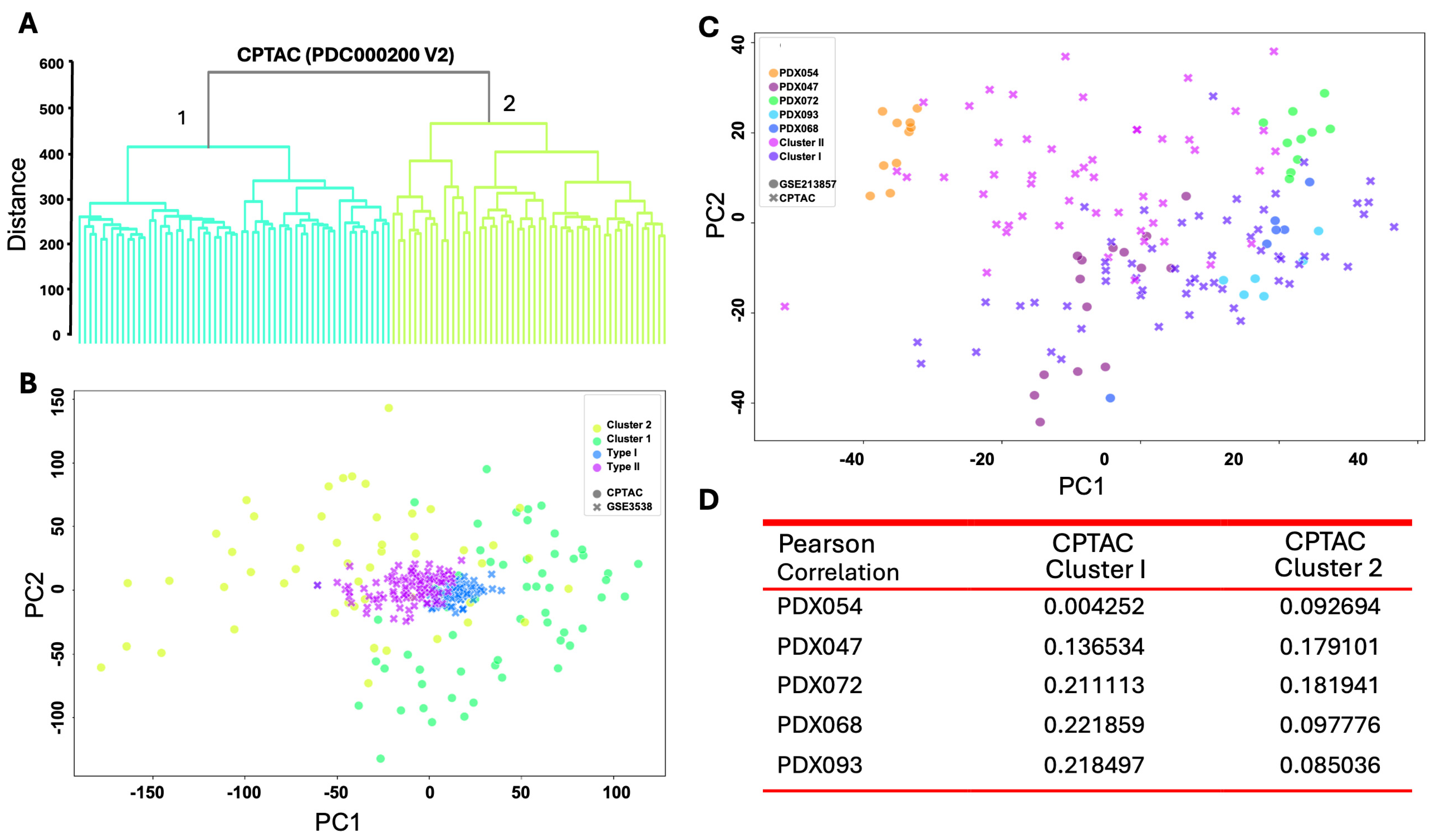
3.5. Proteomic Programs in ccRCC PDXs Resemble Molecular Subtypes of ccRCC Patients
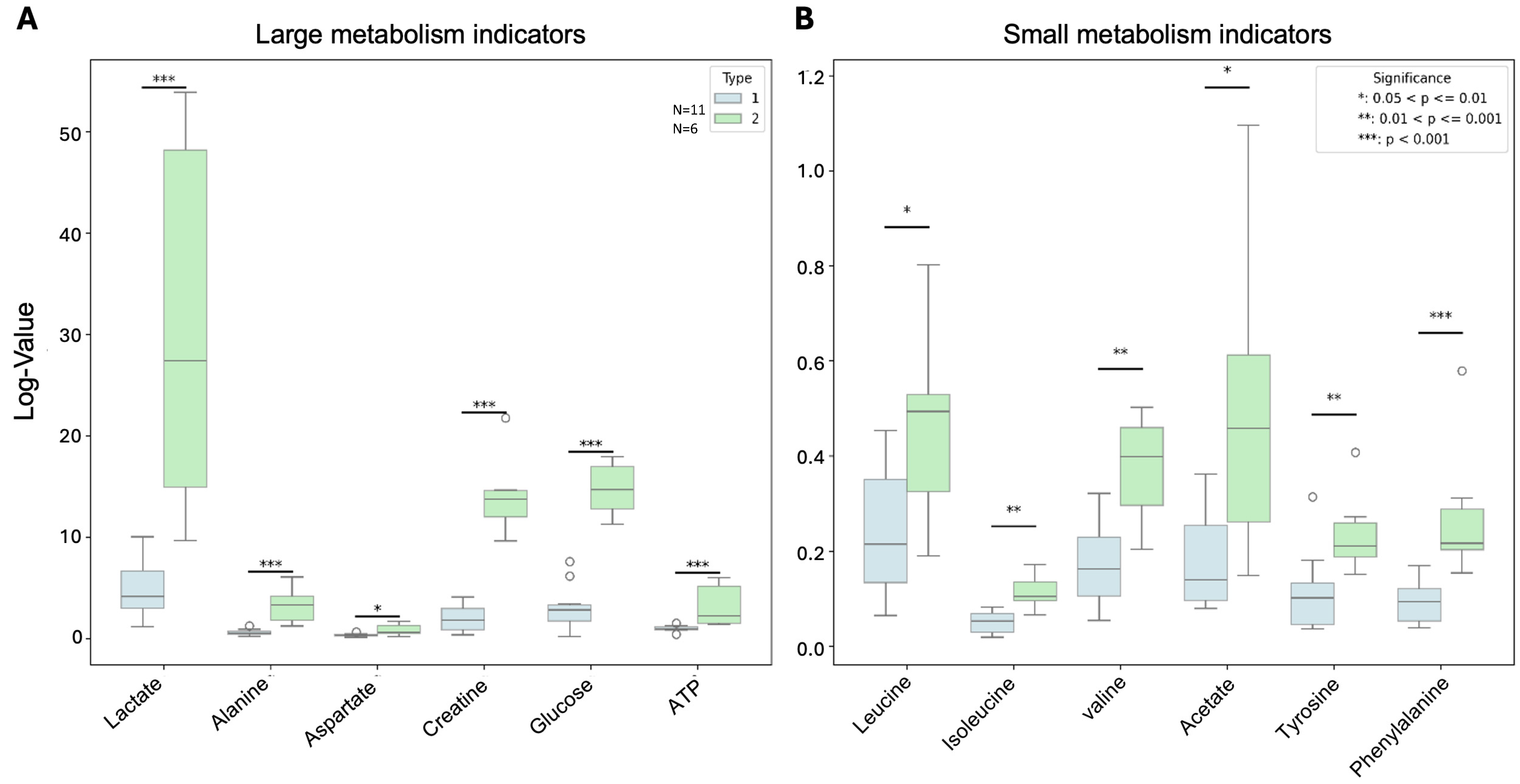
3.6. PDX Subtypes Showed Significant Differences in Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ccRCC | Clear-cell renal cell carcinoma |
| GSEA | Gene Set Enrichment Analysis |
| PCA | Principal Component Analysis |
| PDX | Patient-derived xenograft |
| RCC | Renal cell carcinoma |
| SCLC | Small-cell lung cancer |
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- Rose, T.L.; Kim, W.Y. Renal Cell Carcinoma: A Review. JAMA 2024, 332, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Nezami, B.G.; MacLennan, G.T. Clear Cell Renal Cell Carcinoma: A Comprehensive Review of its Histopathology, Genetics, and Differential Diagnosis. Int. J. Surg. Pathol. 2025, 33, 265–280. [Google Scholar] [CrossRef]
- Liao, C.; Hu, L.; Zhang, Q. Von Hippel-Lindau protein signalling in clear cell renal cell carcinoma. Nat. Rev. Urol. 2024, 21, 662–675. [Google Scholar] [CrossRef]
- Bhat, A.S.; Ahmed, M.; Abbas, K.; Mustafa, M.; Alam, M.; Salem, M.; Tantry, I.Q.; Singh, Y.; Fatima, M.; Usmani, N.; et al. Cancer Initiation and Progression: A Comprehensive Review of Carcinogenic Substances, Anti-Cancer Therapies, and Regulatory Frameworks. Asian J. Res. Biochem. 2024, 14, 111–125. [Google Scholar] [CrossRef]
- Miller, J.W.; Johnson, J.S.; Guske, C.; Mannam, G.; Hatoum, F.; Nassar, M.; Potez, M.; Fazili, A.; Spiess, P.E.; Chahoud, J. Immune-Based and Novel Therapies in Variant Histology Renal Cell Carcinomas. Cancers 2025, 17, 326. [Google Scholar] [CrossRef]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Horswell, S.; Chambers, T.; O’Brien, T.; Lopez, J.I.; Watkins, T.B.K.; Nicol, D.; et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell 2018, 173, 595–610. [Google Scholar] [CrossRef]
- Saliby, R.M.; Labaki, C.; Jammihal, T.R.; Xie, W.; Sun, M.; Shah, V.; Saad, E.; Kane, M.H.; Kashima, S.; Sadak, K.; et al. Impact of renal cell carcinoma molecular subtypes on immunotherapy and targeted therapy outcomes. Cancer Cell 2024, 42, 732–735. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, B.; Yu, Y. Identification and Validation of Cytotoxicity-Related Features to Predict Prognostic and Immunotherapy Response in Patients with Clear Cell Renal Cell Carcinoma. Genet. Res. 2024, 2024, 3468209. [Google Scholar] [CrossRef]
- Miranda-Poma, J.; Trilla-Fuertes, L.; López-Vacas, R.; López-Camacho, E.; García-Fernández, E.; Pertejo, A.; Lumbreras-Herrera, M.I.; Zapater-Moros, A.; Díaz-Almirón, M.; Dittmann, A.; et al. Proteomics Characterization of Clear Cell Renal Cell Carcinoma. J. Clin. Med. 2023, 12, 384. [Google Scholar] [CrossRef]
- Meng, J.; Jiang, A.; Lu, X.; Gu, D.; Ge, Q.; Bai, S.; Zhou, Y.; Zhou, J.; Hao, Z.; Yan, F.; et al. Multiomics characterization and verification of clear cell renal cell carcinoma molecular subtypes to guide precise chemotherapy and immunotherapy. iMeta 2023, 2, e147. [Google Scholar] [CrossRef]
- Qu, Y.; Feng, J.; Wu, X.; Bai, L.; Xu, W.; Zhu, L.; Liu, Y.; Xu, F.; Zhang, X.; Yang, G.; et al. A proteogenomic analysis of clear cell renal cell carcinoma in a Chinese population. Nat. Commun. 2022, 13, 2052. [Google Scholar] [CrossRef]
- Li, Y.; Lih, T.M.; Dhanasekaran, S.M.; Mannan, R.; Chen, L.; Cieslik, M.; Wu, Y.; Lu, R.J.; Clark, D.J.; Kołodziejczak, I.; et al. Histopathologic and proteogenomic heterogeneity reveals features of clear cell renal cell carcinoma aggressiveness. Cancer Cell 2023, 41, 139–163.e17. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zheng, B.; Chen, Z.; Zhang, W.; Chen, X.; Xu, S.; Ji, J.; Fang, X.; Shi, C. Identification of prognostic models for glycosylation-related subtypes and tumor microenvironment infiltration characteristics in clear cell renal cell cancer. Heliyon 2024, 10, e27710. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Lv, Z.; Xing, Z.; Xu, H.; Chand, H.; Wang, J.; Li, Y. Identification of molecular subtypes and diagnostic model in clear cell renal cell carcinoma based on collagen-related genes may predict the response of immunotherapy. Front. Pharmacol. 2024, 15, 1325447. [Google Scholar] [CrossRef]
- Liu, H.; Liang, X.; Tang, G.; Wang, X.; Wang, Z.; Tong, L.; Mao, Q.; Ma, J.; Wu, J. Identifying molecular subtypes and tumor microenvironment infiltration signatures in kidney renal clear cell carcinoma based on stemness-associated disulfidptosis genes by integrating machine learning, single-cell analyses and experimental validation. Heliyon 2024, 10, e26094. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zheng, Z.; Wang, B.; Zhan, C.; Yuan, X.; Lin, X.; Xin, Q.; Zhong, Z.; Qiu, X. Characterization of a G2M checkpoint-related gene model and subtypes associated with immunotherapy response for clear cell renal cell carcinoma. Heliyon 2024, 10, e29289. [Google Scholar] [CrossRef]
- Jiang, A.; Liu, Y.; Zhu, B.; Fang, Y.; Qu, L.; Yang, Q.; Luo, P.; Cai, C.; Wang, L. SPCS, a Novel Classifier System Based on Senescence Axis Regulators Reveals Tumor Microenvironment Heterogeneity and Guides Frontline Therapy for Clear Cell Renal Carcinoma. Clin. Genitourin. Cancer 2024, 22, 497–513. [Google Scholar] [CrossRef]
- Ma, J.; Kang, Z.; Yang, G.; Wang, X.; Si, M.; Wang, Y.; Li, G.; Bai, S.; Zeng, F.; Li, M.; et al. Pyroptosis-Related Subtypes Predict the Response of Clear Cell Renal Cell Carcinoma to Targeted Therapy. Front. Biosci. 2023, 28, 334. [Google Scholar] [CrossRef]
- Pan, X.W.; Chen, W.J.; Xu, D.; Guan, W.B.; Li, L.; Chen, J.X.; Chen, W.J.; Dong, K.Q.; Ye, J.Q.; Gan, S.S.; et al. Molecular subtyping and characterization of clear cell renal cell carcinoma by tumor differentiation trajectories. iScience 2023, 26, 108370. [Google Scholar] [CrossRef]
- Quan, J.; Huang, B. Identification and validation of the molecular subtype and prognostic signature for clear cell renal cell carcinoma based on neutrophil extracellular traps. Front. Cell Dev. Biol. 2022, 10, 1021690. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, H.; Zhu, B.; Xing, Q.; Xie, H. Molecular subtypes based on metabolic genes are potential biomarkers for predicting prognosis and immune responses of clear cell renal cell carcinoma. Eur. J. Immunol. 2023, 53, e2250105. [Google Scholar] [CrossRef] [PubMed]
- Jee, B.; Seo, E.; Park, K.; Kim, Y.R.; Byeon, S.J.; Lee, S.M.; Chung, J.H.; Song, W.; Sung, H.H.; Jeon, H.G.; et al. Molecular Subtypes Based on Genomic and Transcriptomic Features Correlate with the Responsiveness to Immune Checkpoint Inhibitors in Metastatic Clear Cell Renal Cell Carcinoma. Cancers 2022, 14, 2354. [Google Scholar] [CrossRef]
- Zhong, W.; Zhong, H.; Zhang, F.; Huang, C.; Lin, Y.; Huang, J. Characterization of Hypoxia-Related Molecular Subtypes in Clear Cell Renal Cell Carcinoma to Aid Immunotherapy and Targeted Therapy via Multi-Omics Analysis. Front. Mol. Biosci. 2021, 8, 684050. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Terada, N.; Kobayashi, T.; Ogawa, O. Patient-derived xenografts as in vivo models for research in urological malignancies. Nat. Rev. Urol. 2017, 14, 267–283. [Google Scholar] [CrossRef]
- Sobczuk, P.; Brodziak, A.; Khan, M.I.; Chhabra, S.; Fiedorowicz, M.; Wełniak-Kamińska, M.; Synoradzki, K.; Bartnik, E.; Cudnoch-Jędrzejewska, A.; Czarnecka, A.M. Choosing The Right Animal Model for Renal Cancer Research. Transl. Oncol. 2020, 13, 100745. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zheng, G.; Zhong, J.; Sheng, J.; Qin, H. Advances in Renal Cell Carcinoma Drug Resistance Models. Front. Oncol. 2022, 12, 870396. [Google Scholar] [CrossRef]
- Thong, A.E.; Zhao, H.; Ingels, A.; Valta, M.P.; Nolley, R.; Santos, J.; Young, S.R.; Peehl, D.M. Tissue slice grafts of human renal cell carcinoma: An authentic preclinical model with high engraftment rate and metastatic potential. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 43.e23–43.e43. [Google Scholar] [CrossRef][Green Version]
- Ingels, A.; Zhao, H.; Thong, A.E.; Saar, M.; Valta, M.P.; Nolley, R.; Santos, J.; Peehl, D.M. Preclinical trial of a new dual mTOR inhibitor, MLN0128, using renal cell carcinoma tumorgrafts. Int. J. Cancer 2014, 134, 2322–2329. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhao, H.; Tian, L.; Nolley, R.; Diep, A.N.; Ernst, A.; Fuh, K.; Miao, Y.; Eyben, R.; Leppert, J.T.; et al. S100A10 is a critical mediator of GAS6/AXL-induced angiogenesis in renal cell carcinoma. Cancer Res. 2019, 79, 5758–5768. [Google Scholar] [CrossRef]
- Zhao, H.; Nolley, R.; Chan, A.M.W.; Rankin, E.B.; Peehl, D.M. Cabozantinib inhibits tumor growth and metastasis of a patient-derived xenograft model of papillary renal cell carcinoma with MET mutation. Cancer Biol. Ther. 2017, 18, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Agudelo, J.P.; Upadhyay, D.; Zhang, D.; Zhao, H.; Nolley, R.; Sun, J.; Agarwal, S.; Bok, R.A.; Vigneron, D.B.; Brooks, J.D.; et al. Multiparametric Magnetic Resonance Imaging and Metabolic Characterization of Patient-Derived Xenograft Models of Clear Cell Renal Cell Carcinoma. Metabolites 2022, 12, 1117. [Google Scholar] [CrossRef] [PubMed]
- Kluin, R.J.C.; Kemper, K.; Kuilman, T.; de Ruiter, J.R.; Iyer, V.; Forment, J.V.; Cornelissen-Steijger, P.; de Rink, I.; ter Brugge, P.; Song, J.-Y.; et al. XenofilteR: Computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinform. 2018, 19, 366. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ljungberg, B.; Grankvist, K.; Rasmuson, T.; Tibshirani, R.; Brooks, J.D. Gene Expression Profiling Predicts Survival in Conventional Renal Cell Carcinoma. PLoS Med. 2005, 3, e13. [Google Scholar] [CrossRef]
- Wen, R.M.; Stark, J.C.; Marti, G.E.W.; Fan, Z.; Lyu, A.; Garcia Marques, F.J.; Zhang, X.; Riley, N.M.; Totten, S.M.; Bermudez, A.; et al. Sialylated glycoproteins suppress immune cell killing by binding to Siglec-7 and Siglec-9 in prostate cancer. J. Clin. Investig. 2024, 134, e180282. [Google Scholar] [CrossRef]
- Lewis, M.T.; Caldas, C. The Power and Promise of Patient-Derived Xenografts of Human Breast Cancer. Cold Spring Harb. Perspect. Med. 2024, 14, a041329. [Google Scholar] [CrossRef]
- Caeser, R.; Egger, J.V.; Chavan, S.; Socci, N.D.; Jones, C.B.; Kombak, F.E.; Asher, M.; Roehrl, M.H.; Shah, N.S.; Allaj, V.; et al. Genomic and transcriptomic analysis of a library of small cell lung cancer patient-derived xenografts. Nat. Commun. 2022, 13, 2144. [Google Scholar] [CrossRef]
- Xie, D.; Li, G.; Zheng, Z.; Zhang, X.; Wang, S.; Jiang, B.; Li, X.; Wang, X.; Wu, G. The molecular code of kidney cancer: A path of discovery for gene mutation and precision therapy. Mol. Asp. Med. 2025, 101, 101335. [Google Scholar] [CrossRef]
- Ruan, X.; Lai, C.; Li, L.; Wang, B.; Lu, X.; Zhang, D.; Fang, J.; Lai, M.; Yan, F. Integrative analysis of single-cell and bulk multi-omics data to reveal subtype-specific characteristics and therapeutic strategies in clear cell renal cell carcinoma patients. J. Cancer 2024, 15, 6420–6433. [Google Scholar] [CrossRef]
- Cheng, Y.; Guo, L. Lactate metabolism and lactylation in kidney diseases: Insights into mechanisms and therapeutic opportunities. Ren. Fail. 2025, 47, 2469746. [Google Scholar] [CrossRef]
- Liu, R.; Zou, Z.; Chen, L.; Feng, Y.; Ye, J.; Deng, Y.; Zhu, X.; Zhang, Y.; Lin, J.; Cai, S.; et al. FKBP10 promotes clear cell renal cell carcinoma progression and regulates sensitivity to the HIF2α blockade by facilitating LDHA phosphorylation. Cell Death Dis. 2024, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhai, Z.; Duan, J.; Wang, X.; Zhong, J.; Wu, L.; Li, A.; Cao, M.; Wu, Y.; Shi, H.; et al. Lactate: The Mediator of Metabolism and Immunosuppression. Front. Endocrinol. 2022, 13, 901495. [Google Scholar] [CrossRef] [PubMed]
- Akbay, B.; Omarova, Z.; Trofimov, A.; Sailike, B.; Karapina, O.; Molnár, F.; Tokay, T. Double-Edge Effects of Leucine on Cancer Cells. Biomolecules 2024, 14, 1401. [Google Scholar] [CrossRef]
- Xu, E.; Ji, B.; Jin, K.; Chen, Y. Branched-chain amino acids catabolism and cancer progression: Focus on therapeutic interventions. Front. Oncol. 2023, 13, 1220638. [Google Scholar] [CrossRef]
- Li, Z.; Chen, S.; Wu, X.; Liu, F.; Zhu, J.; Chen, J.; Lu, X.; Chi, R. Research advances in branched-chain amino acid metabolism in tumors. Mol. Cell. Biochem. 2024, online ahead of print. [Google Scholar] [CrossRef]
- Ma, Q.; Li, H.; Song, Z.; Deng, Z.; Huang, W.; Liu, Q. Fueling the fight against cancer: Exploring the impact of branched-chain amino acid catalyzation on cancer and cancer immune microenvironment. Metabolism 2024, 161, 156016. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, T.J.; Erfan, S.C.; Ananieva, E.A. The emerging role of the branched chain aminotransferases, BCATc and BCATm, for anti-tumor T-cell immunity. Immunometabolism 2023, 5, e00014. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Zhu, F.; Duan, Q. The role of branched chain amino acids metabolic disorders in tumorigenesis and progression. Biomed. Pharmacother. 2022, 153, 113390. [Google Scholar] [CrossRef]
- Hoge, A.C.H.; Getz, M.; Zimmer, A.; Ko, M.; Raz, L.; Beroukhim, R.; Golub, T.R.; Ha, G.; Ben-David, U. DNA-based copy number analysis confirms genomic evolution of PDX models. NPJ Precis. Oncol. 2022, 6, 30. [Google Scholar] [CrossRef]

| PDX ID | Age | Gender | Furhman Grade | Clinical Stage | Source of Tissue | Previous Treatment | VHL Status |
|---|---|---|---|---|---|---|---|
| PDX047 | 72 | Female | IV | TXNXM1 | Primary tumor | Radiation and chemotherapy | Mutated |
| PDX054 | 44 | Male | IV | pT3aN0M1 | Colon metastasis | Chemotherapy | Mutated |
| PDX068 | 74 | Male | III | pT3aN0M1 | Primary tumor | None | Mutated |
| PDX072 | 65 | Female | IV | TXNXM1 | Lung metastasis | Immunotherapy, radiation, and chemotherapy | Wild type |
| PDX093 | 73 | Female | IV | pT3bNXM1 | Adrenal gland metastasis | None | Mutated |
| Pathway Upregulated in Type I | Rich Factor | Fold Enrichment | FDR | Count |
| PI3K-AKT signaling pathway | 0.110497238 | 1.987157218 | 0.001347240 | 40 |
| Cytoskeleton in muscle cells Type | 0.129310345 | 2.325487865 | 0.000952201 | 30 |
| Calcium signaling pathway | 0.114173228 | 2.053265401 | 0.008726537 | 29 |
| Cell adhesion molecules | 0.151898734 | 2.731712327 | 0.000624878 | 24 |
| Chemical carcinogenesis-receptor activation | 0.111627907 | 2.007490919 | 0.026983909 | 24 |
| Focal adhesion | 0.113300493 | 2.037570319 | 0.026983909 | 23 |
| Protein digestion and absorption | 0.20952381 | 3.768028591 | 7.75207 × 10−6 | 22 |
| Alcoholism | 0.117021277 | 2.104484053 | 0.026983909 | 22 |
| ECM-receptor interaction | 0.235955056 | 4.243362126 | 3.40346 × 10−6 | 21 |
| Drug metabolism-cytochrome P450 | 0.164383562 | 2.956236628 | 0.026983909 | 12 |
| Pathway Upregulated in Type II | Rich Factor | Fold Enrichment | FDR | Count |
| Complement and coagulation cascades | 0.272727273 | 4.944785276 | 9.47878 × 10−9 | 24 |
| Cytokine-cytokine receptor interaction | 0.134228188 | 2.43367508 | l.90717 × 10−5 | 40 |
| TNF signaling pathway | 0.151260504 | 2.742485951 | 0.007931472 | 18 |
| Viral protein interaction with cytokine/cytokine receptor | 0.16 | 2.900940695 | 0.007931472 | 16 |
| Amoebiasis | 0.145631068 | 2.640419322 | 0.029752513 | 15 |
| IL-17 signaling pathway | 0.147368421 | 2.671919061 | 0.033556746 | 14 |
| Cytoskeleton in muscle cells TypeII | 0.107758621 | 1.95375855 | 0.042656078 | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, Z.; Zhang, D.; Garcia-Marques, F.J.; Bermudez, A.; Zhao, H.; Peehl, D.M.; Pitteri, S.J.; Brooks, J.D. Identification of Molecular Subtypes of Clear-Cell Renal Cell Carcinoma in Patient-Derived Xenografts Using Multi-Omics. Cancers 2025, 17, 1361. https://doi.org/10.3390/cancers17081361
Qiu Z, Zhang D, Garcia-Marques FJ, Bermudez A, Zhao H, Peehl DM, Pitteri SJ, Brooks JD. Identification of Molecular Subtypes of Clear-Cell Renal Cell Carcinoma in Patient-Derived Xenografts Using Multi-Omics. Cancers. 2025; 17(8):1361. https://doi.org/10.3390/cancers17081361
Chicago/Turabian StyleQiu, Zhengyuan, Dalin Zhang, Fernando Jose Garcia-Marques, Abel Bermudez, Hongjuan Zhao, Donna M. Peehl, Sharon J. Pitteri, and James D. Brooks. 2025. "Identification of Molecular Subtypes of Clear-Cell Renal Cell Carcinoma in Patient-Derived Xenografts Using Multi-Omics" Cancers 17, no. 8: 1361. https://doi.org/10.3390/cancers17081361
APA StyleQiu, Z., Zhang, D., Garcia-Marques, F. J., Bermudez, A., Zhao, H., Peehl, D. M., Pitteri, S. J., & Brooks, J. D. (2025). Identification of Molecular Subtypes of Clear-Cell Renal Cell Carcinoma in Patient-Derived Xenografts Using Multi-Omics. Cancers, 17(8), 1361. https://doi.org/10.3390/cancers17081361







