Chromone-Based Copper(II) Complexes as Potential Antitumour Agents: Synthesis, Chemical Characterisation and In Vitro Biological Evaluation
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Methods
2.2. Solution-Based Synthesis of the Copper(II) Complexes
2.2.1. Synthesis of [Cu(OH)(C2H5OH)(L)(H2O)2]—Complex (1)
2.2.2. Synthesis of [Cu2(μ-OH)2(L)2(H2O)2]—Complex (2)
2.2.3. Synthesis of [Cu2(μ-OH)2(L)2(H2O)4]—Complex (3)
2.3. Anticancer Activity Evaluation
2.3.1. Cell Lines
2.3.2. Chemicals
2.3.3. MTT Assay
2.3.4. Statistical Analysis
3. Results
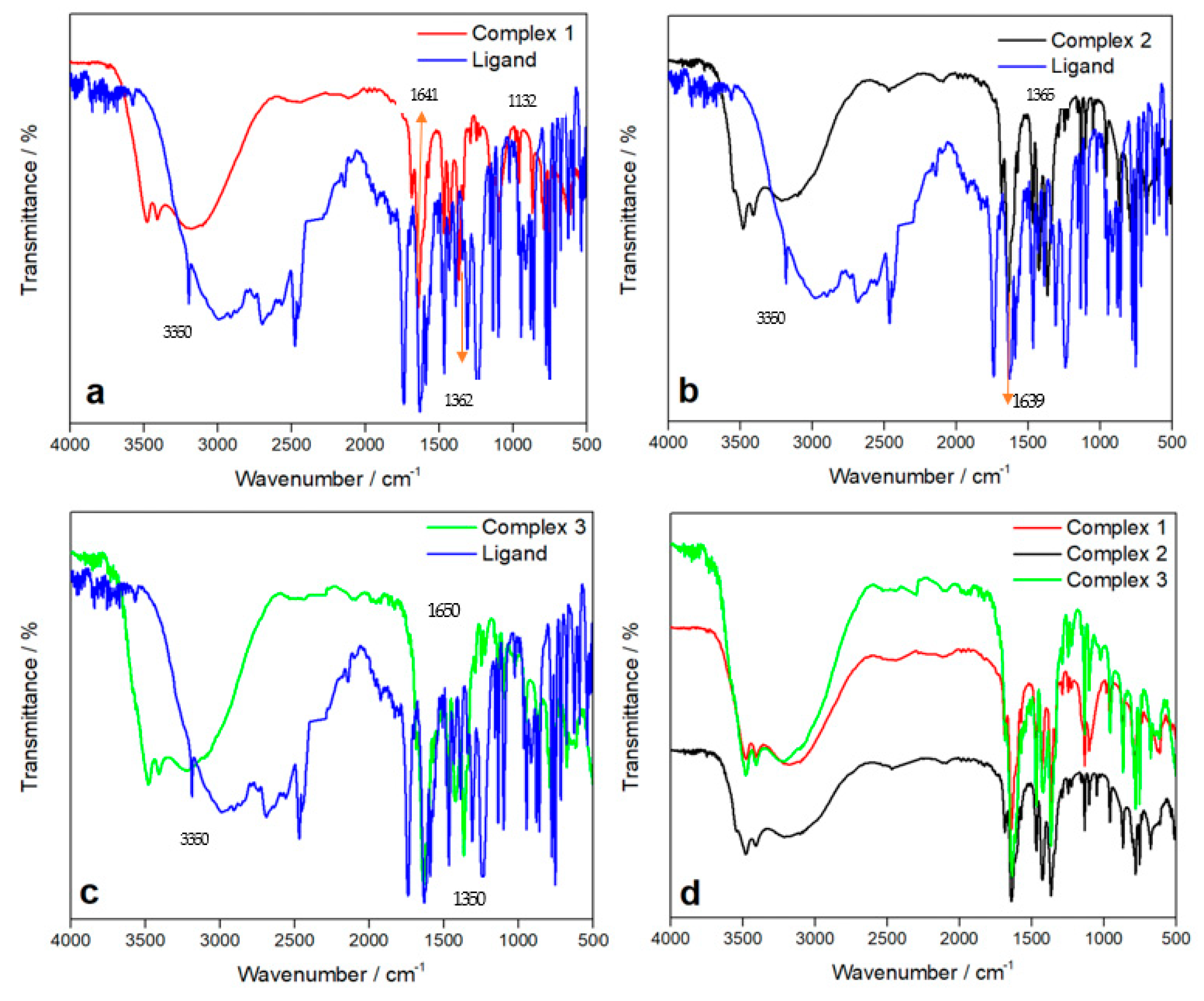
3.1. FT-IR Spectroscopy
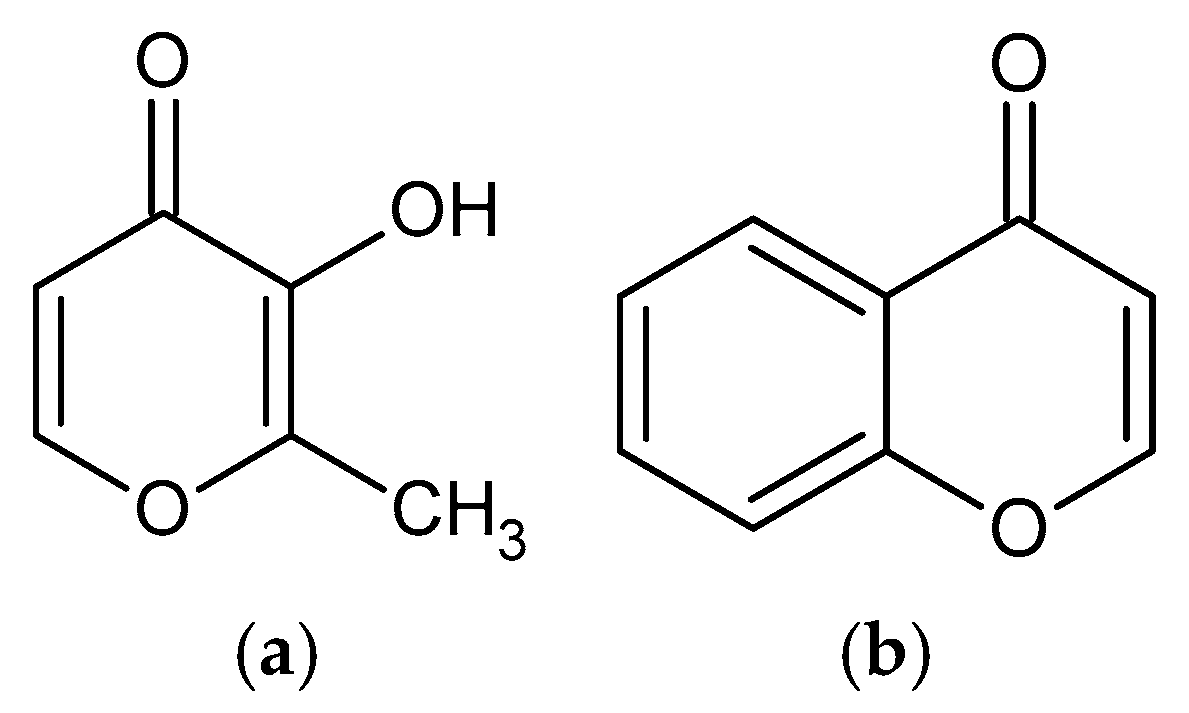
3.1.1. IR Spectrum of Ligand
3.1.2. IR Spectra of Complexes
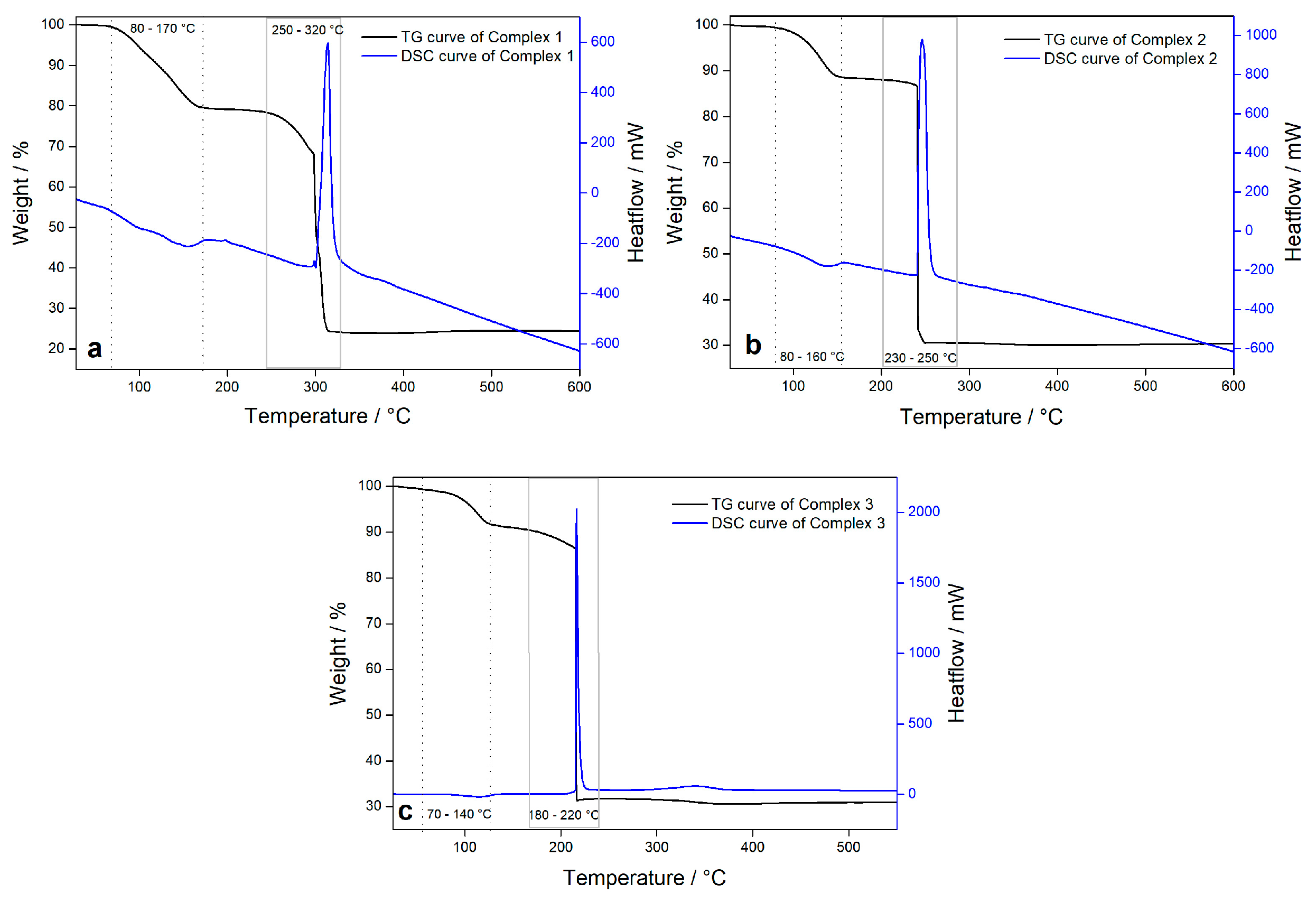
3.2. Thermal Analysis
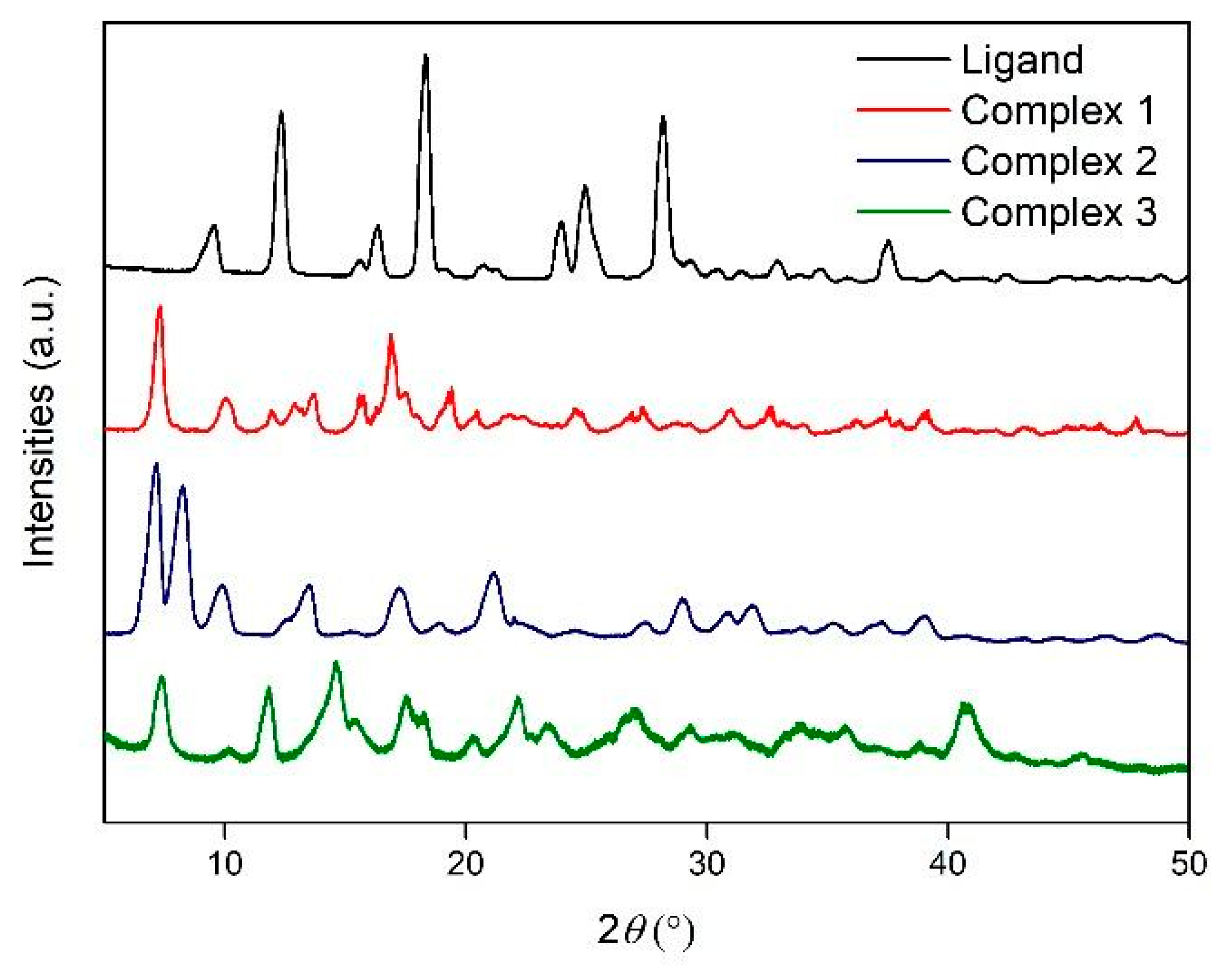
3.3. X-Ray Powder Difraction
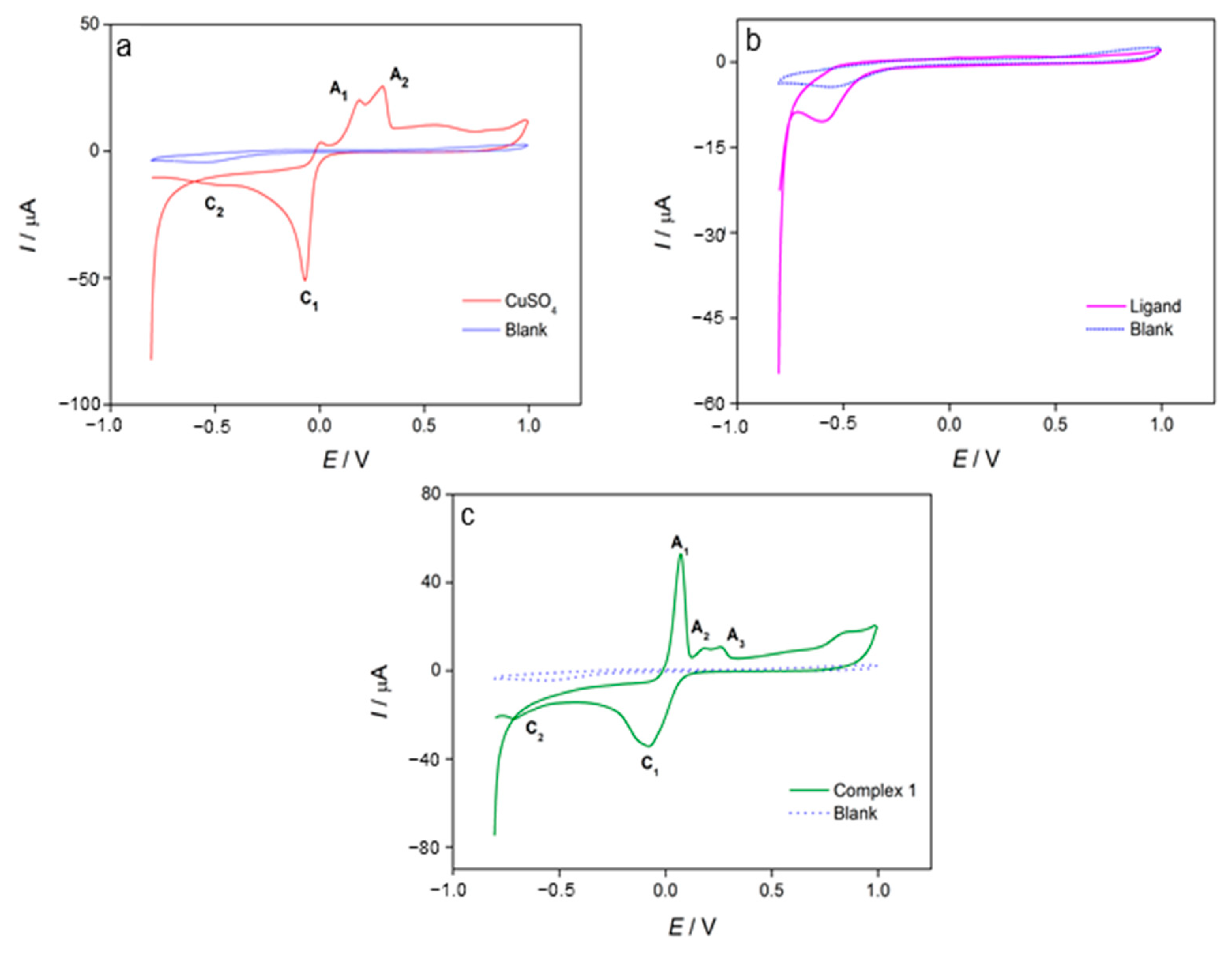
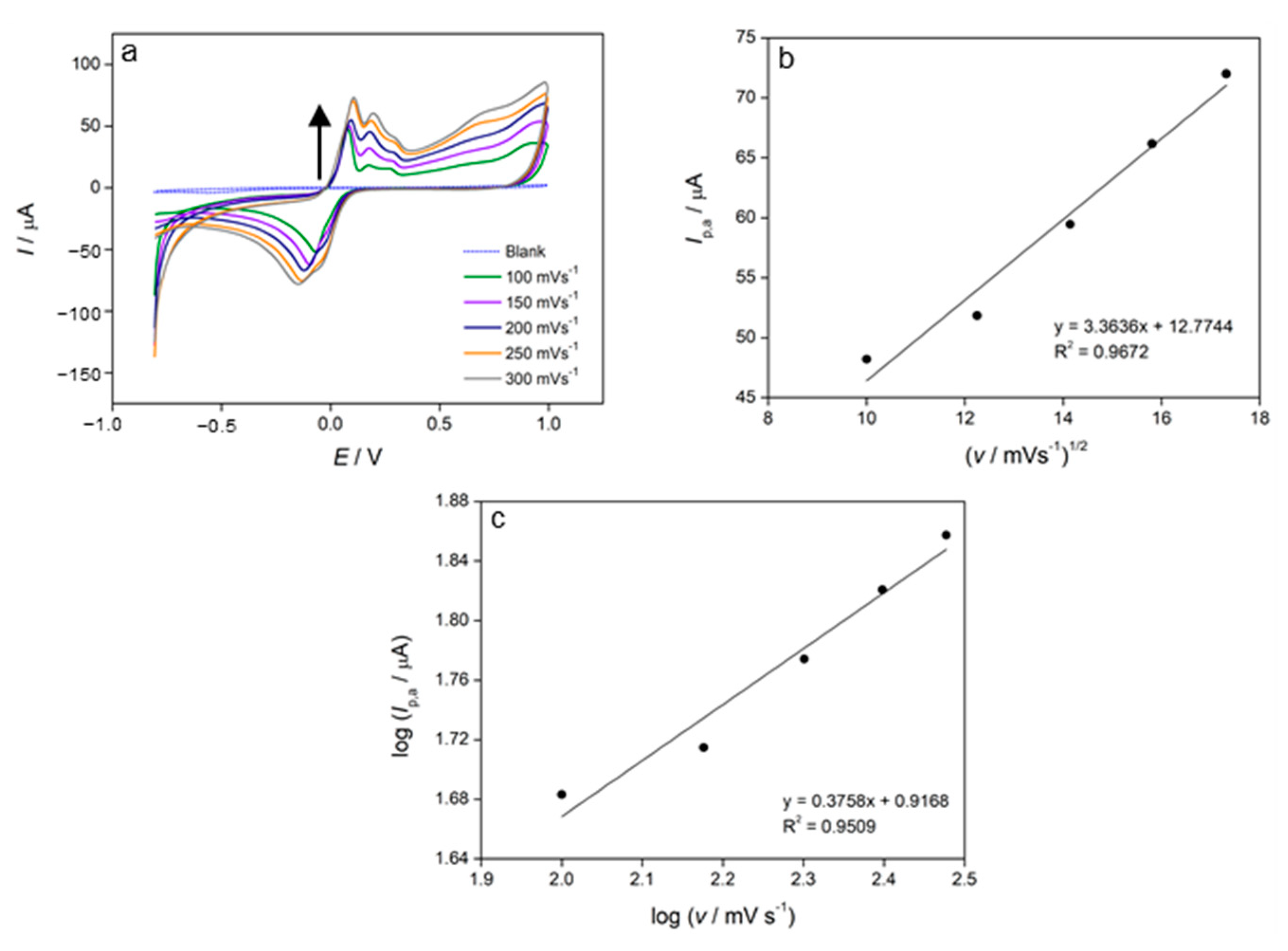
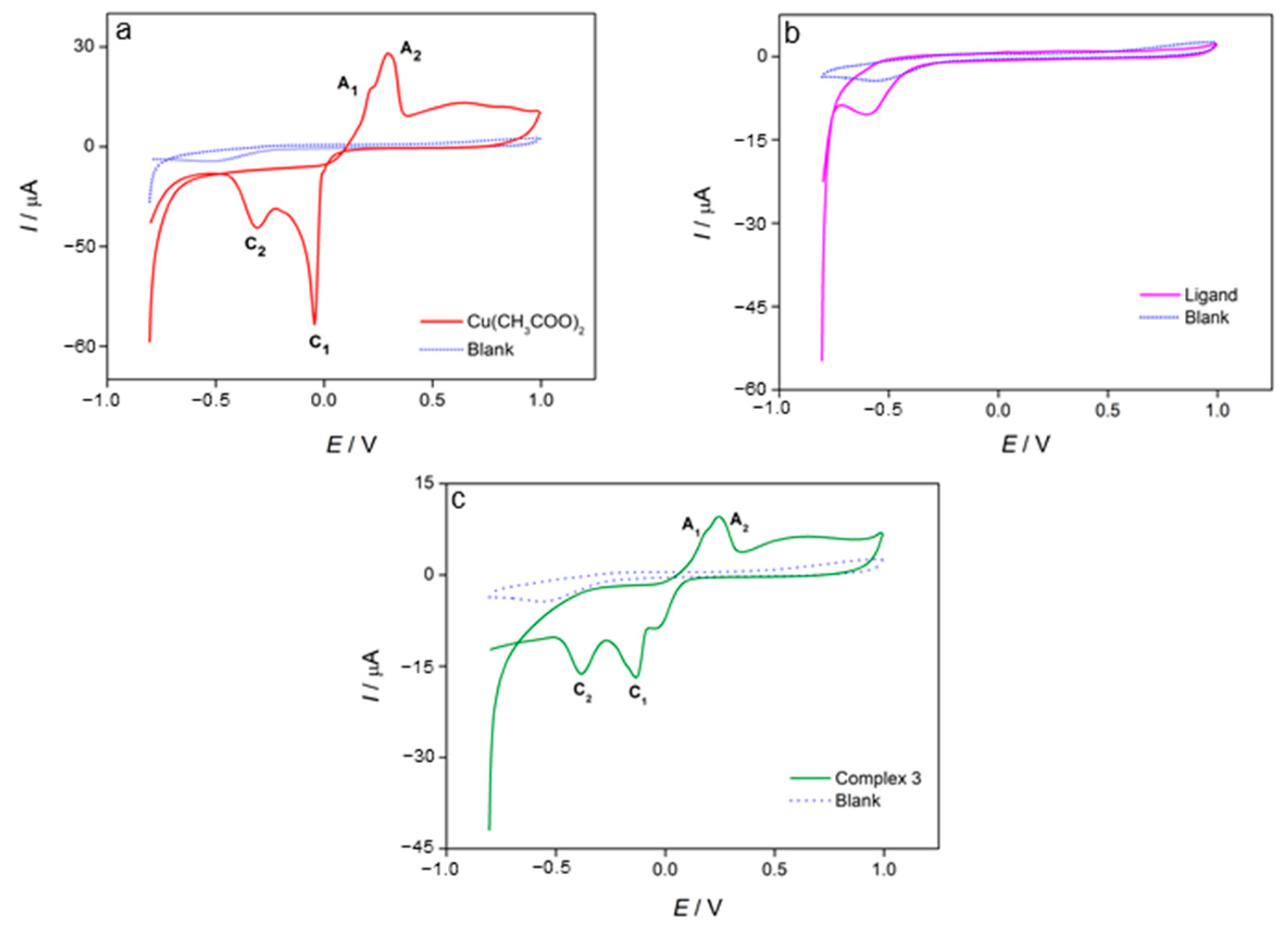
3.4. Cyclic Voltammetry
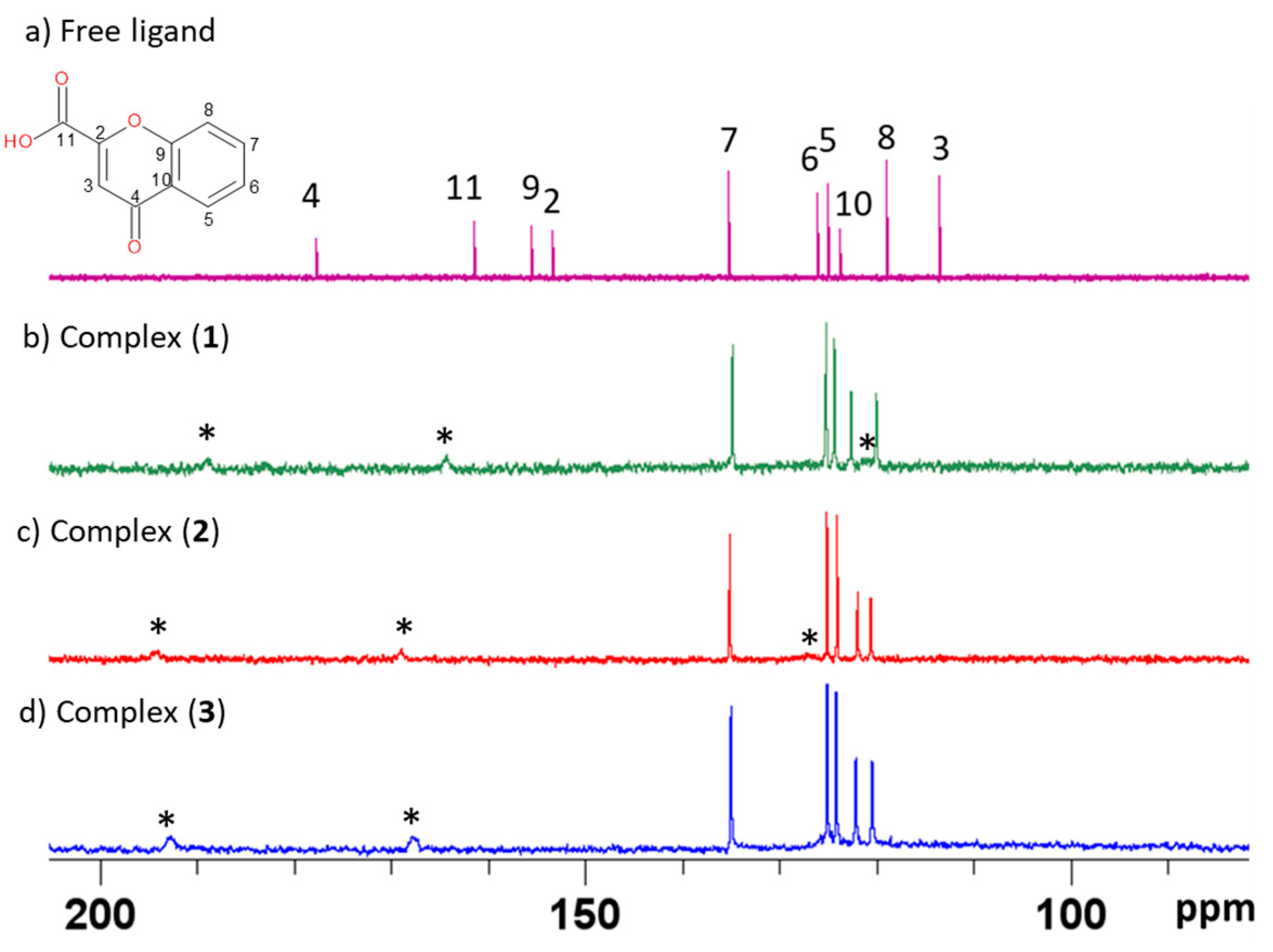
3.5. NMR Spectroscopy
4. Discussion
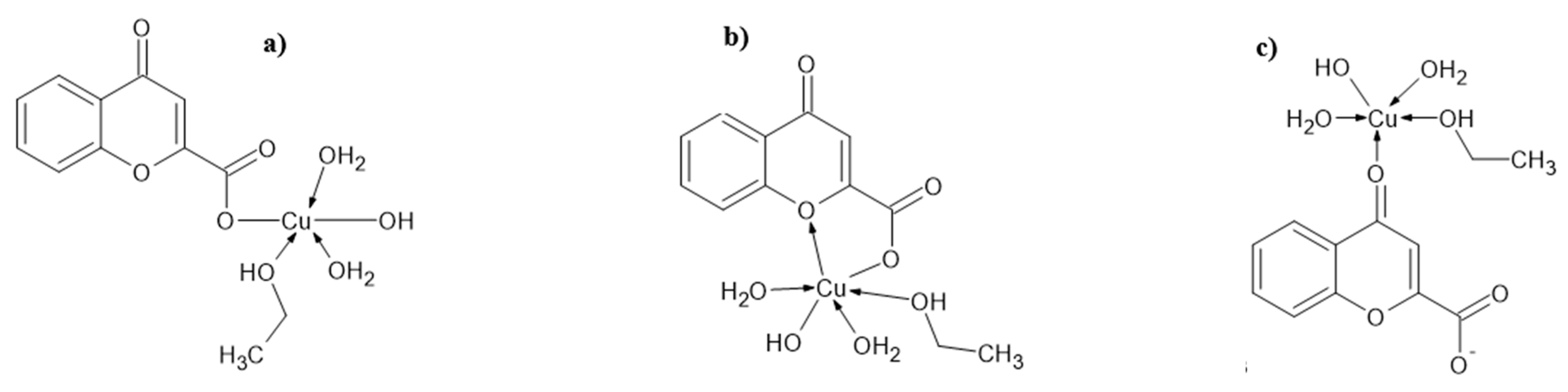
4.1. Proposed Structures of Complexes
4.2. Antitumour Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, Y.-L.; Miller, M.T.; Chao, Y.; Cohen, M.S. Synthesis of hidroxypyrone—Based matrix metalloproteinase inhibitors: Developing a structure—Activity relationship. Bioorg. Medic. Chem. Lett. 2009, 19, 1970–1976. [Google Scholar] [CrossRef] [PubMed]
- Toso, L.; Crisponi, G.; Nurchi, V.M.; Crespo-Alonso, M.; Lachowicz, J.I.; Santos, M.A.; Marques, S.M.; Niclós-Gutiérrez, J.; González-Pérez, J.M.; Domínguez-Martín, A.; et al. A family of hydroxypyrone ligands designed and synthesized as iron chelators. J. Inorg. Biochem. 2013, 127, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.H.; Barta, C.A.; Orvig, C. Metal complexes of maltol and close analogues in medicinal inorganic chemistry. Chem. Soc. Rev. 2006, 35, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Nurchi, V.M.; Crisponi, G.; Lachowicz, J.I.; Murgia, S.; Pivetta, T.; Remelli, M.; Rescigno, A.; Niclós-Gutíerrez, J.; González-Pérez, J.M.; Domínguez-Martín, A.; et al. Iron(III) and aluminum (III) complexes with hydroxypyrone ligands aimed to design kojic acid derivatives with new perspectives. J. Inorg. Biochem. 2010, 104, 560–569. [Google Scholar] [CrossRef]
- Kandioller, W.; Kurzwernhart, A.; Hanif, M.; Meier, S.M.; Henke, H.; Keppler, K.B.; Hartinger, C.G. Pyrone derivatives and metals: From natural products to metal–based drugs. J. Organomet. Chem. 2011, 696, 999–1010. [Google Scholar] [CrossRef]
- Lachowicz, J.I.; Nurchi, V.M.; Crisponi, G.; Jaraquemada-Pelaez, M.; Ostrowska, M.; Jezierska, J.; Gumienna-Kontecka, E.; Peana, M.; Zoroddu, M.A.; Choquesillo-Lazarte, D.; et al. Zinc (II) and copper (II) complexes with hydroxypyrone iron chelators. J. Inorg. Biochem. 2015, 151, 1–13. [Google Scholar] [CrossRef]
- Thompson, K.H.; McNeill, J.H.; Orvig, C. Vanadium compounds as insulin mimics. Chem. Rev. 1999, 99, 2561–2571. [Google Scholar] [CrossRef]
- Marzban, L.; McNeill, J.H. Insulin–like actions of Vanadium: Potential as a therapeutic agent. J. Tr. Elem. 2003, 16, 253–267. [Google Scholar] [CrossRef]
- Tsiani, E.; Fantus, I.G. Vanadium compounds: Biological actions and potential as pharmacological agents. Tr. Endo. Met. 1997, 8, 51–58. [Google Scholar] [CrossRef]
- Srivastava, A.K. Anti–diabetic and toxic effects of vanadium compounds. Mol. Cell. Biochem. 2000, 206, 177–182. [Google Scholar] [CrossRef]
- Thompson, K.H.; Liboiron, B.; Sun, Y.; Bellman, K.; Karunaratne, V.; Rawji, G.; Wheeler, J.; Sutton, K.; Bhanot, S.; Cassidy, C.; et al. Preparation and characterization of vanadyl complexes with bidentate maltol–type ligands: In vivo comparisons of anti–diabetic therapeutic potential. J. Bio. Inorg. Chem. 2003, 8, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Domotor, O.; Aicher, S.; Schmidlehner, M.; Novak, S.M.; Roller, A.; Jakupec, M.A.; Kandioller, W.; Hartinger, C.; Keppler, B.; Enyedy, E. Antitumor pentamethylcyclopentadienyl rhodium complexes of maltol and allomaltol: Synthesis, solution speciation and bioavtivity. J. Inorg. Biochem. 2014, 134, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Carland, M.; Tan, J.K.; White, J.M.; Stephenson, J.; Murraym, V.; Denny, W.A.; McFadyen, W.D. Synthesis, crystal structure and citotoxicity of diamine platinum (II) complexes containing maltol. J. Inorg. Biochem. 2005, 99, 1738–1743. [Google Scholar] [CrossRef]
- Thompson, K.H.; Chiles, J.; Yuen, V.G.; Tse, J.; McNeill, J.H.; Orvig, C. Comparison of anti-hyperglycemic effect amongst vanadium, molybdenum and other metal maltol complexes. J. Inorg. Biochem. 2004, 98, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Livngstone, R. Rodds Chemistry of Carbon Compounds: Six Membered Ring Compounds with One Heteroatom; Elsevier: Amsterdam, The Netherlands, 1975; Chapter 20; pp. 1–397. [Google Scholar]
- Toso, L.; Crisponi, G.; Nurchi, V.M.; Crespo-Alonso, M.; Lachowicz, J.; Mansoori, D.; Arca, M.; Santos, M.A.; Marques, S.M.; Gano, L.; et al. Searching for new aluminium chelating agents: A family of hidroxypyrone ligands. J. Inorg. Biochem. 2014, 130, 112–121. [Google Scholar] [CrossRef]
- Nurchi, V.M. Kojic acid derivatives as powerfull chelators for iron (III) and aluminium(III). Dalton Trans. 2011, 40, 5984–5998. [Google Scholar] [CrossRef]
- Crisponi, G.; Nurchi, V.M.; Zoroddu, M.A. Iron chelating agents for iron overload diseas. Thalass. Rep. 2014, 4, 1–6. [Google Scholar]
- Yang, C.S.; Luo, P.; Zeng, Z.; Wang, H.; Malafa, M.; Suh, N. Vitamin E and cancer prevention: Studies with different forms of tocopherols and tocotrienols. Mol. Carcinog. 2020, 59, 365–389. [Google Scholar] [CrossRef]
- Aggarwal, V.; Kashyap, D.; Sak, K.; Tuli, H.S.; Jain, A.; Chaudhary, A.; Garg, V.K.; Sethi, G.; Yerer, M.B. Molecular Mechanisms of Action of Tocotrienols in Cancer: Recent Trends and Advancements. Int. J. Mol. Sci. 2019, 20, 656. [Google Scholar] [CrossRef]
- Trujillo, M.; Kharbanda, A.; Corley, C.; Simmons, P.; Allen, A.R. Tocotrienols as an Anti-Breast Cancer Agent. Antioxidants 2021, 10, 1383. [Google Scholar] [CrossRef]
- Zhou, T.; Kong, X.-L.; Hider, R.C. Synthesis and iron chelating properties of hydroxypyridinone and hydroxypyranone hexadentate ligands. Dalton Trans. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Hsieh, W.-Y.; Liu, S. Synthesis and Characterization of Cr (III) Complexes with 3-Hydroxy-4-Pyrones and 1,2-Dimethyl-3-Hydroxy-4-Pyridinone (DMHP): X-Ray Crystal Structures of Cr (DMHP)3 · 12H2O and Cr(ma)3. Synth. React. Inorg. Met. Org Nano-Met. Chem. 2005, 35, 61–70. [Google Scholar] [CrossRef]
- Nurchi, V.M.; Crisponi, G. Equilibrium studies of new bis-hidroxypyrone derivatives with Fe3+, Al3+, Cu2+, Zn2+. J. Inorg. Chem. 2018, 189, 103–114. [Google Scholar]
- Gomez-Ruiz, S.; Maksimović-Ivanić, D.; Mijatović, S.; Kaluđerović, G.N. On the discovery, biological effects and use of cisplatin and metallocenes in anticancer chemotherapy. Bioinorg. Chem. Appl. 2012, 20, 140284. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Bernard Tchoounwou, P. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharm. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Alderden, R.A.; Hall, D.M.; Hambley, T.W. The discovery and development of cisplatin. J. Chem. Educ. 2006, 83, 728. [Google Scholar] [CrossRef]
- Tapiero, H.; Townsend, K.D. Trace elements in human physiology and patology. Copper. Biom. Pharm. 2003, 57, 386–398. [Google Scholar] [CrossRef]
- Angelova, M.; Asenova, S.; Nedkova, V.; Koleva-Kolarova, R. Copper in the human organism. Tr. J. Sci. 2011, 9, 88–98. [Google Scholar]
- Pavelkova, M. Biological role of copper as an essential trace element in the human organism. Slov. Farm. 2018, 67, 143–153. [Google Scholar] [CrossRef]
- Mishra, A.; Shukla, D.; Saraf, M. Copper: Its biological role and toxicity. J. Ind. Soc. 2019, 98, 26–36. [Google Scholar]
- Watir, S.M.; Ghobrial, I. Copper deficiency, a new triad: Anemia, leucopenia and myeloneuropathy. J. Com. Hosp. Intern. Med. Persp. 2017, 7, 265–268. [Google Scholar]
- Hart, P.; Galtrey, C.M.; Paviour, D.C.; Htut, M. Clinical Biochemistry: Metabolic and Clinical Aspects, 3rd ed.; Chapter 36, Biochemical Aspects of Nerological Disease; Churcill Livngstone: London, UK, 2014; pp. 683–701. [Google Scholar]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Caillau, K. Cooper complexes as anticancer agents targeting topoisomerases I and II. Cancers 2020, 12, 2863. [Google Scholar] [CrossRef]
- Hussain, A. Copper (II) complexes as potential anticancer and nonsteroidal anti-inflammatory agents: In vivo and in vitro studies. Nature 2019, 9, 5237. [Google Scholar] [CrossRef]
- Marzano, C.; Pellei, M.; Tisato, F.; Santini, C. Copper complexes as anticancer agents. Antican. Agen. Med. Chem. 2009, 9, 185–211. [Google Scholar] [CrossRef] [PubMed]
- Soltys, L.; Olkhovyy, O.; Tatarchuk, T.; Naushad, M. Green Synthesis of Metal and Metal Oxide Nanoparticles: Principles of Green Chemistry and Raw Materials. Magnetochemistry 2021, 7, 145. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Haidere, M.F.; Nurunnabi, M.; Shahriar, S.M.; Ahammad, A.J.S.; Shim, Y.Y.; Reaney, M.J.T.; Cho, J.Y. Green Chemistry Synthesis of Silver Nanoparticles and Their Potential Anticancer Effects. Cancers 2020, 12, 855. [Google Scholar] [CrossRef] [PubMed]
- Cannavacciuolo, C.; Pagliari, S.; Frigerio, J.; Giustra, C.M.; Labra, M.; Campone, L. Natural Deep Eutectic Solvents (NADESs) Combined with Sustainable Extraction Techniques: A Review of the Green Chemistry Approach in Food Analysis. Foods 2023, 12, 56. [Google Scholar] [CrossRef]
- Brza, M.A.; Aziz, S.B.; Anuar, H.; Al Hazza, M.H.F. From Green Remediation to Polymer Hybrid Fabrication with Improved Optical Band Gaps. Int. J. Mol. Sci. 2019, 20, 3910. [Google Scholar] [CrossRef]
- Krasnowskaya, O.; Naumov, A.; Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Cooper coordination compounds as biologically active agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef]
- Liang, J.X.; Zhong, H.J.; Yang, G.; Vellaisamy, K.M.; Leung, C.H. Recent development of transition metal complexes with in vivo tumor activity. J. Inorg. Biochem. 2017, 77, 276–286. [Google Scholar] [CrossRef]
- Gashemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12817. [Google Scholar] [CrossRef]
- Lica, J.J.; Wieczór, M.; Grabe, G.J.; Heldt, M.; Jancz, M.; Misiak, M.; Gucwa, K.; Brankiewicz, W.; Maciejewska, N.; Stupak, A.; et al. Effective Drug Concentration and Selectivity Depends on Fraction of Primitive Cells. Int. J. Mol. Sci. 2021, 22, 4931. [Google Scholar] [CrossRef]
- Bézivin, C.; Tomasi, S.; Lohézic-Le Dévéhat, F.; Boustie, J. Cytotoxic activity of some lichen extracts on murine and human cancer cell lines. Phytomedicine 2003, 10, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-J.; Wang, W.-T.; Wang, M.; Liu, H.; Pan, H.-C. Electrochemical Behavior and Direct Quantitative Determination of Tanshinone IIA in Micro-emulsion. Int. J. Electrochem. Sci. 2016, 11, 5165–5179. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge structural database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, B72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Masood, Z.; Muhammad, H.; Tahiri, I.A. Comparison of Different Electrochemical Methodologies for Electrode Reactions: A Case Study of Paracetamol. Electrochem 2024, 5, 57–69. [Google Scholar] [CrossRef]
- Eubank, K.F.; Kravtsov, V.C.; Eddaoudi, M. Synthesis of Organic Photodimeric Cage Molecules Based on Cycloaddition via Metal−Ligand Directed Assembly. J. Am. Chem. Soc. 2007, 129, 5820–5821. [Google Scholar] [CrossRef]
- Tseberlidis, G.; Intrieri, D.; Caselli, A. Catalytic Applications of Pyridine-Containing Macrocyclic Complexes. Eur. J. Inorg. Chem. 2017, 30, 3589–3603. [Google Scholar] [CrossRef]
- Chaudhary, N.K. Metal Complexes of a Novel Schiff Base Based on Penicillin: Characterization, Molecular Modeling, and Antibacterial Activity Study. Bioinorg. Chem. Appl. 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Deacon, G.B.; Phillips, R.J. Relationship between the carbon-oxygen stretching frequencies of carboxylato complexes and he type of carboxylate coordination. Coord. Chem. Rev. 1980, 33, 227. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Wang, C.; Liu, H.; Zhao, D. Effect of Anions on the Structure and Properties of Copper(II) Complexes with Schiff Base Ligands. J. Mol. Struct. 2024, 1298, 135123. [Google Scholar]
- Shin, J.; Hong, S.; Cho, W.; Kim, J. Dinuclear and Mononuclear Copper(II) Complexes: Structural Control by the Choice of Metal Salt. Inorg. Chim. Acta 2001, 318, 207–214. [Google Scholar]
- El-Faham, A.; El-Gammal, O.; Khalifa, M. Mononuclear versus Dinuclear Copper(II) Complexes: Role of the Metal Salt in Coordination Diversity. J. Mol. Struct. 2021, 1236, 130306. [Google Scholar]
- La, Y.-T.; Yan, Y.-J.; Gan, L.-L.; Xu, L.; Dong, W.-K. Novel phenoxo-bridged di- and tri-nuclear Cu(II) salamo-like complexes driven by various counter-anions. Inorganica Chim. Acta 2023, 546, 121336. [Google Scholar] [CrossRef]
- Díaz, P.; Benet-Buchholz, J.; Vilar, R.; White, A.J. Anion influence on the structures of a series of copper(II) metal-organic frameworks. Inorg. Chem. 2006, 20, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Gatto, C.C.; Dias, L.M.; Paiva, C.A.; da Silva, I.C.R.; Freire, D.O.; Tormena, R.P.I.; Nascimento, É.C.M.; Martins, J.B.L. Effects of changing ions on the crystal design, non-covalent interactions, antimicrobial activity, and molecular docking of Cu(II) complexes with a pyridoxalhydrazone ligand. Front. Chem. 2024, 12, 1347370. [Google Scholar] [CrossRef]
- Fonari, A.; Leonova, E.S.; Yu, M. On justification of Cu(II) environment in mononuclear complexes: Joint X-ray and AIM studies. Polyhedron 2011, 30, 1710–1717. [Google Scholar] [CrossRef]
- Patel, A.K.; Jadeja, R.N.; Roy, H.; Pate, R.N.; Patel, S.K.; Butcher, R.J.; Cortijo, M.; Herrero, S. Copper(II) hydrazone complexes with different nuclearities and geometries: Synthesis, structural characterization, antioxidant SOD activity and antiproliferative properties. Polyhedron 2020, 186, 114624. [Google Scholar] [CrossRef]
- Available online: https://www.ccdc.cam.ac.uk/solutions/software/csd/ (accessed on 15 February 2025).
- Taş, M.; Yeşilel, O.Z.; Büyükgüngör, O. Novel Copper(II) Complexes of N-Phenylanthranilic Acid Containing Ethanol and Hydroxo Ligands. J. Inorg. Organomet. Polym. Mater. 2010, 20, 298–305. [Google Scholar] [CrossRef]
- Vrdoljak, V.; Pavlović, G.; Maltar-Strmečki, N.; Cindrić, M. Copper(ii) hydrazone complexes with different nuclearities and geometries: Synthetic methods and ligand substituent effects. New J. Chem. 2016, 40, 9263–9274. [Google Scholar] [CrossRef]
- Paine, T.K.; Weyhermüller, T.; Wieghardt, K.; Chaudhuri, P. The Methanol−Methanolate CH3OH···OCH3-Bridging Ligand: Tuning of Exchange Coupling by Hydrogen Bonds in Dimethoxo-Bridged Dichromium(III) Complexes. Inorg. Chem. 2002, 41, 6538–6540. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.; Sephton, B.; Sephton, A.; Dugmore, T.; Hickson, M.; Betz, R. Crystal structure of bis((μ2-hydroxo)-aqua-(1,10-phenanthroline-κ2N,N′)-copper(II))bis((μ2-hydroxo)-(carbonato-κO)-(1,10-phenanthroline-κ2N,N′)-copper(II)) tetradecahydrate, C50H64Cu4N8O26. Z. Fur. Krist. New Cryst. Struct. 2015, 230, 168–170. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Zhao, H.; Yang, E.-C.; Liu, Z.-Y.; Shang, Q.; Zhao, X.-J. Triazolate-based 3D frameworks and a 2D layer with centrosymmetric CuII7, CuII5, CuII4 clusters and tunable interlayer/interchain compactness: Hydrothermal syntheses, crystal structures and magnetic properties. Dalton Trans. 2015, 44, 5826–5836. [Google Scholar] [CrossRef]
- Leyva-Porras, C.; Cruz-Alcantar, P.; Espinosa-Solís, V.; Martínez-Guerra, E.; Piñón-Balderrama, C.I.; Compean Martínez, I.; Saavedra-Leos, M.Z. Application of Differential Scanning Calorimetry (DSC) and Modulated Differential Scanning Calorimetry (MDSC) in Food and Drug Industries. Polymers 2020, 12, 5. [Google Scholar] [CrossRef]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Tardito, S.; Luciano, M. Copper Compounds in Anticancer Strategies. Curr. Med. Chem. 2009, 16, 1325–1348. [Google Scholar] [CrossRef] [PubMed]
- Sigman, D.S.; Graham, D.R.; D’Aurora, V.; Stern, A.M. Oxygendependent cleavage of DNA by the 1,10-phenanthroline cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J. Biol. Chem. 1979, 254, 12269–12272. [Google Scholar] [CrossRef]
- Mohsin, N.u.A.; Irfan, M.; Hassan, S.; Sallem, U. Current Strategies in Development of New Chromone Derivatives with Diversified Pharmacological Activities: A Review. Pharm. Chem. J. 2020, 54, 241–257. [Google Scholar] [CrossRef]
- Zhao, K.; Huang, J.; Zhao, Y.; Wang, S.; Xu, J.; Yin, K. Targeting STING in cancer: Challenges and emerging opportunities. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188983. [Google Scholar] [CrossRef]
- Markovic, M.; Ben-Shabat, S.; Dahan, A. Prodrugs for Improved Drug Delivery: Lessons Learned from Recently Developed and Marketed Products. Pharmaceutics 2020, 12, 1031. [Google Scholar] [CrossRef]
- Abet, V.; Filace, F.; Recio, J.; Alvarez-Builla, J.; Burgos, C. Prodrug approach: An overview of recent cases. Eur. J. Med. Chem. 2017, 127, 810–827. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.M. A New Classification of Prodrugs: Regulatory Perspectives. Pharmaceuticals 2009, 2, 77–81. [Google Scholar] [CrossRef]
- Sun, C.; Chen, C.; Xu, S.; Wang, J.; Zhu, Y.; Kong, D.; Tao, H.; Jin, M.; Zheng, P.; Zhu, W. Synthesis and anticancer activity of novel 4-morpholino-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidine derivatives bearing chromone moiety. Bioorg. Med. Chem. 2016, 24, 3862–3869. [Google Scholar] [CrossRef]
- Que, B.G.; Downey, K.M.; So, A.G. Degradation of deoxyribonucleic acid by a 1,10-phenanthroline-copper complex: The role of hydroxyl radicals. Biochemistry 1980, 19, 5987–5991. [Google Scholar] [CrossRef] [PubMed]
- Sigman, D.S. Nuclease activity of 1,10-phenanthroline-copper ion. ACC Chem. Res. 1986, 1, 180–186. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Aruoma, O.I.; Halliwell, B. Modification of bases in DNA by copper ion1,10-phenanthroline complexes. Biochemistry 1990, 29, 8447–8451. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.Y.; Tam, S.C.; Bremner, I.; Burkitt, M.J. Copper-1,10-phenanthroline induces internucleosomal DNA fragmentation in HepG2 cells, resulting from direct oxidation by the hydroxyl radical. Biochem. J. 1996, 317, 13–16. [Google Scholar] [CrossRef]
- Katsarou, M.E.; Efthimiadou, E.K.; Psomas, G.; Karaliota, A.; Vourloumis, D. Novel copper (II) complex of N-propyl-norfloxacinand 1,10-phenanthroline with enhanced antileukemic and DNA nuclease activities. J. Med. Chem. 2008, 51, 470–478. [Google Scholar] [CrossRef]
- Singh, N.K.; Yadav, P.N.; Kumbhar, A.A.; Pokhrel, Y.R. Anticancer potency of copper(II) complexes of thiosemicarbazones. J. Inorg. Biochem. 2020, 210, 111134. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Cong, Y.; Li, N.; Zhang, Z.; Zhao, Z. Cuproptosis: Molecular mechanisms, cancer prognosis, and therapeutic applications. J. Transl. Med. 2025, 23, 104. [Google Scholar] [CrossRef] [PubMed]
- Kahlson, M.A.; Dixon, S.J. Copper-induced cell death. Science 2022, 18, 1231–1232. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Wang, P.; Chen, H.; Xu, Y.; Ge, J.; Tian, Z.; Yan, Z. Potential of Copper and Copper Compounds for Anticancer Applications. Pharmaceuticals 2023, 16, 234. [Google Scholar] [CrossRef] [PubMed]


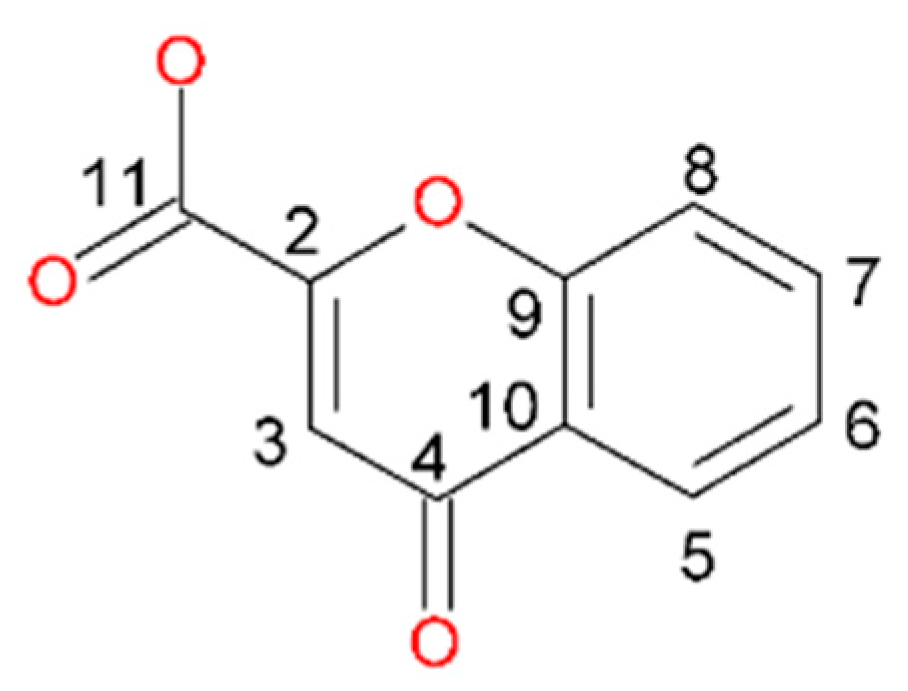



| First Step | Second Step | Third Step | Residue | |
|---|---|---|---|---|
| Temperature Range (°C) | Temperature Range (°C) | Temperature Range (°C) | Temperature Range (°C) | |
| Weight Loss (%) exp./theo. | Weight Loss (%) exp./theo. | Weight Loss (%) exp./theo. | Weight Loss (%) exp./theo. | |
| Complex 1 | 60–120 °C | 120–180 °C | 260–320 °C | at 600 °C |
| Solvent molecules 20.9%/22.2% | H2O 11.3%/10.3% | Ligand molecules 55%/54.1% | CuO 19.5%/18.6% | |
| Complex 2 | 60–180 °C | 220 °C | / | at 600 °C |
| H2O 11.7%/12.1% | Ligand molecules 57%/60% | / | CuO 19.68%/22% | |
| Complex 3 | 60–180 °C | 240 °C | / | at 600 °C |
| H2O 11%/11.7% | Ligand molecules 63.5%/66% | / | CuO 18.4%/20.7% |
| Cell Lines | IC50/µM | |||
|---|---|---|---|---|
| Complex (1) | Complex (2) | Complex (3) | Ligand (L) | |
| MDA-MB 231 | 15.6 (1.6) | 15.7 (2.4) | 16.6 (0.4) | n.s. |
| NCI-H358 | 10.5 (1.3) | 10.9 (0.7) | 11.9 (1.3) | n.s. |
| KATO III | 13.2 (1.8) | 13.4 (1.7) | 11.7 (3.1) | n.s. |
| Hep G2 | 6.1 (0.8) | 7.0 (0.9) | 7.2 (1.2) | n.s. |
| Caco-2 | 18.3 (0.7) | 19.0 (1.3) | 18.4 (2.7) | n.s. |
| HT-29 | 13.5 (2.8) | 10.8 (1.2) | 9.4 (1.5) | n.s. |
| MRC-5 | 9.9 (0.6) | 10.9 (1.9) | 10.3 (1.1) | n.s. |
| Cell Lines | Selectivity Index (SI) | ||
|---|---|---|---|
| Complex (1) | Complex (2) | Complex (3) | |
| MDA-MB 231 | 0.635 | 0.694 | 0.620 |
| NCI-H358 | 0.943 | 1.000 b | 0.866 |
| KATO III | 0.750 | 0.813 | 0.880 c |
| Hep G2 | 1.623 * | 1.557 * | 1.431 * |
| Caco-2 | 0.541 | 0.574 | 0.560 |
| HT-29 | 0.733 b | 1.009 a | 1.096 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filipović, N.; Balić, T.; Medvidović-Kosanović, M.; Goman, D.; Marković, B.; Tatar, D.; Roca, S.; Mišković Špoljarić, K. Chromone-Based Copper(II) Complexes as Potential Antitumour Agents: Synthesis, Chemical Characterisation and In Vitro Biological Evaluation. Crystals 2025, 15, 389. https://doi.org/10.3390/cryst15050389
Filipović N, Balić T, Medvidović-Kosanović M, Goman D, Marković B, Tatar D, Roca S, Mišković Špoljarić K. Chromone-Based Copper(II) Complexes as Potential Antitumour Agents: Synthesis, Chemical Characterisation and In Vitro Biological Evaluation. Crystals. 2025; 15(5):389. https://doi.org/10.3390/cryst15050389
Chicago/Turabian StyleFilipović, Nikolina, Tomislav Balić, Martina Medvidović-Kosanović, Dominik Goman, Berislav Marković, Dalibor Tatar, Sunčica Roca, and Katarina Mišković Špoljarić. 2025. "Chromone-Based Copper(II) Complexes as Potential Antitumour Agents: Synthesis, Chemical Characterisation and In Vitro Biological Evaluation" Crystals 15, no. 5: 389. https://doi.org/10.3390/cryst15050389
APA StyleFilipović, N., Balić, T., Medvidović-Kosanović, M., Goman, D., Marković, B., Tatar, D., Roca, S., & Mišković Špoljarić, K. (2025). Chromone-Based Copper(II) Complexes as Potential Antitumour Agents: Synthesis, Chemical Characterisation and In Vitro Biological Evaluation. Crystals, 15(5), 389. https://doi.org/10.3390/cryst15050389







