Regulatory Roles for Long ncRNA and mRNA
Abstract
:1. Introduction
2. Non-Coding versus Coding RNA

3. Regulatory Functions of lncRNAs and mRNAs
| Function | Interaction | lncRNA^ | mRNA | Mechanism | References | |||
|---|---|---|---|---|---|---|---|---|
| RNA | miRNA | protein | unknown | |||||
| structural | • | SATIII | forms nuclear stress bodies by attracting splicing and transcription factors to SATIII repeats | [79] | ||||
| • | NEAT1 | forms paraspeckles as large foci directly after transcription | [80] | |||||
| • | H2B | forms HLBs and Cajal bodies | [81] | |||||
| • | VegT | integral part of cytoskeleton at vegetal side in X. laevis oocytes | [82] | |||||
| transcriptional control | • | MEG3 | enhances p53 binding to promoters | [83] | ||||
| • | MALAT1 | interacts with splicing factors to influence the localization and action | [84] | |||||
| • | GAS5 | decoy for the glucocorticoid receptor | [54] | |||||
| • | DHFR-minor | prevents DHFR transcription via triple helix formation and TFIIB interaction | [53] | |||||
| • | SRA | co-activator for many nuclear receptors and transcription factors | [85,86,87,88,89,90,91,92,93,94] | |||||
| transcription elongation | • | 7SK | binds and inhibits P-TEFb, thereby blocking RNAPII elongation | [95,96,97,98,99] | ||||
| • | HIC | binds and activates P-TEFb by displacing 7SK RNA from inhibitory complex, allowing RNAPII elongation | [100] | |||||
| miRNA sponge | • | PTEN-P1 | binds miRNAs that also target PTEN, thereby increasing PTEN protein levels | [101] | ||||
| • | HULC | binds amongst others miR-372, thereby increasing PRKACB protein levels | [102] | |||||
| • | VCAN | binds miR-133a, miR-199a*, miR-144 and miR-431, thereby increasing protein levels of CD34 and FN1 | [103] | |||||
| • | CD44 | binds miR-328, miR-512-3p, miR-491 and miR671, thereby increasing protein levels of COL1α1 and FN1 | [104] | |||||
| RNA degradation | • | 1/2sbsRNAs | imperfect base-pairing with Alu elements in UTRs of mRNA, thereby attraction STAU1 and initializing STAU1-mediated decay | [60] | ||||
| • | speculative | imperfect base-pairing between Alu elements in two mRNAs, thereby attraction STAU1 and initializing STAU1-mediated decay | [105] | |||||
| translational control | • | • | lincRNA-p21 | imperfect base-pairing with mRNA can directly impair translation and/or can attract translation inhibitors | [8] | |||
| • | PU.1-antisense | processed RNA binds sense PU.1 transcript and stalls translation | [106] | |||||
| • | BCMA-AS | blocks translation of the sense BCMA transcript | [107] | |||||
| • | BC1 | interacts with eIF4A and PABP and blocks their interaction, thereby repressing the general translation machinery | [108] | |||||
| • | cytoskeletal mRNAs | inhibit translation by interaction with the RNA-binding domain of PKR, resulting in PKR phosphorylation events | [109] | |||||
| • | P23/TCTP | inhibit translation by interaction with the RNA-binding domain of PKR, resulting in PKR phosphorylation events | [110] | |||||
| • | VEGFA, TPM1, IFN-γ, TNF-α | UTR interacts with PKR, thereby inhibiting translation | [111,112,113,114] | |||||
| • | p53 | interacts with MDM2, thereby preventing p53 degradation and promoting p53 translation | [115] | |||||
| unknown | • | PCAT1 | trans-regulates many genes, including BRCA2 | [116] | ||||
| • | PHB | 3' UTR has unknown trans-regulatory role | [117] | |||||
| • | RNR | 3' UTR has unknown trans-regulatory role | [118] | |||||
| • | c-myc P0 | 5' UTR has unknown trans-regulatory role | [119] | |||||
| guide for epigeneticenzymes | • | HOTTIP | interacts with WDR5/MLL complex | [64] | ||||
| • | HOTAIR | interacts with PRC2 and LSD1-CoREST complex | [55] | |||||
| • | ANRIL | interacts with PRC1 and PRC2 complexes | [15,57] | |||||
| HOTAIRM1 | interacts with PRC1 and PRC2 complexes | [120] | ||||||
| • | KCNQ1OT1 | interacts with PRC2 complexes and G9a | [56] | |||||
| • | AIR | interacts with G9a | [121] | |||||
| • | pRNA | recruits DNMT3b to rDNA promoters | [61] | |||||
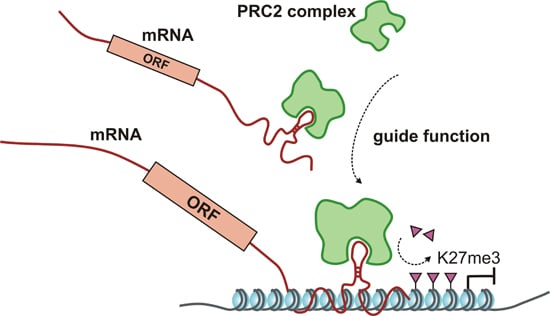
| • | many § | many mRNAs interact with PRC2 complex components | § | |||||
3.1. Structural Function
3.2. Transcriptional Control
3.3. Transcription Elongation
3.4. miRNA Sponge

3.5. RNA Degradation
3.6. Translational Control
3.7. Unknown Function
4. Epigenetic Regulatory Potential of Protein-Coding RNA
| Gene type | % enriched | # enriched | # total examined |
|---|---|---|---|
| lncRNAs | 10.2% | 216 | 2,127 |
| Oncogenes | 44.3% | 182 | 411 |
| Tumor Suppresor Genes | 41.0% | 325 | 793 |
| Imprinted Genes | 41.0% | 34 | 83 |


5. Conclusions
Acknowledgments
References
- Gilbert, W. Origin of life—The RNA world. Nature 1986, 319, 618–618. [Google Scholar] [CrossRef]
- Joyce, G.F. The antiquity of RNA-based evolution. Nature 2002, 418, 214–221. [Google Scholar] [CrossRef]
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry; Freeman: New York, NY, USA, 2003. [Google Scholar]
- The_ENCODE_Project_Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. Gencode: The reference human genome annotation for the encode project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.L.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. Lincrna-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Huarte, M.; Rinn, J.L. Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet. 2010, 19, R152–R161. [Google Scholar] [CrossRef]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer a long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA hotair reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. Malat-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. Gas5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef]
- Zhang, X.; Gejman, R.; Mahta, A.; Zhong, Y.; Rice, K.A.; Zhou, Y.; Cheunsuchon, P.; Louis, D.N.; Klibanski, A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010, 70, 2350–2358. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.D.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Yu, W.Q.; Gius, D.; Onyango, P.; Muldoon-Jacobs, K.; Karp, J.; Feinberg, A.P.; Cui, H.M. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008, 451, 202–206. [Google Scholar] [CrossRef]
- Gibb, E.A.; Vucic, E.A.; Enfield, K.S.; Stewart, G.L.; Lonergan, K.M.; Kennett, J.Y.; Becker-Santos, D.D.; MacAulay, C.E.; Lam, S.; Brown, C.J.; et al. Human cancer long non-coding rna transcriptomes. PLoS One 2011, 6, e25915. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding rnas in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Small regulatory rnas in mammals. Hum. Mol. Genet. 2005, 14, R121–R132. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The gencode v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef]
- Sana, J.; Faltejskova, P.; Svoboda, M.; Slaby, O. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012. [Google Scholar] [CrossRef]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007, 23, 614–622. [Google Scholar] [CrossRef]
- Guil, S.; Soler, M.; Portela, A.; Carrere, J.; Fonalleras, E.; Gomez, A.; Villanueva, A.; Esteller, M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat. Struct. Mol. Biol. 2012, 19, 664–670. [Google Scholar] [CrossRef]
- Furuno, M.; Pang, K.C.; Ninomiya, N.; Fukuda, S.; Frith, M.C.; Bult, C.; Kai, C.; Kawai, J.; Carninci, P.; Hayashizaki, Y.; et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006, 2, 537–553. [Google Scholar]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar]
- Lin, M.F.; Jungreis, I.; Kellis, M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275–i282. [Google Scholar]
- Bánfai, B.; Jia, H.; Khatun, J.; Wood, E.; Risk, B.; Gundling, W.E.; Kundaje, A.; Gunawardena, H.P.; Yu, Y.; Xie, L.; et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012, 22, 1646–1657. [Google Scholar] [CrossRef]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar] [CrossRef]
- Imanishi, T.; Itoh, T.; Suzuki, Y.; O'Donovan, C.; Fukuchi, S.; Koyanagi, K.O.; Barrero, R.A.; Tamura, T.; Yamaguchi-Kabata, Y.; Tanino, M.; et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004, 2, 856–875. [Google Scholar]
- Dinger, M.E.; Gascoigne, D.K.; Mattick, J.S. The evolution of RNAs with multiple functions. Biochimie 2011, 93, 2013–2018. [Google Scholar] [CrossRef] [Green Version]
- Prasanth, K.V.; Spector, D.L. Eukaryotic regulatory RNAs: An answer to the “genome complexity” conundrum. Genes Dev. 2007, 21, 11–42. [Google Scholar] [CrossRef]
- Frith, M.C.; Forrest, A.R.; Nourbakhsh, E.; Pang, K.C.; Kai, C.; Kawai, J.; Carninci, P.; Hayashizaki, Y.; Bailey, T.L.; Grimmond, S.M. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006, 2, 515–528. [Google Scholar]
- Odermatt, A.; Taschner, P.E.; Scherer, S.W.; Beatty, B.; Khanna, V.K.; Cornblath, D.R.; Chaudhry, V.; Yee, W.C.; Schrank, B.; Karpati, G.; et al. Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with serca1: Absence of structural mutations in five patients with brody disease. Genomics 1997, 45, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Klaudiny, J.; von der Kammer, H.; Scheit, K.H. Characterization by cdna cloning of the mRNA of a highly basic human protein homologous to the yeast ribosomal protein yl41. Biochem. Biophys. Res. Commun. 1992, 187, 901–906. [Google Scholar]
- Galindo, M.I.; Pueyo, J.I.; Fouix, S.; Bishop, S.A.; Couso, J.P. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007, 5, 1052–1062. [Google Scholar]
- Clamp, M.; Fry, B.; Kamal, M.; Xie, X.H.; Cuff, J.; Lin, M.F.; Kellis, M.; Lindblad-Toh, K.; Lander, E.S. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. USA 2007, 104, 19428–19433. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Garber, M.; Zuk, O.; Lin, M.F.; Parker, B.J.; Washietl, S.; Kheradpour, P.; Ernst, J.; Jordan, G.; Mauceli, E.; et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 2011, 478, 476–482. [Google Scholar] [CrossRef] [Green Version]
- Duret, L.; Chureau, C.; Samain, S.; Weissenbach, J.; Avner, P. The xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 2006, 312, 1653–1655. [Google Scholar]
- Brosch, M.; Saunders, G.I.; Frankish, A.; Collins, M.O.; Yu, L.; Wright, J.; Verstraten, R.; Adams, D.J.; Harrow, J.; Choudhary, J.S.; et al. Shotgun proteomics aids discovery of novel protein-coding genes, alternative splicing, and “resurrected” pseudogenes in the mouse genome. Genome Res. 2011, 21, 756–767. [Google Scholar] [CrossRef]
- Niazi, F.; Valadkhan, S. Computational analysis of functional long noncoding rnas reveals lack of peptide-coding capacity and parallels with 3' UTRs. RNA 2012, 18, 825–843. [Google Scholar] [CrossRef]
- Wan, Y.; Qu, K.; Ouyang, Z.; Kertesz, M.; Li, J.; Tibshirani, R.; Makino, D.L.; Nutter, R.C.; Segal, E.; Chang, H.Y. Genome-wide measurement of RNA folding energies. Mol. Cell 2012, 48, 169–181. [Google Scholar] [CrossRef]
- Hannon, G.J.; Rivas, F.V.; Murchison, E.P.; Steitz, J.A. The expanding universe of noncoding RNAs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 551–564. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding rnas. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef]
- Qiu, M.T.; Hu, J.W.; Yin, R.; Xu, L. Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol. 2013. [Google Scholar] [CrossRef]
- Martianov, I.; Ramadass, A.; Serra Barros, A.; Chow, N.; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007, 445, 666–670. [Google Scholar]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA anril is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St. Laurent, G., 3rd; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. LncRNAs transactivate STAU1-mediated mrna decay by duplexing with 3' UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Schmitz, K.M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3B and silencing of rRNA genes. Genes Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef]
- Aguilera, A.; Garcia-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef]
- Jeon, Y.; Lee, J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 2011, 146, 119–133. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Baldassarre, A.; Masotti, A. Long non-coding RNAs and p53 regulation. Int. J. Mol. Sci. 2012, 13, 16708–16717. [Google Scholar] [CrossRef]
- Da Sacco, L.; Baldassarre, A.; Masotti, A. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int. J. Mol. Sci. 2012, 13, 97–114. [Google Scholar]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. Lncrnadisease: A database for long-non-coding Rna-associated diseases. Nucleic Acids Res. 2013, 41, D983–D986. [Google Scholar] [CrossRef]
- Chen, J.Z.; Yang, T.; Yu, H.; Sun, K.; Shi, Y.; Song, W.H.; Bai, Y.Y.; Wang, X.J.; Lou, K.J.; Song, Y.; et al. A functional variant in the 3'-UTR of angiopoietin-1 might reduce stroke risk by interfering with the binding efficiency of microRNA 211. Hum. Mol. Genet. 2010, 19, 2524–2533. [Google Scholar] [CrossRef]
- Delay, C.; Calon, F.; Mathews, P.; Hebert, S.S. Alzheimer-specific variants in the 3' UTR of amyloid precursor protein affect microrna function. Mol. Neurodegener. 2011, 6. [Google Scholar] [CrossRef]
- Wilkie, G.S.; Dickson, K.S.; Gray, N.K. Regulation of mrna translation by 5'- and 3'-UTR-binding factors. Trends Biochem. Sci. 2003, 28, 182–188. [Google Scholar] [CrossRef]
- Kochetov, A.V.; Ischenko, I.V.; Vorobiev, D.G.; Kel, A.E.; Babenko, V.N.; Kisselev, L.L.; Kolchanov, N.A. Eukaryotic mrnas encoding abundant and scarce proteins are statistically dissimilar in many structural features. FEBS Lett. 1998, 440, 351–355. [Google Scholar] [CrossRef]
- Pickering, B.M.; Willis, A.E. The implications of structured 5' untranslated regions on translation and disease. Semin. Cell Dev. Biol. 2005, 16, 39–47. [Google Scholar] [CrossRef]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microrna targets containing simultaneous 5'-UTR and 3'-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef]
- Zhang, L.N.; Liu, Y.X.; Song, F.J.; Zheng, H.; Hu, L.M.; Lu, H.; Liu, P.F.; Hao, X.S.; Zhang, W.; Chen, K.X. Functional SNP in the microrna-367 binding site in the 3' UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. USA 2011, 108, 13653–13658. [Google Scholar] [CrossRef]
- Valgardsdottir, R.; Chiodi, F.; Giordano, M.; Cobianchi, F.; Riva, S.; Biamonti, G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell 2005, 16, 2597–2604. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: Neat1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Shevtsov, S.P.; Dundr, M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011, 13, 167–173. [Google Scholar] [CrossRef]
- Kloc, M.; Wilk, K.; Vargas, D.; Shirato, Y.; Bilinski, S.; Etkin, L.D. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of xenopus oocytes. Development 2005, 132, 3445–3457. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Colley, S.M.; Leedman, P.J. Sra and its binding partners: An expanding role for RNA-binding coregulators in nuclear receptor-mediated gene regulation. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 25–33. [Google Scholar] [CrossRef]
- Lanz, R.B.; McKenna, N.J.; Onate, S.A.; Albrecht, U.; Wong, J.M.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999, 97, 17–27. [Google Scholar] [CrossRef]
- Deblois, G.; Giguere, V. Ligand-independent coactivation of er alpha AF-1 by steroid receptor RNA activator (SRA) via MAPK activation. J. Steroid Biochem. Mol. Biol. 2003, 85, 123–131. [Google Scholar] [CrossRef]
- Zhao, X.S.; Patton, J.R.; Davis, S.L.; Florence, B.; Ames, S.J.; Spanjaard, R.A. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor rna activator. Mol. Cell 2004, 15, 549–558. [Google Scholar]
- Hatchell, E.C.; Colley, S.M.; Beveridge, D.J.; Epis, M.R.; Stuart, L.M.; Giles, K.M.; Redfern, A.D.; Miles, L.E.C.; Barker, A.; MacDonald, L.M.; et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol. Cell 2006, 22, 657–668. [Google Scholar] [CrossRef]
- Caretti, G.; Schiltz, R.L.; Dilworth, F.J.; Di Padova, M.; Zhao, P.; Ogryzko, V.; Fuller-Pace, F.V.; Hoffman, E.P.; Tapscott, S.J.; Sartorelli, V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell 2006, 11, 547–560. [Google Scholar] [CrossRef]
- Hube, F.; Velasco, G.; Rollin, J.; Furling, D.; Francastel, C. Steroid receptor rna activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011, 39, 513–525. [Google Scholar] [CrossRef]
- Watanabe, M.; Yanagisawa, J.; Kitagawa, H.; Takeyama, K.; Ogawa, S.; Arao, Y.; Suzawa, M.; Kobayashi, Y.; Yano, T.; Yoshikawa, H.; et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor ALPHA coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001, 20, 1341–1352. [Google Scholar] [CrossRef]
- Zhao, X.S.; Patton, J.R.; Ghosh, S.K.; Fischel-Ghodsian, N.; Shen, L.; Spanjaard, R.A. Pus3p-and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol. Endocrinol. 2007, 21, 686–699. [Google Scholar]
- Lanz, R.B.; Razani, B.; Goldberg, A.D.; O'Malley, B.W. Distinct RNA motifs are important for coactivation of steroid hormone receptors by steroid receptor RNA activator (SRA). Proc. Natl. Acad. Sci. USA 2002, 99, 16081–16086. [Google Scholar]
- Yik, J.H.; Chen, R.; Nishimura, R.; Jennings, J.L.; Link, A.J.; Zhou, Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 2003, 12, 971–982. [Google Scholar] [CrossRef]
- Egloff, S.; van Herreweghe, E.; Kiss, T. Regulation of polymerase ii transcription by 7SK snRNA: Two distinct rna elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 2006, 26, 630–642. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Kiss, T.; Michels, A.A.; Bensaude, O. 7SK small nuclear rna binds to and inhibits the activity of CDK9/Cyclin T complexes. Nature 2001, 414, 322–325. [Google Scholar] [CrossRef]
- Barboric, M.; Kohoutek, J.; Price, J.P.; Blazek, D.; Price, D.H.; Peterlin, B.M. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-Tefb. EMBO J. 2005, 24, 4291–4303. [Google Scholar] [CrossRef]
- He, N.; Jahchan, N.S.; Hong, E.; Li, Q.; Bayfield, M.A.; Maraia, R.J.; Luo, K.; Zhou, Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 2008, 29, 588–599. [Google Scholar] [CrossRef]
- Young, T.M.; Tsai, M.; Tian, B.; Mathews, M.B.; Pe’ery, T. Cellular mrna activates transcription elongation by displacing 7SK RNA. PLoS One 2007, 2, e1010. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.W.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar]
- Wang, J.Y.; Liu, X.F.; Wu, H.C.; Ni, P.H.; Gu, Z.D.; Qiao, Y.X.; Chen, N.; Sun, F.Y.; Fan, Q.S. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar]
- Fang, L.; Du, W.W.; Yang, X.; Chen, K.; Ghanekar, A.; Levy, G.; Yang, W.; Yee, A.J.; Lu, W.Y.; Xuan, J.W.; et al. Versican 3'-untranslated region (3'-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating mirna activity. FASEB J. 2013, 27, 907–919. [Google Scholar] [CrossRef]
- Rutnam, Z.J.; Yang, B.B. The non-coding 3' UTR of CD44 induces metastasis by regulating extracellular matrix functions. J. Cell Sci. 2012, 125, 2075–2085. [Google Scholar]
- Wang, P.; Yin, S.; Zhang, Z.; Xin, D.; Hu, L.; Kong, X.; Hurst, L.D. Evidence for common short natural trans sense-antisense pairing between transcripts from protein coding genes. Genome Biol. 2008, 9, R169. [Google Scholar] [CrossRef] [Green Version]
- Ebralidze, A.K.; Guibal, F.C.; Steidl, U.; Zhang, P.; Lee, S.; Bartholdy, B.; Jorda, M.A.; Petkova, V.; Rosenbauer, F.; Huang, G.; et al. Pu.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008, 22, 2085–2092. [Google Scholar] [CrossRef]
- Hatzoglou, A.; Deshayes, F.; Madry, C.; Lapree, G.; Castanas, E.; Tsapis, A. Natural antisense RNA inhibits the expression of BCMA, a tumour necrosis factor receptor homologue. BMC Mol. Biol. 2002, 3, 4. [Google Scholar] [CrossRef]
- Wang, H.; Iacoangeli, A.; Lin, D.; Williams, K.; Denman, R.B.; Hellen, C.U.; Tiedge, H. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 2005, 171, 811–821. [Google Scholar] [CrossRef]
- Nussbaum, J.M.; Gunnery, S.; Mathews, M.B. The 3'-untranslated regions of cytoskeletal muscle mrnas inhibit translation by activating the double-stranded rna-dependent protein kinase PKR. Nucleic Acids Res. 2002, 30, 1205–1212. [Google Scholar] [CrossRef]
- Bommer, U.A.; Borovjagin, A.V.; Greagg, M.A.; Jeffrey, I.W.; Russell, P.; Laing, K.G.; Lee, M.; Clemens, M.J. The mRNA of the translationally controlled tumor protein p23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR. RNA 2002, 8, 478–496. [Google Scholar] [CrossRef]
- Masuda, K.; Teshima-Kondo, S.; Mukaijo, M.; Yamagishi, N.; Nishikawa, Y.; Nishida, K.; Kawai, T.; Rokutan, K. A novel tumor-promoting function residing in the 5' non-coding region of vascular endothelial growth factor mRNA. PLoS Med. 2008, 5, e94. [Google Scholar] [CrossRef]
- Ben-Asouli, Y.; Banai, Y.; Pel-Or, Y.; Shir, A.; Kaempfer, R. Human interferon-gamma mrna autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 2002, 108, 221–232. [Google Scholar] [CrossRef]
- Davis, S.; Watson, J.C. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3' untranslated regions of human alpha-tropomyosin. Proc. Natl. Acad. Sci. USA 1996, 93, 508–513. [Google Scholar] [CrossRef]
- Osman, F.; Jarrous, N.; Ben-Asouli, Y.; Kaempfer, R. A cis-acting element in the 3'-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 1999, 13, 3280–3293. [Google Scholar] [CrossRef]
- Candeias, M.M.; Malbert-Colas, L.; Powell, D.J.; Daskalogianni, C.; Maslon, M.M.; Naski, N.; Bourougaa, K.; Calvo, F.; Fahraeus, R. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol. 2008, 10, 1098–1105. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef]
- Manjeshwar, S.; Branam, D.E.; Lerner, M.R.; Brackett, D.J.; Jupe, E.R. Tumor suppression by the prohibitin gene 3' untranslated region RNA in human breast cancer. Cancer Res. 2003, 63, 5251–5256. [Google Scholar]
- Fan, H.; Villegas, C.; Huang, A.; Wright, J.A. Suppression of malignancy by the 3' untranslated regions of ribonucleotide reductase R1 and R2 messenger RNAs. Cancer Res. 1996, 56, 4366–4369. [Google Scholar]
- Blume, S.W.; Miller, D.M.; Guarcello, V.; Shrestha, K.; Meng, Z.; Snyder, R.C.; Grizzle, W.E.; Ruppert, J.M.; Gartland, G.L.; Stockard, C.R.; et al. Inhibition of tumorigenicity by the 5'-untranslated RNA of the human c-myc P0 transcript. Exp. Cell Res. 2003, 288, 131–142. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. Lincrnas act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef]
- Alastalo, T.P.; Hellesuo, M.; Sandqvist, A.; Hietakangas, V.; Kallio, M.; Sistonen, L. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J. Cell Sci. 2003, 116, 3557–3570. [Google Scholar] [CrossRef]
- Biamonti, G.; Caceres, J.F. Cellular stress and RNA splicing. Trends Biochem. Sci. 2009, 34, 146–153. [Google Scholar] [CrossRef]
- Fox, A.H.; Lam, Y.W.; Leung, A.K.L.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A novel nuclear domain. Curr. Biol. 2002, 12, 13–25. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007, 8, 39. [Google Scholar]
- Sasaki, Y.T.F.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. Men epsilon/beta noncoding rnas are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar]
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(a) tail. Nat. Rev. Genet. 2008, 9, 843–854. [Google Scholar] [CrossRef]
- Cioce, M.; Lamond, A.I. Cajal bodies: A long history of discovery. In Annual Review of Cell and Developmental Biology; Annual Reviews: Palo Alto, CA, USA, 2005; Volume 21, pp. 105–131. [Google Scholar]
- Matera, A.G.; Izaguire-Sierra, M.; Praveen, K.; Rajendra, T.K. Nuclear bodies: Random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell 2009, 17, 639–647. [Google Scholar] [CrossRef]
- Heasman, J.; Wessely, O.; Langland, R.; Craig, E.J.; Kessler, D.S. Vegetal localization of maternal mRNAs is disrupted by vegt depletion. Dev. Biol. 2001, 240, 377–386. [Google Scholar] [CrossRef]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. MicroRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef]
- Benetatos, L.; Hatzimichael, E.; Dasoula, A.; Dranitsaris, G.; Tsiara, S.; Syrrou, M.; Georgiou, I.; Bourantas, K.L. CPG methylation analysis of the MEG3 and snRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk. Res. 2010, 34, 148–153. [Google Scholar] [CrossRef]
- Yamada, K.; Kano, J.; Tsunoda, H.; Yoshikawa, H.; Okubo, C.; Ishiyama, T.; Noguchi, M. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006, 97, 106–112. [Google Scholar] [CrossRef]
- Lin, R.; Maeda, S.; Liu, C.; Karin, M.; Edgington, T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 2007, 26, 851–858. [Google Scholar] [CrossRef]
- Tano, K.; Mizuno, R.; Okada, T.; Rakwal, R.; Shibato, J.; Masuo, Y.; Ijiri, K.; Akimitsu, N. Malat-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010, 584, 4575–4580. [Google Scholar] [CrossRef]
- Kawashima, H.; Takano, H.; Sugita, S.; Takahara, Y.; Sugimura, K.; Nakatani, T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: Expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem. J. 2003, 369, 163–171. [Google Scholar] [CrossRef]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 2000, 49, 341–351. [Google Scholar] [CrossRef]
- Shi, Y.H.; Downes, M.; Xie, W.; Kao, H.Y.; Ordentlich, P.; Tsai, C.C.; Hon, M.; Evans, R.M. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001, 15, 1140–1151. [Google Scholar] [CrossRef]
- Emberley, E.; Huang, G.J.; Hamedani, M.K.; Czosnek, A.; Ali, D.; Grolla, A.; Lu, B.; Watson, P.H.; Murphy, L.C.; Leygue, E. Identification of new human coding steroid receptor RNA activator isoforms. Biochem. Biophys. Res. Commun. 2003, 301, 509–515. [Google Scholar] [CrossRef]
- Hussein-Fikret, S.; Fuller, P.J. Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Mol. Cell. Endocrinol. 2005, 229, 149–160. [Google Scholar] [CrossRef]
- Lanz, R.B.; Chua, S.S.; Barron, N.; Soder, B.M.; DeMayo, F.; O’Malley, B.W. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell. Biol. 2003, 23, 7163–7176. [Google Scholar] [CrossRef]
- Leygue, E.; Dotzlaw, H.; Watson, P.H.; Murphy, L.C. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999, 59, 4190–4193. [Google Scholar]
- Hube, F.; Guo, J.M.; Chooniedass-Kothari, S.; Cooper, C.; Hamedani, M.K.; Dibrov, A.A.; Blanchard, A.A.A.; Wang, X.M.; Deng, G.; Myal, Y.; et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006, 25, 418–428. [Google Scholar] [CrossRef]
- Cooper, C.; Guo, J.M.; Yan, Y.; Chooniedass-Kothari, S.; Hube, F.; Hamedani, M.K.; Murphy, L.C.; Myal, Y.; Leygue, E. Increasing the relative expression of endogenous non-coding steroid receptor RNA activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009, 37, 4518–4531. [Google Scholar] [CrossRef]
- Murphy, L.C.; Simon, S.L.R.; Parkes, A.; Leygue, E.; Dotzlaw, H.; Snell, L.; Troup, S.; Adeyinka, A.; Watson, P.H. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res. 2000, 60, 6266–6271. [Google Scholar]
- Chooniedass-Kothari, S.; Hamedani, M.K.; Troup, S.; Hube, F.; Leygue, E. The steroid receptor RNA activator protein is expressed in breast tumor tissues. Int. J. Cancer 2006, 118, 1054–1059. [Google Scholar] [CrossRef]
- Faust, T.; Frankel, A.; D’Orso, I. Transcription control by long non-coding RNAs. Transcription 2012, 3, 78–86. [Google Scholar] [CrossRef]
- Wassarman, D.A.; Steitz, J.A. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 1991, 11, 3432–3445. [Google Scholar]
- Marz, M.; Donath, A.; Verstraete, N.; Nguyen, V.T.; Stadler, P.F.; Bensaude, O. Evolution of 7SK RNA and its protein partners in metazoa. Mol. Biol. Evol. 2009, 26, 2821–2830. [Google Scholar] [CrossRef]
- Krueger, B.J.; Jeronimo, C.; Roy, B.B.; Bouchard, A.; Barrandon, C.; Byers, S.A.; Searcey, C.E.; Cooper, J.J.; Bensaude, O.; Cohen, E.A.; et al. Larp7 is a stable component of the 7SK snRNP while P-Tefb, hexim1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008, 36, 2219–2229. [Google Scholar] [CrossRef]
- Markert, A.; Grimm, M.; Martinez, J.; Wiesner, J.; Meyerhans, A.; Meyuhas, O.; Sickmann, A.; Fischer, U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008, 9, 569–575. [Google Scholar] [CrossRef]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of micrornas on protein output. Nature 2008, 455, 64–71. [Google Scholar]
- Bartel, D.P. Micrornas: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Ventura, A.; Jacks, T. MicroRNAs and cancer: Short RNAs go a long way. Cell 2009, 136, 586–591. [Google Scholar] [CrossRef]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010, 20, R858–R861. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar]
- Cazalla, D.; Yario, T.; Steitz, J.A. Down-regulation of a host microRNA by a herpesvirus saimiri noncoding RNA. Science 2010, 328, 1563–1566. [Google Scholar] [CrossRef]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar] [CrossRef]
- Almeida, M.I.; Reis, R.M.; Calin, G.A. Decoy activity through microRNAs: The therapeutic implications. Expert Opin. Biol. Ther. 2012, 12, 1153–1159. [Google Scholar] [CrossRef] [Green Version]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating cells express mrnas with shortened 3' untranslated regions and fewer microRNA target sites. Science 2008, 320, 1643–1647. [Google Scholar] [CrossRef]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar]
- Matsui, K.; Nishizawa, M.; Ozaki, T.; Kimura, T.; Hashimoto, I.; Yamada, M.; Kaibori, M.; Kamiyama, Y.; Ito, S.; Okumura, T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology 2008, 47, 686–697. [Google Scholar]
- Yanagida, S.; Taniue, K.; Sugimasa, H.; Nasu, E.; Takeda, Y.; Kobayashi, M.; Yamamoto, T.; Okamoto, A.; Akiyama, T. ASBEL, an ANA/BTG3 antisense transcript required for tumorigenicity of ovarian carcinoma. Sci. Rep. 2013, 3, 1305. [Google Scholar]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2012. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Kindler, S.; Wang, H.; Richter, D.; Tiedge, H. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 2005, 21, 223–245. [Google Scholar] [CrossRef]
- Hollstein, M.; Sidransky, D.; Vogelstein, B.; Harris, C.C. P53 mutations in human cancers. Science 1991, 253, 49–53. [Google Scholar]
- Soussi, T.; Wiman, K.G. Shaping genetic alterations in human cancer: The p53 mutation paradigm. Cancer Cell 2007, 12, 303–312. [Google Scholar] [CrossRef]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef]
- Vousden, K.H.; Prives, C. Blinded by the light: The growing complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef]
- Riley, T.; Sontag, E.; Chen, P.; Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008, 9, 402–412. [Google Scholar] [CrossRef]
- Kubbutat, M.H.G.; Jones, S.N.; Vousden, K.H. Regulation of p53 stability by MDM2. Nature 1997, 387, 299–303. [Google Scholar] [CrossRef]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. MDM2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef]
- Gajjar, M.; Candeias, M.M.; Malbert-Colas, L.; Mazars, A.; Fujita, J.; Olivares-Illana, V.; Fahraeus, R. The p53 mRNA-MDM2 interaction controls MDM2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell 2012, 21, 25–35. [Google Scholar] [CrossRef]
- Rastinejad, F.; Blau, H.M. Genetic complementation reveals a novel regulatory role for 3' untranslated regions in growth and differentiation. Cell 1993, 72, 903–917. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding rnas associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Bertani, S.; Sauer, S.; Bolotin, E.; Sauer, F. The noncoding RNA mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell 2011, 43, 1040–1046. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse x chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef]
- Karapetyan, A.R.; Kuiper, R.A.; Coolen, M.W. Department of Human Genetics, Nijmegen Centre for Molecular Life Sciences (NCMLS), Radboud University Nijmegen Medical Centre, P.O. Box 9101, Nijmegen 6500 HB, The Netherlands. Unpublished work, 2013.
- Hamamoto, R.; Furukawa, Y.; Morita, M.; Iimura, Y.; Silva, F.P.; Li, M.; Yagyu, R.; Nakamura, Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004, 6, 731–740. [Google Scholar] [CrossRef]
- Hamamoto, R.; Silva, F.P.; Tsuge, M.; Nishidate, T.; Katagiri, T.; Nakamura, Y.; Furukawa, Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006, 97, 113–118. [Google Scholar] [CrossRef]
- St. Laurent, G.; Shtokalo, D.; Tackett, M.R.; Yang, Z.; Eremina, T.; Wahlestedt, C.; Urcuqui-Inchima, S.; Seilheimer, B.; McCaffrey, T.A.; Kapranov, P. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics 2012, 13, 504. [Google Scholar]
- Kanhere, A.; Viiri, K.; Araujo, C.C.; Rasaiyaah, J.; Bouwman, R.D.; Whyte, W.A.; Pereira, C.F.; Brookes, E.; Walker, K.; Bell, G.W.; et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell 2010, 38, 675–688. [Google Scholar] [CrossRef]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Karapetyan, A.R.; Buiting, C.; Kuiper, R.A.; Coolen, M.W. Regulatory Roles for Long ncRNA and mRNA. Cancers 2013, 5, 462-490. https://doi.org/10.3390/cancers5020462
Karapetyan AR, Buiting C, Kuiper RA, Coolen MW. Regulatory Roles for Long ncRNA and mRNA. Cancers. 2013; 5(2):462-490. https://doi.org/10.3390/cancers5020462
Chicago/Turabian StyleKarapetyan, Armen R., Coen Buiting, Renske A. Kuiper, and Marcel W. Coolen. 2013. "Regulatory Roles for Long ncRNA and mRNA" Cancers 5, no. 2: 462-490. https://doi.org/10.3390/cancers5020462
APA StyleKarapetyan, A. R., Buiting, C., Kuiper, R. A., & Coolen, M. W. (2013). Regulatory Roles for Long ncRNA and mRNA. Cancers, 5(2), 462-490. https://doi.org/10.3390/cancers5020462