Abstract
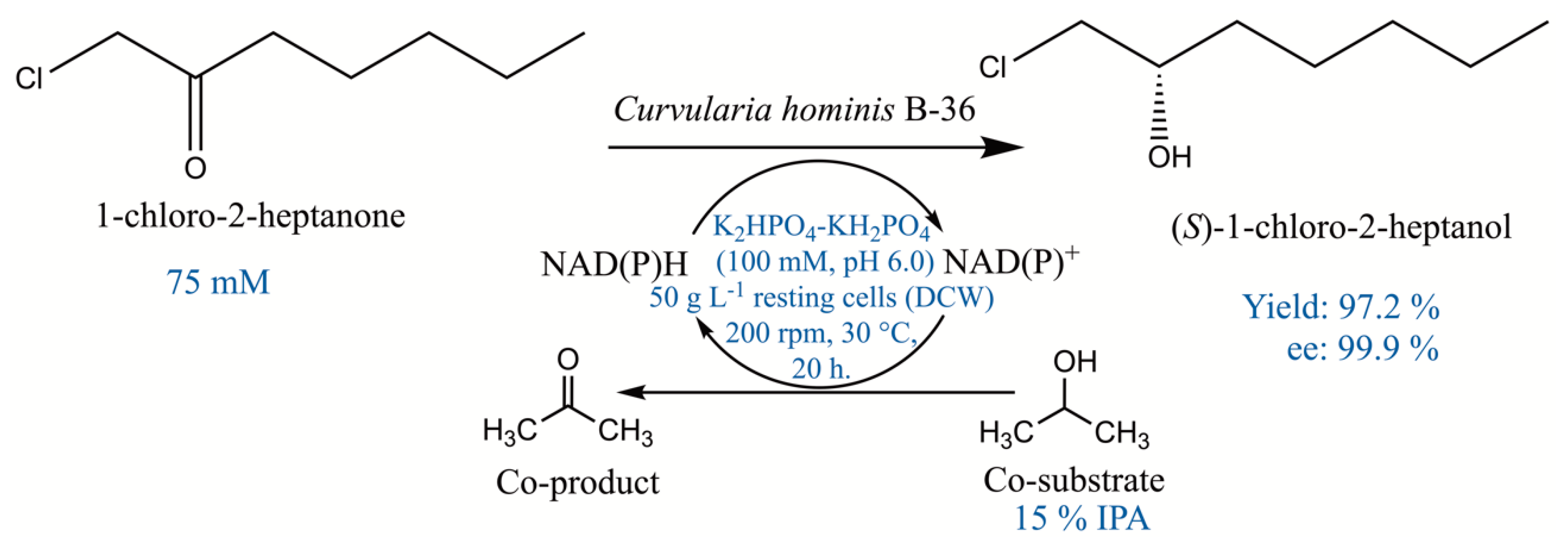
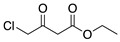
(S)-1-chloro-2-heptanol is an enantiopure chemical of great value that can synthesize Treprostinil for treating primary pulmonary hypertension. In this work, a new strain B-36, capable of asymmetric reduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol, was screened and identified as Curvularia hominis B-36 (CCTCC M 2017654) based on the morphological and internally transcribed spacer (ITS) sequence. The reductive capacity of Curvularia hominis B-36 was investigated as a whole-cell biocatalyst in the bioreduction, and the excellent yield (97.2%) and enantiomeric excess (ee) value (99.9%) were achieved under the optimal conditions as follows: 75 mM 1-chloro-2-heptanone, K2HPO4-KH2PO4 (100 mM, pH 6.0), 50 g L−1 resting cells (dry cell weight; DCW), 15% (v/v) isopropanol as co-substrate, 200 rpm, 30 °C, 20 h. The scaled-up biocatalytic process was accomplished at a bioreactor in a 1.5 L working volume, showing superb yield (~97%) and selectivity (99.9%). The product (S)-1-chloro-2-heptanol was purified and characterized by NMR. Curvularia hominis B-36 is a novel catalyst and the asymmetric synthesis route is benign and eco-friendly.
1. Introduction
In the pharma industry, chiral alcohols are important structural and functional chemicals of multiple active pharmaceutical ingredients (APIs) of drug candidates [1,2,3]. The synthesis pathway of asymmetric reduction of ketones to their corresponding chiral alcohols in a chemical reaction requires the chiral specific ligands and metals or high temperature and pressure [4,5]. Biosynthesis has been described as a distinguished and simple pathway to synthesize active enantiomerically pharmaceutical intermediates, accessing high stereoselectivity (ee > 99%), 100% of theoretical yield, room reaction temperature, metal-free catalysts, etc. [6,7]. Biocatalysis, as an environmentally benign technology, offers a pollution-free approach to asymmetric synthesis [8,9].
(S)-1-chloro-2-heptanol is a critical chiral intermediate for synthesizing Treprostinil, which has been used for the treatment of primary pulmonary hypertension (PPH) [10,11]. Two approaches to synthesize (S)-1-chloro-2-heptanol are chemical and biosynthetic methods. The chemical method for synthesizing (S)-1-Chloro-2-heptanone refers to lower reaction temperature (−78 °C) and Grignard reagent [12,13]. Mostly, asymmetry biocatalytic reduction was carried out at room temperature [14,15]. In 1992, Sakai used a bakers’ yeast as biocatalyst to synthesize (S)-1-chloro-2-heptanone at room temperature with 56% of yield and 65% of ee [16]. However, the lower ee value (65%) cannot meet the requirement of asymmetric synthesis. Enzymes and whole cells are commonly employed as catalysts in biocatalytic reactions. In the bioreduction, enzyme catalysis required additional co-factors (NAD(P)H) for coenzyme regeneration [17]. The extra supplementation of NAD(P)H is expensive, putting a burden on the commercial application [18,19]. Whole-cell-mediated bioreduction can establish a coenzyme regeneration system without feeding additional NAD(P)H, so whole-cell catalysis is an efficient, green, economical, and environmentally friendly synthetic route for asymmetric reductions [20,21]. Many microorganisms have been developed as biocatalysts in the bioreductions, such as Weissella cibaria N9 for bioreduction of trans-4-phenylbut-3-en-2-one [22]; Arthrobacter sp. SUK 1201 for bioreduction of hexavalent chromium [23]; Leuconostoc pseudomesenteroides N13 for bioreduction of cyclohexyl phenyl ketone [24]; Lactobacillus senmaizukei for asymmetric reduction of prochiral aromatic and hetero aromatic ketones, etc. [15]. Therefore, screening and discovery of new biocatalysts are of great significance in asymmetric synthesis.
In this work, a soil microbe B-36 was screened and isolated, exhibiting an outstanding enantiomeric excess (ee > 99%) for efficient bioreduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol. B-36 cells were identified by analyzing the sequence of the internal transcribed spacer (ITS) region and named Curvularia hominis B-36. It is a novel biocatalyst for asymmetric reduction and was reported for the first time for synthesizing (S)-1-chloro-2-heptanol. Some indispensable reaction parameters were systematically investigated and established an efficient and environmentally friendly approach for asymmetric bioreduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol in a scale-up process.
2. Results and Discussion
2.1. Isolation and Characterization of Strain B-36
Asymmetric biocatalytic reactions have evolved increasingly broad applications in pharmaceutical synthesis [25,26]. Screening biocatalysts with high ee value and yield is quite principal in biocatalysis [7]. After multiple cycles of screening, a fungal strain B-36 exhibited the highest catalytic efficiency, showing excellent enantioselectivity (>99% ee) and yield (~90.7%) to produce (S)-1-chloro-2-heptanol. Therefore, it was identified as the best strain for further evaluation. B-36 were incubated on PDA agar plates at 25 °C for a week. The colonies are circular, attaining 70–73 mm diam and spread around. The mycelia are white to dark green, with a fimbriate margin.
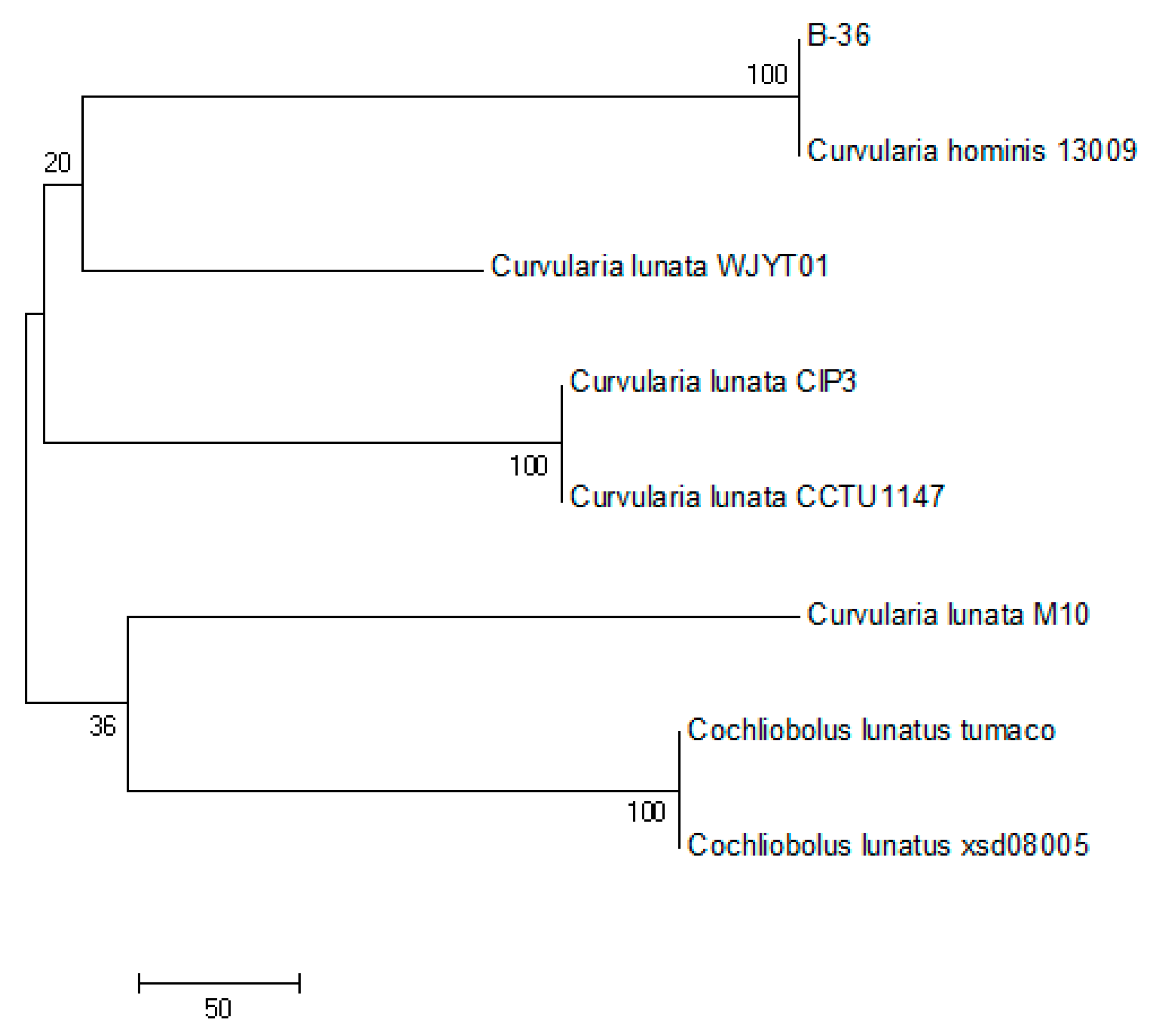
The partial ITS sequence of B-36 (575 bp) was verified and deposited in the GenBank database under accession no. MH656705.1. The ITS sequence alignment and phylogenetic analysis illustrated that strain B-36 was closely clustered with Curvularia hominis 13009 (GenBank accession no. LC494370.1), sharing a high similarity of 99% (Figure 1). Therefore, the strain B-36 was identified as Curvularia hominis B-36 and has been deposited in the China Center for Type Culture Collection (CCTCC M 2017654).
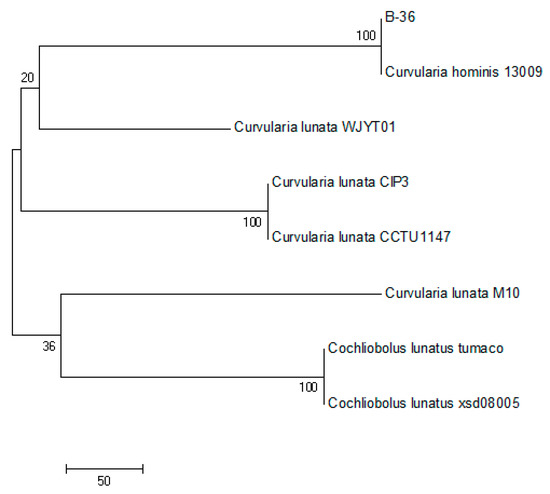
Figure 1.
Phylogenetic tree between B-36 and other related species based on ITS sequences.
2.2. Effects of Key Factors for Synthesizing (S)-1-chloro-2-heptanol by Curvularia hominis B-36
2.2.1. Co-Substrate
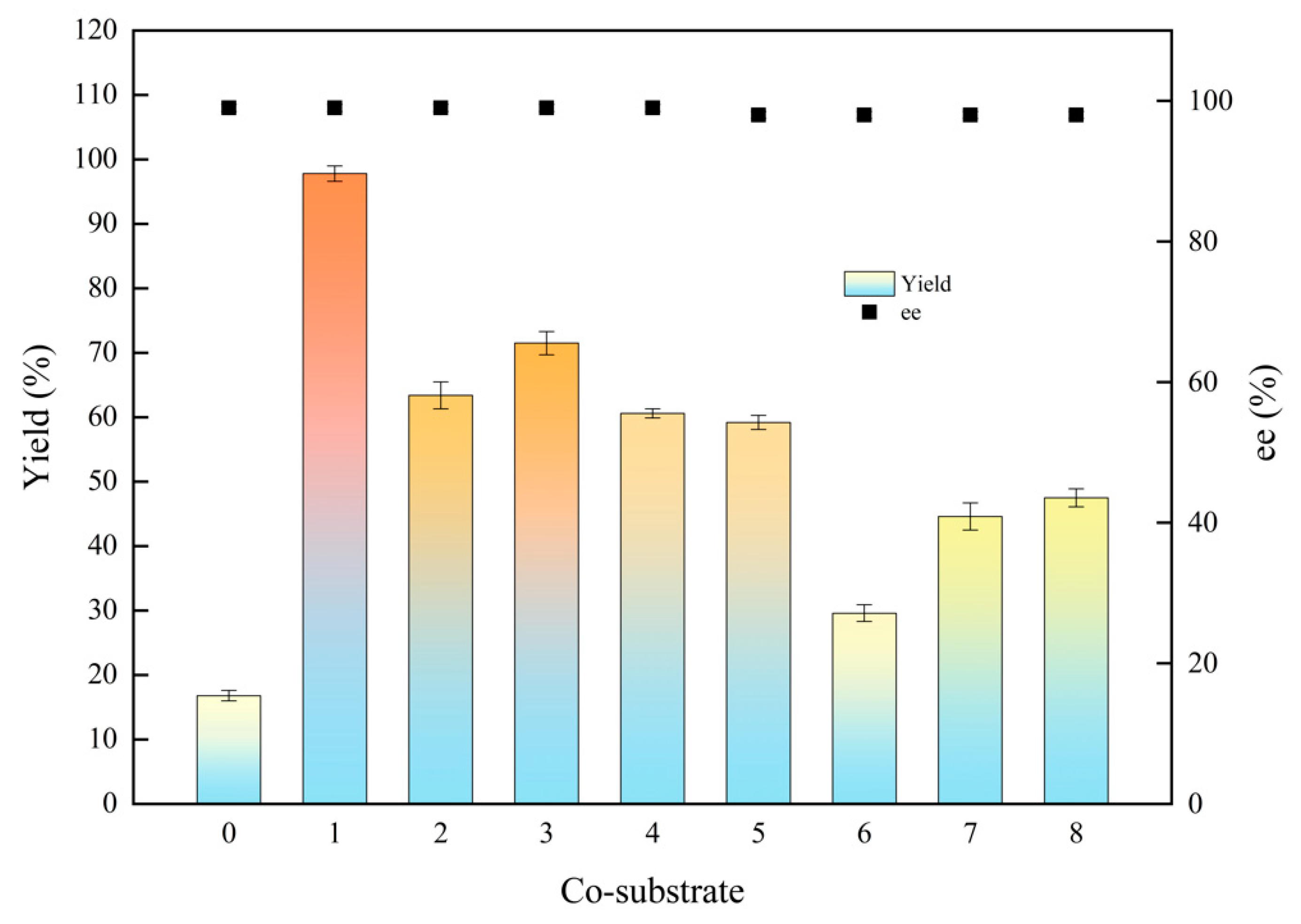
Asymmetric reduction is a significant approach to synthesizing chiral alcohols. Carbonyl reductase requires nicotinamide cofactor NAD(P)H as a hydrogen donor to participate in the coenzyme regeneration system. Carbohydrates and alcohols are commonly employed as proton donors [27]. In this section, the effect of four carbohydrates on product yield and stereoselectivity (ee) was investigated, including glucose, sucrose, fructose, and lactose, four kinds of alcohols were also measured such as isopropanol, ethanol, methanol, and glycerol. As shown in Figure 2, the lowest yield of 16.8% was obtained without the addition of co-substrate. Among the four tested carbohydrates, the highest yield reached 59.2% while employing glucose as co-substrate. Isopropanol is a good co-substrate among the tested alcohols to enhance the reaction efficiency, achieving 97.8% of the yield. The ee value of the (S)-1-chloro-2-heptanol was 99% with alcohols as co-substrate, and 98% with carbohydrates. Mostly, it is probable that alcohols may enhance the solubility of the substrate, thereby accelerating the enzymatic reaction rate and increasing the ee value [28].
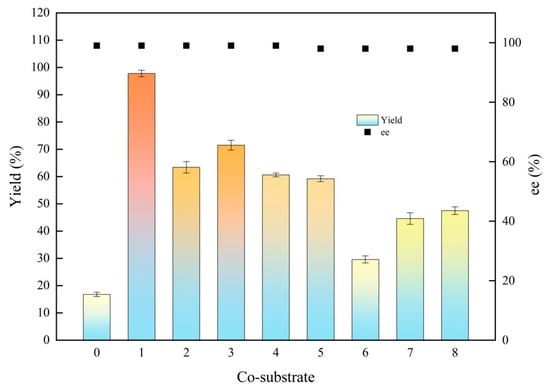
Figure 2.
Effects of co-substrates on the biocatalytic reduction. Reaction conditions: 20.0 mL phosphate buffer (100 mM, pH 6.0), 1.0 g resting cells (DCW), 50 mM 1-chloro-2-heptanone, 15% (v/v) various co-substrate, 200 rpm, 30 °C, 24 h. Symbols: 0: control (without co-substrate); 1: isopropanol; 2: ethanol; 3: methanol; 4: glycerol; 5: glucose; 6: sucrose; 7: fructose; 8: lactose.
2.2.2. Isopropanol Concentration
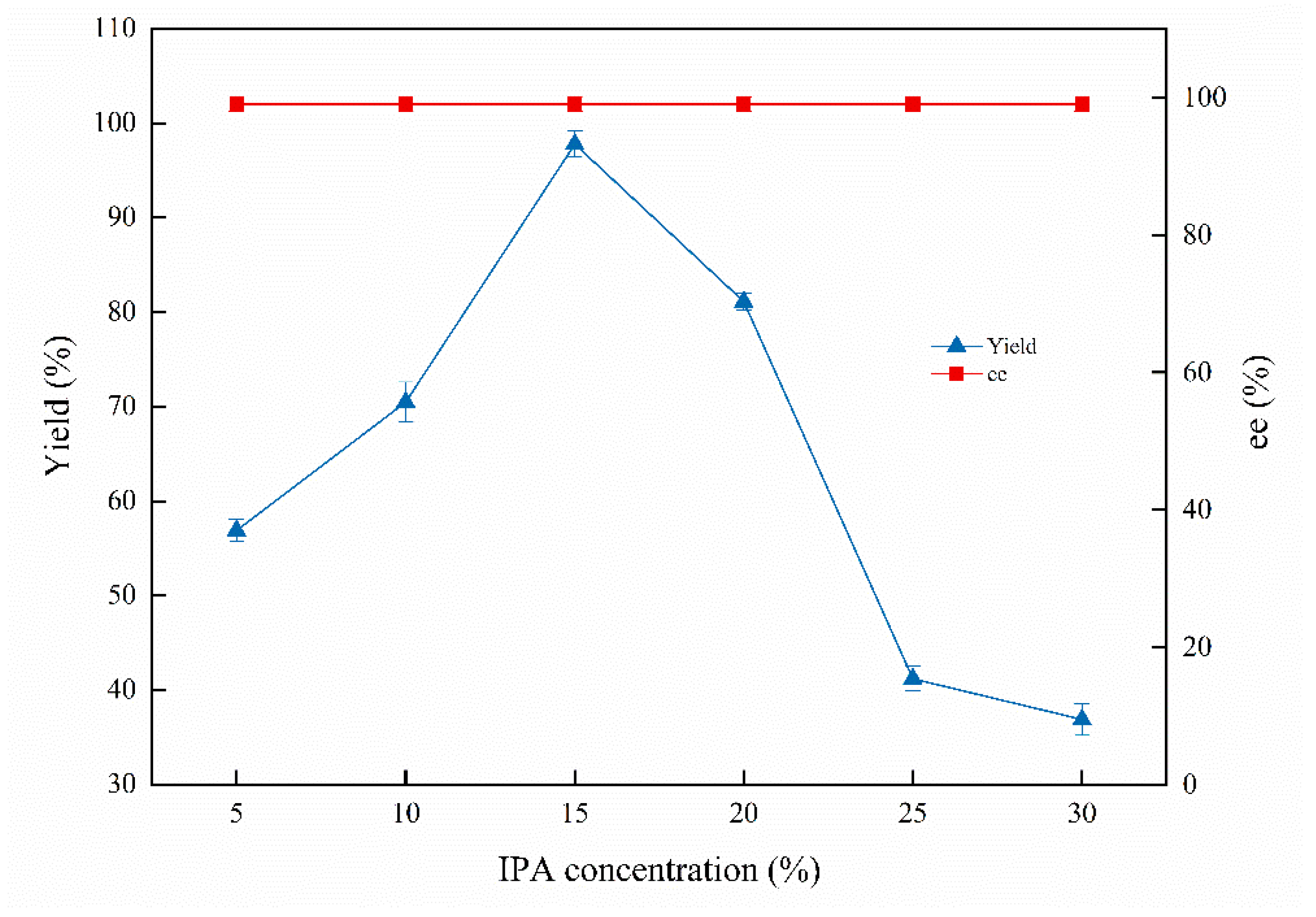
Isopropanol (IPA) was employed as a satisfactory co-substrate for the asymmetric bioreduction. IPA concentration, as a crucial factor, was examined and is shown in Figure 3. The highest product yield was 97.8%, while the IPA concentration reached 15%. The product yield was improved with the IPA concentration increased to 15%, which declined along with the increased IPA concentration to 20%. The yield of (S)-1-chloro-2-heptanol was sharply decreased with continuously increased IPA concentration from 25% to 30%. It is demonstrated that the appropriate concentration of IPA can accelerate the asymmetric bioreduction efficiency, and the higher IPA concentration (25–30%) may unbalance the osmotic pressure of the cells and the enzyme rush in the reaction system [7]. The original existing coenzyme regeneration system in whole cells has been destroyed; therefore, the biocatalytic reduction is terminated, resulting in the yield sharply decreasing [29]. In addition, the ee value has not been affected by the IPA concentration and is kept constant at 99%.
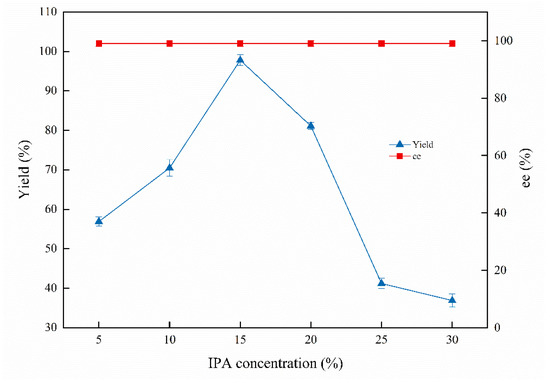
Figure 3.
Effects of IPA concentrations on the biocatalytic reduction. Reaction conditions: 20.0 mL phosphate buffer (100 mM, pH 6.0), 1.0 g resting cells (DCW), 50 mM 1-chloro-2-heptanone, different concentrations of IPA, 200 rpm, 30 °C, 24 h.
2.2.3. pH
pH is an essential factor in the enzyme-mediated asymmetric reaction, which may affect the activity and product ee value [30]. The effect of pH 4.0–8.0 was measured on the bioreduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol. As shown in Table 1, Curvularia hominis B-36 is effective at pH 4.0–8.0 and obtained higher yields of 86.9–94.5%, making it suitable for acidic, neutral, and alkaline conditions. It differs from the most reported whole cells which have a narrow pH tolerance range and is speculated to belong to a new family of carbonyl reductases. Furthermore, different buffer systems at pH 6.0 were also investigated on the bioreduction, and the yields of (S)-1-chloro-2-heptanol were 93.7–97.8% at four tested phosphate buffer systems involving Na2HPO4-NaH2PO4, Na2HPO4-KH2PO4, Na2HPO4-Citric acid, and K2HPO4-KH2PO4. However, distilled water is not an efficient medium for the reaction catalyzed by Curvularia hominis B-36; probably, phosphate buffer can maintain the configuration and stability of the enzyme in contrast to the distilled water [31]. The ee value kept constant above 99% in various pH conditions. In conclusion, K2HPO4-KH2PO4 (pH = 6.0) is selected as the best buffer system for the bioreduction of 1-chloro-2-heptanone, owing to its highest product yield.

Table 1.
Effect of pH on the bioreduction of 1-chloro-2-heptanone.
2.2.4. Temperature
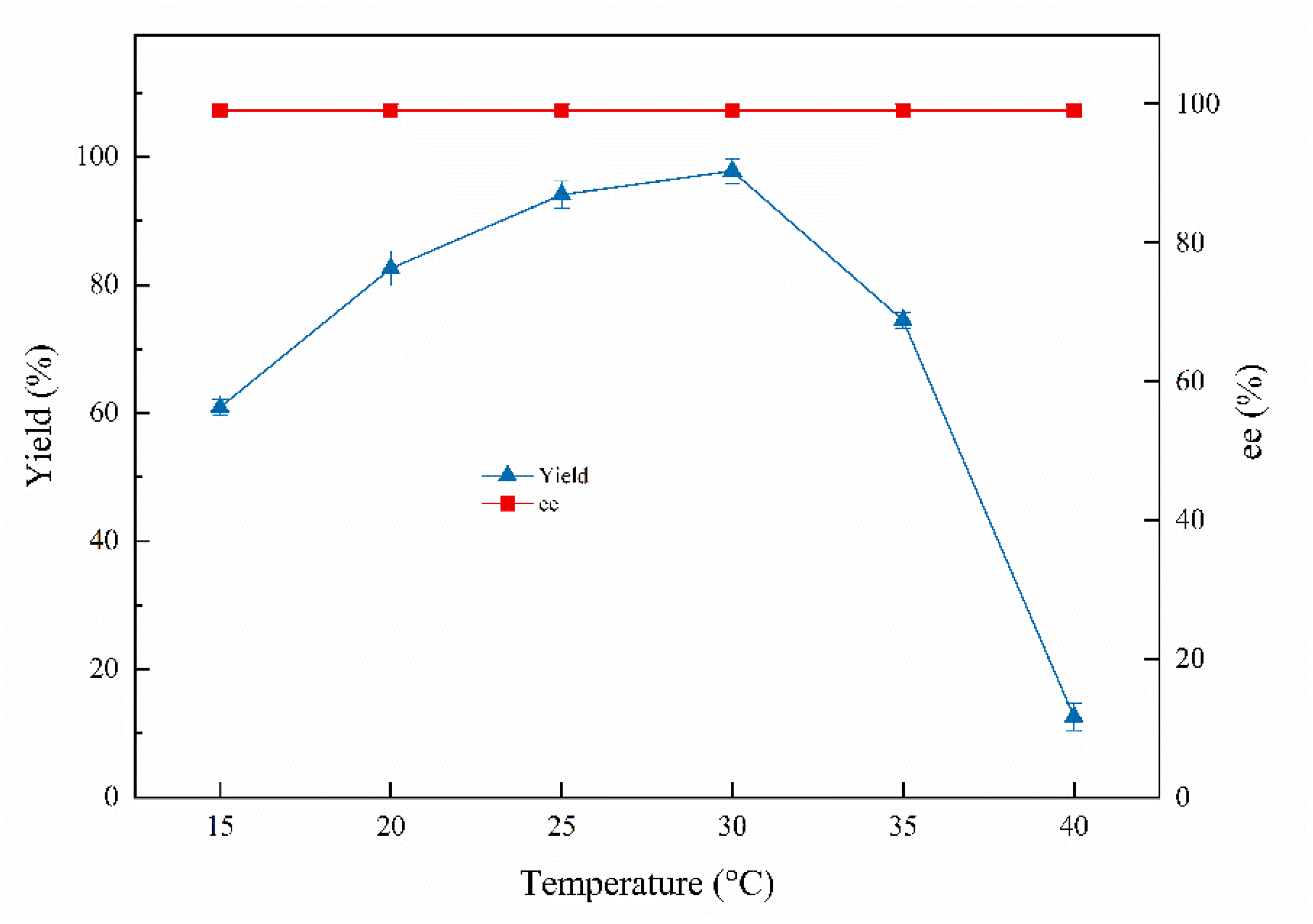
Temperature is a crucial indicator of the biosynthetic process; moderately elevated temperatures can accelerate enzymatic reaction rates but enzymes will become inactive at higher temperatures [32]. As shown in Figure 4, approximately 97.8% of the highest yield was reached at 30 °C and 12.6% of the lowest yield was obtained at 40 °C, indicating that Curvularia hominis B-36 shows poor activity at high temperature (40 °C). The ee values of the products were all above 99% at 15–40 °C.
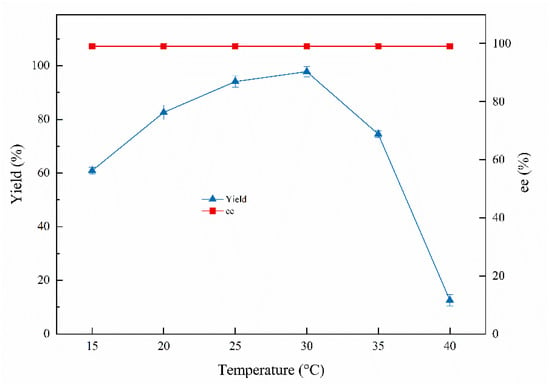
Figure 4.
Effects of temperatures on the biocatalytic reduction. Reaction conditions: 20.0 mL K2HPO4-KH2PO4 (100 mM, pH 6.0), 1.0 g resting cells (DCW), 50 mM 1-chloro-2-heptanone, 15% (v/v) IPA, different reaction temperature, 200 rpm, 24 h.
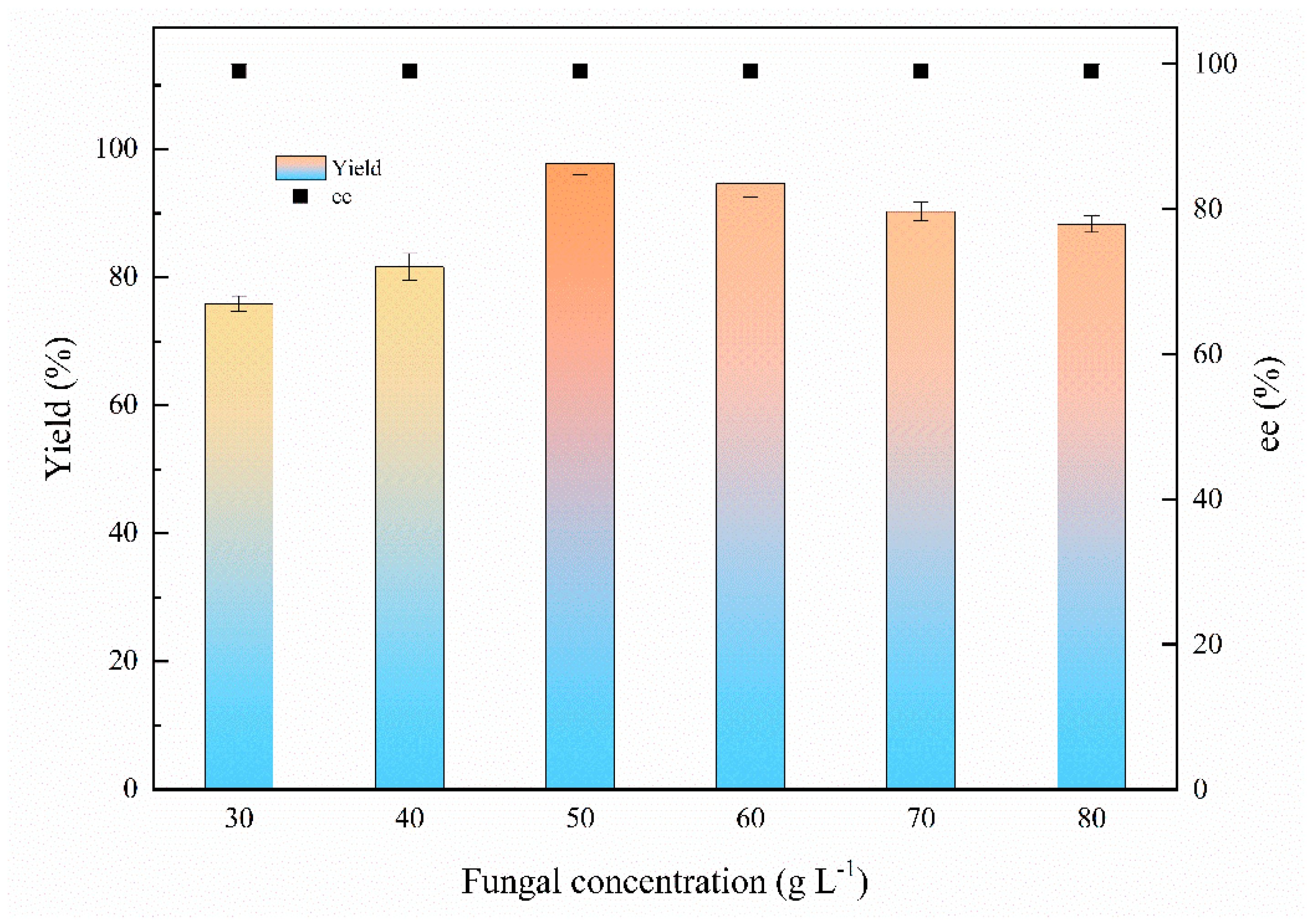
2.2.5. Fungal Concentration
Fungal concentration plays a vital role in biocatalysis; the fungal concentrations varied from 30 to 80 g L−1 (DCW) were assayed to determine the optimum biocatalyst feeding strategy for the bioreduction. Figure 5 demonstrates that the yield for (S)-1-chloro-2-heptanol was increased from 75.9% to 97.8% with 30–50 g L−1 of fungus feeding. However, the yield was slightly decreased approximately from 97.8% to 88.3% with 50–80 g L−1 of the fungal concentration because the excessive fungal concentration may affect the mass transfer [7]. The ee value was above 99% at tested fungal concentrations. Therefore, the optimal fungal concentration is 50 g L−1.
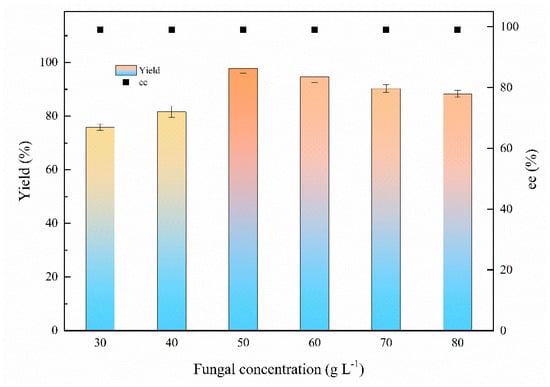
Figure 5.
Effects of fungal concentration on the biocatalytic reduction. Reaction conditions: 20.0 mL K2HPO4-KH2PO4 (100 mM, pH 6.0), 30–80 g L−1 resting cells (DCW), 50 mM 1-chloro-2-heptanone, 15% (v/v) IPA, 200 rpm, 30 °C, 24 h.
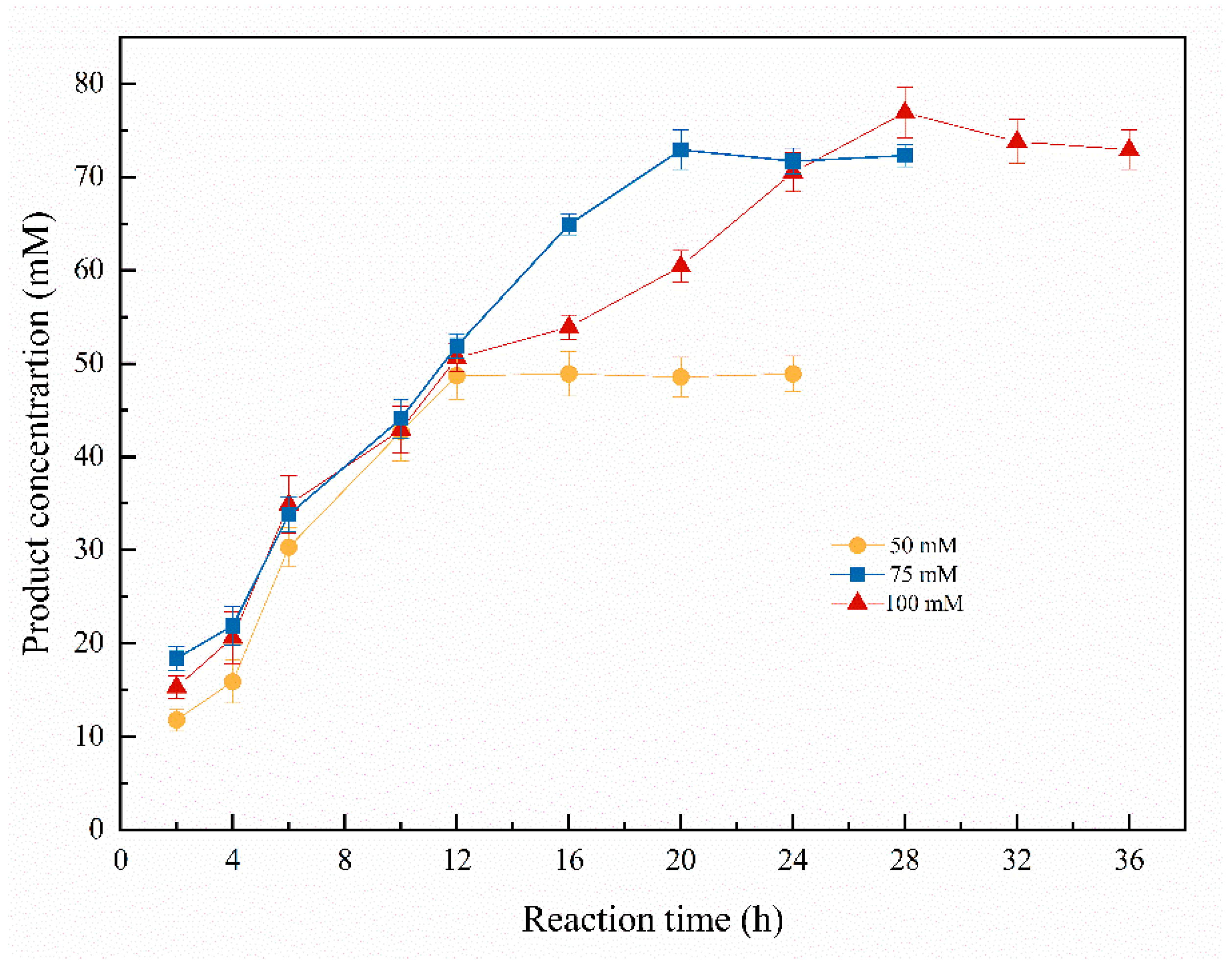
2.3. Time Course of Bioreduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol Catalyzed by Curvularia hominis B-36
Time course of the asymmetric biosynthesis of (S)-1-chloro-2-heptanol was studied at 50–100 mM of 1-chloro-2-heptanone. As shown in Figure 6, when the substrate concentration was 50 mM, the highest (S)-1-chloro-2-heptanol concentration of 48.9 mM (97.8% of yield) was obtained within 16 h. Notably, the product concentrations were kept at 48.6–48.9 mM, with a 12–24 h reaction time. When the substrate concentration was 75 mM, the highest product concentration of 72.9 mM (97.2% of yield) was attained within the reaction for 20 h. Continually increasing the substrate concentration to 100 mM, the highest product concentration is 76.8 mM (76.8% of yield) within a reaction for 28 h, and no increased product concentration was observed while extending reaction time to 32–36 h. Therefore, the suitable substrate concentration is 75 mM due to its relatively higher yield of 97.2%. Asymmetric reduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol catalyzed by Curvularia hominis B-36 whole cells under the optimal reaction condition is summarized in Scheme 1.
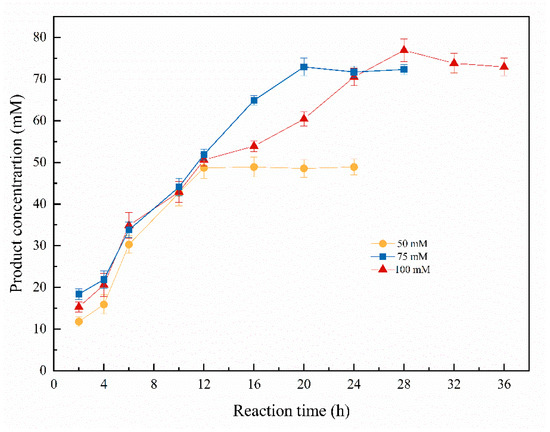
Figure 6.
Time course of asymmetric biosynthesis of (S)-1-chloro-2-heptanol. Reaction conditions: 20.0 mL K2HPO4-KH2PO4 (100 mM, pH 6.0), 1.0 g resting cells (DCW), 50 mM 1-chloro-2-heptanone, 15% (v/v) IPA, 200 rpm, 30 °C.
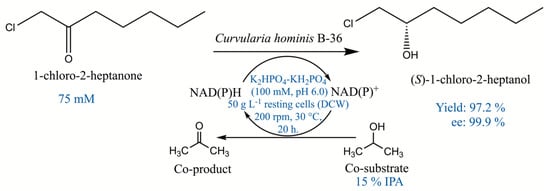
Scheme 1.
Asymmetric reduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol catalyzed by Curvularia hominis B-36 under the optimal reaction conditions.
2.4. Scale-up Asymmetric Reduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol
The preparative scale bioreduction of 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol catalyzed by Curvularia hominis B-36 was established in a 5.0 L bioreactor (Sartorius stedimTM BIOSTATB2) containing 1.5 L reaction mixture. The best yield of ~97% and 99.9% of ee were accomplished after reaction for 20 h at 800 rpm and 30 °C. The product was purified by silica gel column chromatography (eluent: petroleum ether/ethyl acetate, 10:1, v/v). The isolated yield of (S)-1-chloro-2-heptanol is 93.54%. The purified (S)-1-chloro-2-heptanol was characterized by 1H and 13C NMR (see Figures S3 and S4). 1H NMR (500 MHz, DMSO-d6) δ 4.90 (s, 1H), 3.61 (s, 1H), 3.45 (d, J = 5.0 Hz, 2H), 1.55–1.22 (m, 8H), 0.87 (t, J = 7.0 Hz, 3H); 13C NMR (126 MHz, DMSO-d6) δ 70.52 (s), 49.88 (s), 34.30 (s), 31.82 (s), 25.12 (s), 22.58 (s), 14.12 (s); = +2.32 (c = 1.0, CHCl3).
2.5. Stereoselective Reduction of Various Ketones
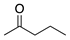
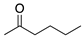
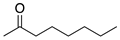
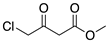
To broaden the application of Curvularia hominis B-36, various aliphatic and aromatic ketones were evaluated for bioreduction under the optimal reaction conditions of 1-chloro-2-heptanone (Scheme 1). As shown in Table 2, Curvularia hominis B-36 exhibited S-enantioselectivity for all aliphatic ketones, including Butan-2-one, Pentan-2-one, Hexan-2-one, Octan-2-one, Methyl 4-chloro-3-oxobutanoate, and Ethyl 4-chloro-3-oxobutanoate, affording ee values of above 99% and yields of above 90%. For aromatic ketones, Curvularia hominis B-36 shows R-enantioselectivity and a satisfactory ee value (ee > 99%). The relatively lower yield of 54% for acetophenone and 31% for 1-(3,5-Bis(trifluoromethyl)phenyl)ethan-1-one might be enhanced by optimizing the reaction systems. Therefore, Curvularia hominis B-36 shows a broad substrate spectrum for asymmetric reduction.

Table 2.
Asymmetric reduction of different ketones by Curvularia hominis B-36.
3. Experimental
3.1. Chemicals
1-chloro-2-heptanone, (R)-1-chloro-2-heptanol, and (S)-1-chloro-2-heptanol (HPLC purity, >98%) were synthesized and supplied by InnoChem Co., Ltd., Beijing, China. Substrate 1-chloro-2-heptanone was characterized by 1H and 13C NMR (see Figures S1 and S2). 1H NMR (400 MHz, DMSO-d6) δ 4.38 (s, 2H), 2.56 (t, J = 7.4 Hz, 2H), 1.64—1.48 (m, 2H), 1.28 (ddd, J = 14.8, 13.2, 7.2 Hz, 4H), 0.89 (t, J = 7.0 Hz, 3H); 13C NMR (101 MHz, DMSO-d6) δ 201.96 (s), 48.94 (s), 39.33 (s), 31.23 (s), 23.19 (s), 22.43 (s), 13.87 (s). The other chemicals used in this study are of analytical grade.
3.2. Screening of the Microorganisms and Cultural Conditions
Microbes were screened from various soil samples collected from Zhejiang Province (Hangzhou, China) based on their efficiency in converting 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol. Microorganisms were isolated based on the substrate-oriented screening strategy, using an enrichment medium supplemented with 3 g L−1 of 1-chloro-2-heptanone as the sole source of carbon. The enrichment medium contained (g L−1): 1-chloro-2-heptanone 3.0, (NH4)2SO4 5.0, KH2PO4 4.0, and NaCl 2.5, pH 6.0. The enrichment cultured liquid was diluted appropriately, and then plated over the screening medium plates. Screening medium plates were prepared with an enrichment medium supplemented with 15 g L−1 of agar. Single colonies were picked and cultured. The cells were collected after centrifugation and evaluated for the efficiency in asymmetric reduction of 1-chloro-2-heptanone.
The protocol for strain B-36 cultivation was as follows: strain B-36 was inoculated overnight at 30 °C and 200 rpm in a flask containing 100 mL of potato dextrose agar (PDA). Subsequently, 10% (v/v) of the seeding culture was removed into a 150 mL (fermentation medium)/500 mL flask and further cultured for 24 h at 30 °C and 200 rpm. The fermentation medium was composed of (g L−1): glucose 17.0, peptone 25.5, CaCl2 5.0, and (NH4)2SO4 15.5, pH 6.5. Finally, the culture medium was removed by centrifugation (12,000× g, 4 °C, 10 min), the mycelia were washed with normal saline, and harvested by centrifugation (12,000× g, 4 °C, 10 min).
3.3. Sequence Analysis of the Strain B-36
The genomic DNA of strain B-36 was extracted with a fungi genomic DNA extraction kit (SK8259). The ITS genes were amplified by PCR with the universal primer ITS1: (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4: (5′-TCCTCCGCTTAT TGATATGC-3′). The thermal profile of PCR was as follows: 4 min at 94 °C, 30 cycles of 45 s at 94 °C, 45 s at 55 °C, and 1 min at 72 °C, and extension at 72 °C for 10 min. The PCR products were purified and verified. The sequence alignment was determined from the GenBank database (NCBI) using the BLAST tool. A phylogenetic tree was established based on the neighbor-joining method using MEGA software 7.0.
3.4. Asymmetric Bioreduction Process
The reaction system is composed of 50 g L−1 (dry cell weight; DCW) mycelia (biocatalysts), 15% (v/v) of isopropanol (co-substrate), K2HPO4-KH2PO4 (100 mM, pH 6.0), and 75 mM 1-chloro-2-heptanone (substrate). The reaction mixture was agitated at 30 °C and 200 rpm. The biomass was removed by centrifugation. The substrate and product in the reaction mixture were extracted by ethyl acetate thrice. The ethyl acetate layer was dried by using anhydrous MgSO4 and analyzed by GC.
The preparative scale bioreduction system (1.5 L working volume) is composed of 50 g L−1 (DCW) mycelia, 15% (v/v) of isopropanol, K2HPO4-KH2PO4 (100 mM, pH 6.0), and 75 mM 1-chloro-2-heptanone, with reaction for 20 h at 800 rpm and 30 °C.
3.5. Analytical Methods
The reaction mixtures were analyzed by GC-7890A, and CP-Chirasil-Dex CB column (25 m × 0.25 mm × 0.25 μm, df = 0.25) was used to determine the chirality of reduced alcohols. Dodecane was employed as an internal standard. The procedure was set up as follows: carrier gas: N2; flow rate: 1.0 mL/min; inlet temperature: 250 °C; detector temperature: 250 °C; injection volume: 1 μL; and split ratio: 15:1. The column temperature was as follows: 100 °C for 2 min, increasing the temperature to 200 °C at a rate of 4 °C/min. The peak time of substrate 1-chloro-2-heptanone was 4.328 min; (R)-1-chloro-2-heptanol and (S)-1-chloro-2-heptanol were 8.531 and 8.793 min, respectively.
The yield and ee value were calculated as follows:
CP: the final molar concentration of the product;
CS: the initial molar concentration of the substrate.
CS: the molar concentrations of (S)-1-chloro-2-heptanol;
CR: the molar concentrations of (R)-1-chloro-2-heptanol.
4. Conclusions
Curvularia hominis B-36, a new fungus, was screened and characterized by molecular biological identification. This is the first report about Curvularia hominis B-36 that can reduce 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol with excellent yield (97.2%) and ee value (99.9%); it has applied for a patent (CN111925949B). The optimal reaction conditions were determined to be K2HPO4-KH2PO4 (100 mM, pH 6.0), 50 g L−1 resting cells (DCW), and 15% (v/v) IPA as co-substrate, 200 rpm, 30 °C, 20 h. A search about biocatalytic 1-chloro-2-heptanone to (S)-1-chloro-2-heptanol in SciFinder® (CAS) with one hit was documented. Absidia-repens-mediated bioreduction attained 14 % yield and 60% ee [33], which was much lower than that of newly isolated Curvularia hominis B-36. Furthermore, Curvularia hominis B-36 could asymmetrically reduce a variety of aliphatic and aromatic ketones, possessing excellent enantioselectivity. In conclusion, Curvularia hominis B-36 is a brilliant biocatalyst and has the potential to synthesize various pharmaceutical chiral intermediates.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/catal13010052/s1, Figure S1: 1H NMR of substrate 1-chloro-2-heptanone, Figure S2: 13C NMR of substrate 1-chloro-2-heptanone, Figure S3: 1H NMR of product (S)-1-chloro-2-heptanol, Figure S4: 13C NMR of product (S)-1-chloro-2-heptanol, Figure S5: 1H NMR of (S)-butan-2-ol, Figure S6: 1H NMR of (S)-pentan-2-ol, Figure S7: 1H NMR of (S)- hexan-2-ol, Figure S8: 1H NMR of (S)- octan-2-ol, Figure S9: 1H NMR of methyl (S)-4-chloro-3-hydroxybutanoate, Figure S10: 1H NMR of ethyl (S)-4-chloro-3-hydroxybutanoate, Figure S11: 1H NMR of ethyl (R)-1-phenylethan-1-ol, Figure S12: 1H NMR of (R)-1-(3,5-bis(trifluoromethyl)phenyl)ethan-1-ol, Figure S13: GC graph for bioreduction of butan-2-one to (S)-butan-2-ol, Figure S14: GC graph for bioreduction of pentan-2-one to (S)- pentan-2-ol, Figure S15: GC graph for bioreduction of hexan-2-one to (S)- hexan-2-ol, Figure S16: GC graph for bioreduction of octan-2-one to (S)- octan-2-ol, Figure S17: GC graph for bioreduction of methyl 4-chloro-3-oxobutanoate to methyl (S)-4-chloro-3-hydroxybutanoate, Figure S18: GC graph for bioreduction of ethyl 4-chloro-3-oxobutanoate to ethyl (S)-4-chloro-3-hydroxybutanoate, Figure S19: GC graph for bioreduction of acetophenone to ethyl (R)-1-phenylethan-1-ol, Figure S20: GC graph for bioreduction of 1-(3,5-bis(trifluoromethyl)phenyl) ethan-1-one to (R)-1-(3,5-bis(trifluoromethyl)phenyl)ethan-1-ol.
Author Contributions
Data curation, W.C.; Formal analysis, P.T.; Investigation, R.L. and S.Y.; Methodology, Y.S.; Project administration, W.D.; Supervision, J.L.; Writing—original draft, S.X.; Writing—review and editing, Q.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Zhejiang Provincial Natural Science Foundation under Grant No. LGF20B060001 and No. LY21H300003; Basic Scientific Research Funds of Department of Education of Zhejiang Province under Grant No. KYZD202009 and No. KYYB202006; and National Innovation and Entrepreneurship Training Program for College Students under Grant No. 202213023014.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic synthesis for industrial applications. Angew. Chem. Int. Edit. 2021, 60, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Fryszkowska, A.; Devine, P.N. Biocatalysis in drug discovery and development. Curr. Opin. Chem. Biol. 2020, 55, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.P.; Brown, M.J.; Diaz-Rodriguez, A.; Lloyd, R.C.; Roiban, G.D. Biocatalysis: A pharma perspective. Adv. Synth. Catal. 2019, 361, 2421–2432. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, L.; Qiu, J.; Yang, K.; Song, Q. Rh-Catalyzed diastereoselective addition of arylboronic acids to α-keto N-tert-butanesulfinyl aldimines: Synthesis of α-amino ketones. Org. Chem. Front. 2022, 9, 1016–1022. [Google Scholar] [CrossRef]
- Bi, X.; Feng, J.; Xue, X.; Gu, Z. Construction of axial chirality and silicon-stereogenic center via Rh-catalyzed asymmetric ring-opening of silafluorenes. Org. Lett. 2021, 23, 3201–3206. [Google Scholar] [CrossRef] [PubMed]
- Zhi, L.; Xue, Y.F.; Liang, X.W.; Wang, J.; Lin, S.J.; Tao, J.; You, S.L.; Liu, W. Oxidative Indole Dearomatization for Asymmetric Furoindoline Synthesis by a Flavin-Dependent Monooxygenase Involved in the Biosynthesis of Bicyclic Thiopeptide Thiostrepton. Angew. Chem. Int. Edit. 2021, 60, 8401–8405. [Google Scholar]
- Xiong, L.; Kong, X.; Liu, H.; Wang, P. Efficient biosynthesis of (S)-1-[2-(trifluoromethyl) phenyl] ethanol by a novel isolate Geotrichum silvicola ZJPH1811 in deep eutectic solvent/cyclodextrin-containing system. Bioresour. Technol. 2021, 329, 124832. [Google Scholar] [CrossRef]
- Shajahan, R.; Sarang, R.; Saithalavi, A. Polymer Supported Proline-Based Organocatalysts in Asymmetric Aldol Reactions: A Review. Curr. Organocatal. 2022, 9, 124–146. [Google Scholar]
- Saravanan, A.; Kumar, P.S.; Vo, D.V.N.; Jeevanantham, S.; Karishma, S.; Yaashikaa, P.R. A review on catalytic-enzyme degradation of toxic environmental pollutants: Microbial enzymes. J. Hazard. Mater. 2021, 419, 126451. [Google Scholar] [CrossRef]
- Behr, J. Inhaled Treprostinil in Pulmonary Hypertension in the Context of Interstitial Lung Disease: A Success, Finally. Am. J. Resp. Crit. Care 2022, 205, 144–145. [Google Scholar] [CrossRef]
- Stubbe, B.; Opitz, C.F.; Halank, M.; Habedank, D.; Ewert, R. Intravenous prostacyclin-analogue therapy in pulmonary arterial hypertension–A review of the past, present and future. Resp. Med. 2021, 179, 106336–106347. [Google Scholar] [CrossRef]
- Jana, S.; Sarpe, V.A.; Kulkarni, S.S. Total synthesis of emmyguyacins A and B, potential fusion Inhibitors of Influenza Virus. Org. Lett. 2018, 20, 6938–6942. [Google Scholar] [CrossRef]
- García-Lacuna, J.; Domínguez, D.; Blanco-Urgoiti, J.; Pérez-Castells, J. Synthesis of treprostinil: Key Claisen rearrangement and catalytic Pauson–Khand reactions in continuous flow. Org. Biomol. Chem. 2019, 17, 9489. [Google Scholar] [CrossRef]
- Venkataraman, S.; Chadha, A. Whole Cells Mediated Biocatalytic Reduction of Alpha-Keto Esters: Preparation of Optically Enriched Alkyl 2-hydroxypropanoates. Curr. Trends Biotechnol. Pharm. 2022, 16, 111–122. [Google Scholar]
- Çolak, N.S.; Kalay, E.; Şahin, E. Asymmetric reduction of prochiral aromatic and hetero aromatic ketones using whole-cell of Lactobacillus senmaizukei biocatalyst. Synth. Commun. 2021, 51, 2305–2315. [Google Scholar] [CrossRef]
- Sakai, T.; Wada, K.; Murakami, T.; Kohra, K.; Imajo, N.; Ooga, Y.; Tsuboi, S.; Takeda, A.; Utaka, M. A systematic study on the bakers’ yeast reduction of 2-oxoalkyl benzoates and 1-chloro-2-alkanones. Bull. Chem. Soc. Jpn. 1992, 65, 631–638. [Google Scholar] [CrossRef]
- Kalay, E.; Dertli, E.; Şahin, E. Biocatalytic asymmetric synthesis of (S)-1-indanol using Lactobacillus paracasei BD71. Biocatal. Biotransfor. 2021, 40, 386–392. [Google Scholar] [CrossRef]
- Liao, Q.; Liu, W.; Meng, Z. Strategies for overcoming the limitations of enzymatic carbon dioxide reduction. Biotechnol. Adv. 2022, 60, 108024. [Google Scholar] [CrossRef]
- Hollmann, F.; Opperman, D.J.; Paul, C.E. Biocatalytic reduction reactions from a chemist’s perspective. Angew. Chem. Int. Edit. 2021, 60, 5644–5665. [Google Scholar] [CrossRef]
- Bi, S.; Liu, H.; Lin, H.; Wang, P. Integration of natural deep-eutectic solvent and surfactant for efficient synthesis of chiral aromatic alcohol mediated by Cyberlindnera saturnus whole cells. Biochem. Eng. J. 2021, 172, 108053. [Google Scholar] [CrossRef]
- Vivek, H.; Parhi, P.; Mohan, B.; Hazeena, S.H.; Kumar, A.N.; Gullón, B.; Srivastava, A.; Nair, L.M.; Alphy, M.P.; Sindhu, R.; et al. Valorization of renewable resources to functional oligosaccharides: Recent trends and future prospective. Bioresour. Technol. 2021, 346, 126590. [Google Scholar]
- Kalay, E.; Şahin, E. Regioselective asymmetric bioreduction of trans-4-phenylbut-3-en-2-one by whole-cell of Weissella cibaria N9 biocatalyst. Chirality 2021, 33, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Paul, A.K. In-vitro bioreduction of hexavalent chromium by viable whole cells of Arthrobacter sp. SUK 1201. J. Microb. Biotechnol. Food Sci. 2014, 4, 19–23. [Google Scholar] [CrossRef][Green Version]
- Özdemir, A.; Şahin, E. Efficient bioreduction of cyclohexyl phenyl ketone by Leuconostoc pseudomesenteroides N13 biocatalyst using a distance-based design-focused optimization model. Mol. Catal. 2022, 528, 112474. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, H.; Ang, E.L.; Zhao, H. Biocatalysis for the synthesis of pharmaceuticals and pharmaceutical intermediates. Bioorg. Med. Chem. 2018, 26, 1275–1284. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, P.; Gao, S.; Wang, Z.; Luan, P.; González-Sabín, J.; Jiang, Y. Construction of chemoenzymatic cascade reactions for bridging chemocatalysis and Biocatalysis: Principles, strategies and prospective. Chem. Eng. J. 2021, 420, 127659. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Brady, D. Streamlining design, engineering, and applications of enzymes for sustainable biocatalysis. ACS Sustain. Chem. Eng. 2021, 9, 8032–8052. [Google Scholar] [CrossRef]
- Li, J.; Qian, F.; Wang, P. Exploiting benign ionic liquids to effectively synthesize chiral intermediate of NK-1 receptor antagonists catalysed by Trichoderma asperellum cells. Biocatal. Biotransfor. 2021, 39, 124–129. [Google Scholar] [CrossRef]
- Li, J.; Wang, P.; He, Y.S.; Zhu, Z.R.; Huang, J. Toward Designing a Novel Oligopeptide-Based Deep Eutectic Solvent: Applied in Biocatalytic Reduction. ACS Sustain. Chem. Eng. 2019, 7, 1318–1326. [Google Scholar] [CrossRef]
- Alberto, M.; Núria, S.; Mercè, T.; Ramon, C.G. Biocatalytic transformation of 5-hydroxymethylfurfural into 2,5-di(hydroxymethyl)furan by a newly isolated Fusarium striatum strain. Catalysts 2021, 11, 216. [Google Scholar]
- Morellon-Sterling, R.; Carballares, D.; Arana-Peña, S.; Siar, E.H.; Braham, S.A.; Fernandez-Lafuente, R. Advantages of supports activated with divinyl sulfone in enzyme coimmobilization: Possibility of multipoint covalent immobilization of the most stable enzyme and immobilization via ion exchange of the least stable enzyme. ACS Sustain. Chem. Eng. 2021, 9, 7508–7518. [Google Scholar] [CrossRef]
- Ramos, M.D.; Miranda, L.P.; Giordano, R.L.; Fernandez-Lafuente, R.; Kopp, W.; Tardioli, P.W. 1,3-Regiospecific ethanolysis of soybean oil catalyzed by crosslinked porcine pancreas lipase aggregates. Biotechnol. Progr. 2018, 34, 910–920. [Google Scholar] [CrossRef]
- Schwarz, S.; Truckenbrodt, G.; Schick, H.; Depner, J. Microbial transformation of chloroheptan-2-one; preliminary results of the synthesis of a chiral phosphonium salt for prostaglandin synthesis. Chem. Inf. 1982, 13. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).