Abstract
Macrophages are mainly active cells of the immune system and play a role in the defense of pathogens. However, the overactivation of macrophages by fatal pathogens can result in toxic responses. 2-hydroxyethyl methacrylate (HEMA), which is a hydrophilic monomer, is used in dental adhesive reagents and composite resins as well as biocompatible hydrogels. The mechanisms underlying the genotoxicity engendered by HEMA-induced apoptosis that leads to cytotoxicity remain unclear. Accordingly, this study was conducted to clarify such mechanisms. The results showed that HEMA induced cell toxicity in RAW264.7 macrophages depending on the concentration. A higher HEMA concentration was associated with a higher level of apoptosis and genotoxicity. Moreover, HEMA induced a concentration-dependent increase in mitochondrial dysfunction and the intrinsic caspase pathway, including the activation of caspase-3 and caspase-9. HEMA was also found to upregulate intracellular reactive oxygen species generation and to decrease the activity of antioxidant enzymes, including superoxide dismutase and catalase. Taken together, the mitochondrial-dependent intrinsic caspase pathway and intracellular reactive oxygen species accumulation were found to mediate HEMA-induced genotoxicity and apoptosis, leading to cytotoxicity in RAW264.7 macrophages.
1. Introduction
Macrophages are the active cells of the innate immune system. They phagocytose bacteria and secrete pro-inflammatory mediators, thus contributing to the defense against pathogens [1]. Macrophages are present in all vertebrate tissues and are involved in hemostasis. Resident macrophages in the liver, brain, lung, and epidermis are called Kupffer cells, microglia, alveolar macrophages, and Langerhans cells, respectively [2]. When faced with dangerous pathogens that appear foreign, macrophages are activated to impart toxic effects to self- and peripheral tissues [3,4]. The formation of atherosclerotic plaque, viral infection, inflammation, and sepsis are associated with macrophage death [5]. Pathological pathways regulate macrophage cytotoxicity, including apoptosis and necrosis via genotoxicity [5,6,7]. The molecular mechanisms underlying these toxic effects involve caspase activation, mitochondrial dysfunction, the generation of reactive oxygen species (ROS), and the depletion of antioxidative enzymes (AOEs) in macrophages [5,6,7,8].
2-hydroxyethyl methacrylate (HEMA), a hydrophilic monomer, can be used to create poly(2-hydroxyethylmethacrylate), a polymer, as well as to produce biomaterials such as dental adhesive reagents, dental composite resins, and biocompatible hydrogels [9,10]. HEMA is released from the aforementioned biomaterials in the presence of differential solvents on the first day after polymerization [11]. Because dental biomaterials are highly permeable, the peripheral tissues and cells can be harmed by the leached HEMA [12]. HEMA induces cytotoxicity and apoptosis in macrophages, macrophage-like osteoclasts, and alveolar macrophages [13,14,15,16]. The overgeneration of ROS and the depletion of AOEs are the main factors affecting HEMA-induced apoptosis through caspase-3 activation in macrophages [16,17,18,19]. However, the mechanisms underlying the genotoxicity engendered by HEMA-induced apoptosis, which ultimately leads to cytotoxicity, remain unknown. Consequently, the objective of the present study was to evaluate whether the pathways that depend on mitochondrial disruption and the accumulation of ROS within the RAW264.7 macrophages are involved in the cytotoxicity and genotoxicity caused by apoptosis after HEMA treatment.
2. Materials and Methods
2.1. Materials
Fetal bovine serum (FBS), antibiotic–antimycotic solution, Dulbecco’s modified Eagle’s medium (DMEM), and phosphate-buffered saline (PBS) were obtained from Gibco BRL, Life Technologies (Grand Island, NY, USA). The Annexin V-FITC apoptosis-detection kit, JC-1, HEMA, dichlorofluorescin diacetate (DCFH-DA) and other chemical reagents, unless specifically stated, were purchased from Sigma-Aldrich (St. Louis, MO, USA). The caspase-3 fluorometric assay kit, caspase-8 fluorometric assay kit, and caspase-9 fluorometric assay kit were obtained from Enzo Life Sciences (Farmingdale, NY, USA). The catalase (CAT) assay kit and superoxide dismutase (SOD) assay kit were obtained from Cayman Chemical (Ann Arbor, MI, USA). The HEMA solution was derived from dimethyl sulfoxide (DMSO), with the final concentration being less than 0.5% (v/v) nontoxic.
2.2. Cell Culture and Treatment
RAW264.7 cells (BCRC No.6001) are murine macrophage cells derived from BALB/c mice extracted from the Bioresources Collection and Research Center in Hsinchu, Taiwan. The RAW264.7 cells were cultured in DMEM supplemented with 10% FBS, 1 mM sodium pyruvate, and 1% antibiotic–antimycotic solution—which contains penicillin, streptomycin, and Amphotericin B—and incubated at 37 °C in a humidified atmosphere of 5% CO2 [20]. After seeding for the night, the cells were incubated in HEMA for 24 h at concentrations of 0, 0.5, 1, 5, and 10 mM. The cells were then harvested and used for further experiments.
2.3. Cell Viability Assay
The effect of HEMA on the viability of the RAW 264.7 cells was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay, as described in our previous study [20]. In brief, the 1 × 106 cells were incubated with various concentrations of HEMA (0, 0.5, 1, 5, and 10 mM) for 24 h, then 0.5 mg/mL of MTT were added to each well. After incubation for 4 h, the reaction was completed by adding the DMSO solution. The absorption was detected with a 570 nm microplate reader (Synergy HT, BioTek, Winooski, VT, USA). The percentage of cell survival was calculated by using the following formula: (absorbance of cells treated with HEMA − absorbance of blank well/absorbance of cells treated without HEMA − absorbance of blank well) × 100%.
2.4. Flow Cytometric Analysis of Apoptosis and Necrosis
The effects of HEMA on apoptosis and necrosis were determined using the Annexin V-FITC apoptosis-detection kit, as described previously [20]. After 24 h of incubation with different HEMA concentrations, the cells were collected via trypsinization and washed with PBS. The 1 × 106 cells were dyed for 30 min at room temperature with the 100 μL binding buffer that contained the 5 μL of Annexin V–FITC and 5 μL of PI. The acquisition of the RAW264.7 cells and the data analysis were measured by the Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA). The percentages of cells in the viable, necrotic, and apoptotic cells were presented as annexin-V-FITC-negative and PI-negative, PI-positive, and annexin-V-FITC-positive, respectively.
2.5. Micronucleus Assay
Micronucleus (MN) formation, as the marker of DNA damage, was measured by the alkaline cytokinesis–block MN assay, as described above [21]. In short, the 1 × 106 cells were incubated with different concentrations of HEMA and 3 mg/mL of cytochalasin B for 24 h. After incubation, the cells were washed, incubated with 75 mM KCl, fixed in the methanol/acetic acid mixture, and stained with the 3% Giemsa solution. The MN were observed using light microscopy.
2.6. Single-Cell Gel Electrophoresis
DNA damage was analyzed using the alkaline single-cell gel electrophoresis (Comet) assay, as described above [21]. After treatment, 1 × 106 cells were mixed with the solutions of low-melting-point agarose and placed in a microscopic slide that was pre-loaded with normal-melting-point agarose after treatment. The slides were incubated with the alkaline lysis buffer, which contained 2.5 M NaCl, 100 mM EDTA, 10 mM Tris pH 10, 1% Triton X-100, 200 mM NaOH, 34.1 mM N-Lauroyl-Sarcosine, and 10% DMSO, at 4 °C for 1 h. The slides were then washed, electrophoresed, neutralized, stained with ethidium bromide, and finally analyzed using the image-analysis software Comet v. 3 (Kinetic Imaging Ltd., Liverpool, UK).
2.7. Flow Cytometric Analysis of Mitochondrial Dysfunction
Mitochondrial dysfunction induced by HEMA was evaluated using JC-1 staining, as described previously [21]. In brief, after incubation with HEMA, the cells were collected and stained with 10 mg/mL JC-1 at 37 °C for 30 min. The cells were washed with PBS, and the collected cells and data were analyzed using the BD Accuri C6 flow cytometer with C6 software.
2.8. Caspase Activation Assay
The activation of caspase-3, -8, and -9 induced by HEMA was determined by the caspase fluorometric assay kits, as described above [21]. The lysis buffer and reaction buffer were obtained from caspase fluorometric assay kits. In short, the cells were collected and lysed with lysis buffer after treatment with HEMA. An equal amount of protein was extracted from each sample and incubated with the fluorogenic substrates including DEVD-AFC, IETD-AFC, and LEHD-AFC for caspase-3, caspase-8, and caspase-9 in reaction buffer, respectively. After incubation at 37 °C for 2 h and upon excitation at 485 nm, the fluorescence intensity was measured at 505 nm using a fluorescence microplate reader (BioTek Instruments, Winooski, VT, USA).
2.9. AOE Activition Assay
The effect of HEMA on the activation of AOEs, including CAT and SOD, was evaluated by CAT and SOD assay kits in accordance with the manufacturer’s protocol and previous studies, respectively [20].
2.10. Intracellular ROS-Generation Measurement
The intracellular levels of ROS induced by HEMA were evaluated using the DCFH-DA assay, as described above [20]. In brief, the cells were incubated with 5 μM DCFH-DA for 0.5 h at 37 °C in the dark after treatment with HEMA. Fluorescence was determined at 485/530 nm using a fluorescence microplate reader (BioTek Instruments).
2.11. Statistical Analysis
All experiments were carried out at least three times. The data are represented as an average of the standard deviation (SD). Statistical comparisons were made using the one-way analysis of variance followed by the Bonferroni post-hoc test. Data were analyzed using SPSS software (IBM, Armonk, NY, USA). The p-value was considered statistically significant if less than 0.05.
3. Results
3.1. HEMA’s Effects on the Cellular Viability of RAW264.7 Cells
To assess the cell survival of HEMA in RAW264.7 macrophage cells, the cells were treated for 24 h with HEMA at different concentrations. The results of the MTT assay (Figure 1) revealed that HEMA significantly reduced the function of RAW264.7 cells, depending on the concentration. Significant decreases in cell viability were observed at the concentration of 1 mM (p < 0.05).
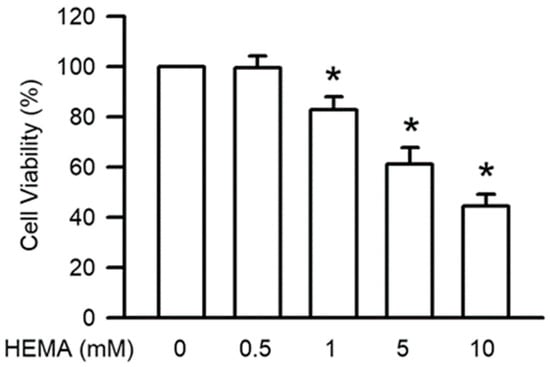
Figure 1.
HEMA reduced cell viability in RAW264.7 cells. Cell survival was measured by the MTT test after 24 h of HEMA incubation. The value is expressed as the percentage of control cells treated with the vehicle. Results are expressed as means ± SD. The number of parallel measurements is 5. * means that p < 0.05 compared with HEMA at the concentration of 0 mM.
3.2. HEMA’s Effects on Necrosis or Apoptosis of RAW264.7 Cells
The RAW264.7 cells labeled with annexin V-FITC and PI were used to identify the necrosis or apoptosis induced by HEMA. As illustrated in Figure 2 and Table 1, HEMA induced apoptosis in the RAW264.7 cells in a concentration-dependent manner. Significant increases in apoptosis were observed at the concentration of 1 mM (p < 0.05). On the other hand, necrosis was not caused by HEMA at the concentration of 10 mM in the RAW264.7 macrophages.
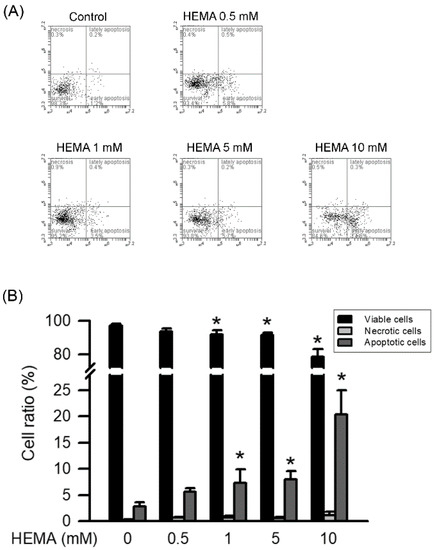
Figure 2.
HEMA induced apoptosis in RAW264.7 cells. After treatment, RAW264.7 cells were collected, stained with Annexin V-FITC and PI, and then analyzed by dual-staining flow cytometry. (A) Representative FACS plot of RAW264.7 macrophages incubated with HEMA. (B) Quantitatively, the percentage of viable cells (black column), necrotic cells (gray column), and apoptotic cells (dark gray column) was calculated and analyzed. Data are expressed as mean ± SD. The number of parallel measurements is 4. * means that p < 0.05 compared with HEMA at the concentration of 0 mM.

Table 1.
The parameters of cell responses in RAW264.7 macrophages exposed to the different concentrations of HEMA for 24 h in Annexin V-FITC apoptosis-detection kit.
3.3. HEMA’s Effects on Genotoxicity in RAW264.7 Macrophages
Genotoxicity, which also means DNA damage, is the major inducer of apoptosis. As shown in Figure 3, HEMA-induced genotoxicity was measured by the Comet and MN assays. The results of the Comet assay indicated that compared to those of the control cells, the tail moment and tail length values in the HEMA-treated cells were significantly increased in a concentration-dependent manner (starting from 1 mM; p < 0.05; Figure 3A). According to the MN assay results, MN formation was significantly increased in the HEMA-treated cells compared with that in the control cells in a concentration-dependent manner (starting from 1 mM; p < 0.05; Figure 3B).
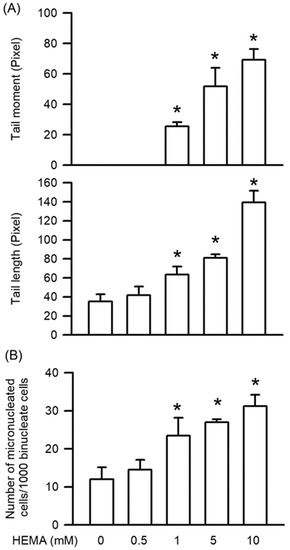
Figure 3.
HEMA induced genotoxicity in RAW264.7 cells. (A) After the Comet assay, genotoxicity was quantified as tail moment and tail length. (B) After the MN assay, genotoxicity was quantified as MN formation. Data are expressed as mean ± SD. The number of parallel measurements is 4. * means that p < 0.05 compared with HEMA at the concentration of 0 mM.
3.4. HEMA’s Effects on the Activation of Caspase-3, Caspase-8, and Caspase-9 in RAW264.7 Macrophages
Caspase-3, caspase-8, and caspase-9 play an important role in apoptosis induced by genotoxicity. After treatment for 24 h, the HEMA-induced activation of caspase-3 and caspase-9 was significantly dependent on concentration (starting from 1 mM; p < 0.05; Figure 4). Moreover, significant HEMA-induced caspase-8 activity was observed (starting from 10 mM; p < 0.05; Figure 4).
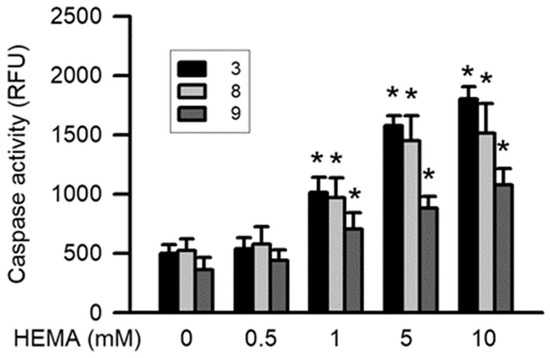
Figure 4.
HEMA induced activation of caspase-3, caspase-8, and caspase-9 in RAW264.7 macrophages. Activation of caspase-3 (black column), caspase-8 (gray column), and caspase-9 (dark gray column) was measured by the caspase fluorometric assay kits after 24 h of HEMA incubation. Data are expressed as means ± SD. The number of parallel measurements is 5. * means that p < 0.05 compared with HEMA at the concentration of 0 mM.
3.5. Effects of HEMA on Mitochondrial Dysfunction in RAW264.7 Macrophages
Mitochondrial dysfunction leads to apoptosis by disrupting the mitochondrial integrity of cells. According to the results of the JC-1 staining (Figure 5 and Table 2), HEMA-induced mitochondrial dysfunction was concentration-dependent, and significant effects were observed starting from 1 mM (p < 0.05).
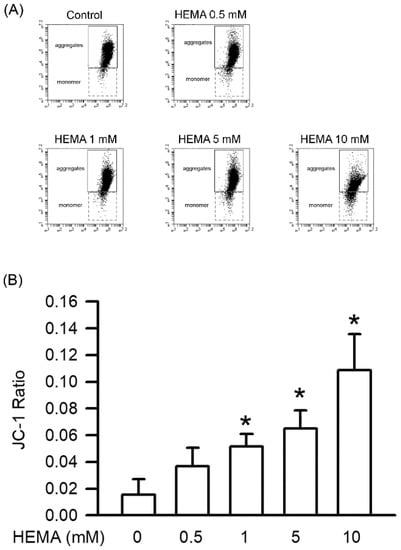
Figure 5.
HEMA caused mitochondrial dysfunction. The mitochondrial dysfunction was measured with JC-1 after 24 h of HEMA treatment. (A) The original flow cytometry plot showing the evaluation of the mitochondrial dysfunction in RAW264.7 macrophages incubated with HEMA. (B) Quantitatively, the JC-1 ratio of the ratios of monomers/aggregates is shown. Data are expressed as mean ± SD. The number of parallel measurements is 4. * means that p < 0.05 compared with HEMA at the concentration of 0 mM.

Table 2.
The parameters of mitochondrial dysfunction in RAW264.7 macrophages exposed to the different concentrations of HEMA for 24 h via JC-1 staining.
3.6. Effects of HEMA on Intracellular ROS Generation in RAW264.7 Cells
Intracellular ROS generation plays a major role in apoptosis through mitochondrial dysfunction. As displayed in Figure 6, HEMA induced significant intracellular ROS generation in the RAW264.7 cells in a concentration-dependent manner (starting from 1 mM; p < 0.05).
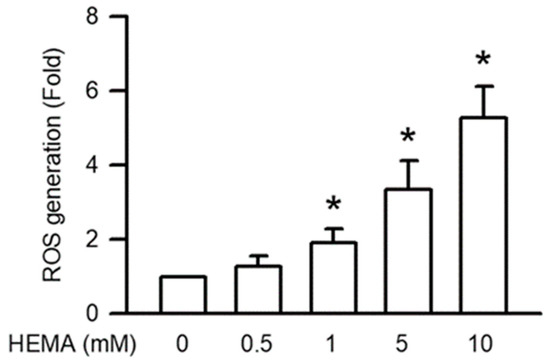
Figure 6.
Generation of ROS was induced by HEMA in RAW264.7 cells. After 24 h of treatment with HEMA, DCFH-DA measured ROS generation. Data are expressed as mean ± SD. The number of parallel measurements is 4. * means that p < 0.05 compared with HEMA at the concentration of 0 mM.
3.7. Effects of HEMA on AOE Activity in RAW264.7 Cells
The activation of AOEs including SOD and CAT after 24 h of exposure to HEMA at different concentrations was monitored by the SOD and CAT activity assay kit. It was found that after 24 h of exposure to HEMA at different concentrations, the activity of AOEs (including SOD and CAT) significantly decreased depending on the concentration (starting from 1 mM; p < 0.05; Figure 7).
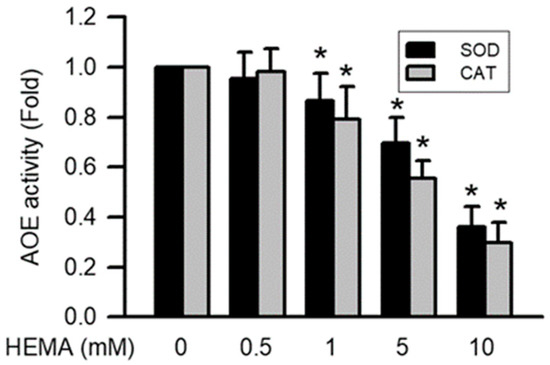
Figure 7.
Activation of AOEs was reduced by HEMA in RAW264.7 macrophages. Data are expressed as mean ± SD. The number of parallel measurements is 4. * means that p < 0.05 compared with HEMA at the concentration of 0 mM.
4. Discussion
HEMA, a water-soluble, low-molecular-weight monomer, enhances the wetting properties and penetration efficacy of demineralized bone matrices of HEMA-based polymers; this property has led to its widespread application in the manufacture of biomaterials for dentistry, orthopedics, and ophthalmology [9,10,22,23]. Previous reports have revealed that HEMA can form polymerized biomaterials in the presence of various solvents and in optimal incubation durations. It can also be eluted from adhesive systems [24,25,26]. Furthermore, HEMA is a derivative of methacrylic acid and acrylic acid, which cause allergic symptoms such as rubefaction, pruritus, persistent paresthesia, and induration [27,28]. HEMA has been reported to induce cytotoxicity in differential macrophages, including alveolar macrophages, J774A.1 macrophages, RAW264.7 macrophages, and human peripheral blood mononuclear cells through apoptosis and not necrosis [15,16,17,18,19,29]. Our findings corroborate those of this previous report; specifically, we observed that HEMA induced cytotoxicity in RAW264.7 macrophages in a concentration-dependent manner through apoptosis and not necrosis.
Apoptosis is usually caused by genotoxicity in macrophages [7,21]. Genotoxicity leads to destructive effects and hampers the integrity of double-stranded or single-stranded DNA [30,31]. The Comet and MN assays are single-cell microgel electrophoresis techniques that help detect single- or double-stranded breaks in the DNA at the individual cell level [30,31]. Genotoxicity was evaluated using HEMA in a concentration-dependent manner in human lymphocytes, gingival fibroblasts, and bronchial epithelial cells [32,33,34]. In addition, HEMA induced DNA fragmentation in RAW264.7 macrophages and human peripheral blood mononuclear cells [29]. In the present study, using the Comet and MN assays, we successfully demonstrated that HEMA induced genotoxicity in macrophages. These results indicate that genotoxicity is the primary process through which HEMA induces cytotoxicity through apoptosis in macrophages.
Caspases are a family of cysteinyl proteases and play a critical role in the regulation of genotoxicity [35]. Caspases are subclassified according to their mechanism of action as follows: extrinsic initiator caspase-8, intrinsic initiator caspase-9, and executor caspase-3 [36]. Extrinsic pathway-initiated apoptosis includes caspase-8 activation mediated by death receptors. Intrinsic pathway-initiated apoptosis includes mitochondrial dysfunction-mediated caspase-9 activation. The activation of caspase-8 and caspase-9 results in the activation of caspase-3 [35,36]. HEMA induces caspase-3 activation in RAW 264.7 macrophages, human gingival fibroblasts, and human dental pulp cells [19,37,38]. HEMA has been reported to induce the activation of caspase-9, and not caspase-8, in rat submandibular salivary gland acinar cells [39]. The results of the present study demonstrate that the activation of caspase-3 and caspase-9 is more sensitive than that of caspase-8 in RAW264.7 cells treated with HEMA. Furthermore, this study revealed that mitochondrial dysfunction, which is the initiation factor of the intrinsic pathway, is induced by HEMA in a concentration-dependent manner in macrophages. On the basis of these findings, we propose that HEMA-induced genotoxicity occurs mainly through the intrinsic caspase pathway, which includes caspase-3 and caspase-9, in macrophages.
Cell cycle progression, cell proliferation, cell differentiation, and other normal physiological functions in mammalian cells are regulated by ROS, including superoxide, nitric oxide, hydrogen peroxide, hydroxyl radicals, and relative free radicals [40,41]. In addition, ROS play a major role in the immune system by defending against pathogens. The overgeneration of ROS is harmful to the peripheral tissues [41,42]. The generation of intracellular ROS is the main reason for mitochondrial dysfunction [43]. Moreover, the accumulation of intracellular ROS is induced by the detoxification system of AOE, which does not maintain low activity [41,43]. The upregulation of ROS generation considerably increases caspase activation and mitochondrial disruption [41]. Previous studies have reported that HEMA induces intracellular ROS generation in macrophages, gingival fibroblasts, and odontoblast-like cells [16,17,18,44]. HEMA also induces the downregulation of SOD expression and the upregulation of CAT expression in macrophages [16,17,18]. We also demonstrated that HEMA induces intracellular ROS generation. Furthermore, the activation of AOEs, including SOD and CAT, is inhibited by HEMA in macrophages. Accordingly, we can infer that HEMA-induced intracellular ROS accumulation results in caspase activation.
5. Conclusions
In summary, the present study demonstrated the deleterious effects of HEMA in RAW264.7 macrophages. Exposure to HEMA resulted in decreased cell viability through apoptosis and not necrosis. Moreover, the apoptotic effect of HEMA was found to be associated with genotoxicity. The molecular mechanisms underlying HEMA-induced genotoxicity involve the mitochondrial-dependent intrinsic caspase pathway and intracellular ROS accumulation. Our findings provide a clearer understanding of the mechanisms underlying the cytotoxicity of HEMA via apoptosis and genotoxicity in the macrophages.
Author Contributions
Conceptualization, C.-Y.L.; Data curation, C.-Y.L. and Y.-C.H.; Formal analysis, Y.-C.L.; Funding acquisition, M.-Y.L. and Y.-H.K.; Investigation, C.-Y.L. and S.-S.L.; Methodology, C.-Y.L., Y.-C.H., S.-S.L. and Y.-C.L.; Supervision, Y.-H.K.; Visualization, Y.-C.H.; Writing—original draft, C.-Y.L., Y.-C.H., S.-S.L., Y.-C.L. and M.-Y.L.; Writing—review & editing, Y.-H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Chung Shan Medical University Hospital research program of Taiwan grant number CSH-2020-C-006. This research was funded by Ministry of Science and Technology of Taiwan grant number MOST 109-2320-B-040-MY3.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on reasonable request from the corresponding author.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
| AOE | antioxidative enzyme |
| CAT | Catalase |
| Comet | alkaline single-cell gel electrophoresis |
| DCFH-DA | dichlorofluorescin diacetate |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DMSO | dimethyl sulfoxide |
| FBS | Fetal bovine serum |
| HEMA | 2-Hydroxyethyl methacrylate |
| MN | Micronucleus |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide |
| ROS | reactive oxygen species |
References
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef]
- Watanabe, S.; Alexander, M.; Misharin, A.V.; Budinger, G.R.S. The role of macrophages in the resolution of inflammation. J. Clin. Investig. 2019, 129, 2619–2628. [Google Scholar] [CrossRef]
- Laskin, D.L.; Sunil, V.R.; Gardner, C.R.; Laskin, J.D. Macrophages and tissue injury: Agents of defense or destruction? Annu. Rev. Pharmacol. Toxicol. 2011, 51, 267–288. [Google Scholar] [CrossRef]
- Vannella, K.M.; Wynn, T.A. Mechanisms of Organ Injury and Repair by Macrophages. Annu. Rev. Physiol. 2017, 79, 593–617. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.; Ganesan, R.; Hegedűs, C.; Kovács, K.; Kufer, T.A.; Virág, L. Programmed necrotic cell death of macro-phages: Focus on pyroptosis, necroptosis, and parthanatos. Redox Biol. 2019, 26, 101239. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.-K.; Wu, S.-W.; Chiang, C.-Y.; Lee, M.-W.; Chen, H.-Y.; Chen, W.-Y.; Chen, C.-J.; Yang, S.-F.; Yeh, C.-B.; Kuan, Y.-H. Evaluation of cytotoxicity, apoptosis, and genotoxicity induced by indium chloride in macrophages through mitochondrial dysfunction and reactive oxygen species generation. Ecotoxicol. Environ. Saf. 2020, 193, 110348. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-Y.; Chiang, C.-Y.; Chiang, Y.-W.; Lee, M.-W.; Lee, C.-Y.; Chen, H.-Y.; Lin, H.-W.; Kuan, Y.-H. Toxic Effects of Urethane Dimethacrylate on Macrophages Through Caspase Activation, Mitochondrial Dysfunction, and Reactive Oxygen Species Generation. Polymers 2020, 12, 1398. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.-M.; Chang, Y.-C.; Lee, S.-S.; Ho, Y.-C.; Yang, M.-L.; Lin, H.-W.; Kuan, Y.-H. Bisphenol A exhibits cytotoxic or genotoxic potential via oxidative stress-associated mitochondrial apoptotic pathway in murine macrophages. Food Chem. Toxicol. 2018, 122, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Babić Radić, M.M.; Filipović, V.V.; Vukomanović, M.; Nikodinović Runić, J.; Tomić, S.L. Degradable 2-Hydroxyethyl Methacrylate/Gelatin/Alginate Hydrogels Infused by Nanocolloidal Graphene Oxide as Promising Drug Delivery and Scaffolding Biomaterials. Gels 2021, 8, 22. [Google Scholar] [CrossRef]
- Pandey, S.A.; Lokhande, M.T.; Gulve, M.N.; Kolhe, S.J.; Aher, G.B. Shear bond strength of composite resin to res-in-modified glass ionomer cement using 2-hydroxyethyl methacrylate-based and 2-hydroxyethyl methacrylate-free adhesive system. J. Conserv. Dent. 2019, 22, 292–295. [Google Scholar] [CrossRef]
- Yoshii, E. Cytotoxic effects of acrylates and methacrylates: Relationships of monomer structures and cytotoxicity. J. Biomed. Mat. Res. 1997, 37, 517–524. [Google Scholar] [CrossRef]
- Krifka, S.; Spagnuolo, G.; Schmalz, G.; Schweikl, H. A review of adaptive mechanisms in cell responses towards oxidative stress caused by dental resin monomers. Biomaterials 2013, 34, 4555–4563. [Google Scholar] [CrossRef]
- Murakami, Y.; Kawata, A.; Suzuki, S.; Fujisawa, S. Cytotoxicity and Pro-/Anti-inflammatory Properties of Cinnamates, Acrylates and Methacrylates Against RAW264.7 Cells. In Vivo 2018, 32, 1309–1322. [Google Scholar] [CrossRef]
- Inamitsu, H.; Okamoto, K.; Sakai, E.; Nishishita, K.; Murata, H.; Tsukuba, T. The dental resin monomers HEMA and TEGDMA have inhibitory effects on osteoclast differentiation with low cytotoxicity. J. Appl. Toxicol. 2017, 37, 817–824. [Google Scholar] [CrossRef]
- Becher, R.; Kopperud, H.; Al, R.; Samuelsen, J.; Morisbak, E.; Dahlman, H.; Lilleaas, E.; Dahl, J. Pattern of cell death after in vitro exposure to GDMA, TEGDMA, HEMA and two compomer extracts. Dent. Mater. 2006, 22, 630–640. [Google Scholar] [CrossRef]
- Krifka, S.; Hiller, K.A.; Spagnuolo, G.; Jewett, A.; Schmalz, G.; Schweikl, H. The influence of glutathione on redox regulation by antioxidant proteins and apoptosis in macrophages exposed to 2-hydroxyethyl methacrylate (HEMA). Biomaterials 2012, 33, 5177–5186. [Google Scholar] [CrossRef]
- Gallorini, M.; Petzel, C.; Bolay, C.; Hiller, K.-A.; Cataldi, A.; Buchalla, W.; Krifka, S.; Schweikl, H. Activation of the Nrf2-regulated antioxidant cell response inhibits HEMA-induced oxidative stress and supports cell viability. Biomaterials 2015, 56, 114–128. [Google Scholar] [CrossRef]
- Schweikl, H.; Godula, M.; Petzel, C.; Bolay, C.; Hiller, K.; Buchalla, W. Critical role of superoxide anions and hydroxyl radicals in HEMA-induced apoptosis. Dent. Mater. 2016, 33, 110–118. [Google Scholar] [CrossRef]
- Schweikl, H.; Petzel, C.; Bolay, C.; Hiller, K.-A.; Buchalla, W.; Krifka, S. 2-Hydroxyethyl methacrylate-induced apoptosis through the ATM- and p53-dependent intrinsic mitochondrial pathway. Biomaterials 2014, 35, 2890–2904. [Google Scholar] [CrossRef]
- Su, C.H.; Chen, S.P.; Chen, L.Y.; Yang, J.J.; Lee, Y.C.; Lee, S.S.; Chen, H.H.; Ng, Y.Y.; Kuan, Y.H. 3-Bromofluoranthene-induced cardiotoxicity of zebrafish and apoptosis in the vascular endothelial cells via intrinsic and extrinsic caspase-dependent pathways. Ecotoxicol. Environ. Saf. 2021, 228, 112962. [Google Scholar] [CrossRef]
- Wu, S.-W.; Su, C.-H.; Ho, Y.-C.; Huang-Liu, R.; Tseng, C.-C.; Chiang, Y.-W.; Yeh, K.-L.; Lee, S.-S.; Chen, W.-Y.; Chen, C.-J.; et al. Genotoxic effects of 1-nitropyrene in macrophages are mediated through a p53-dependent pathway involving cytochrome c release, caspase activation, and PARP-1 cleavage. Ecotoxicol. Environ. Saf. 2021, 213, 112062. [Google Scholar] [CrossRef]
- Xi, Y.; Sharma, P.K.; Kaper, H.J.; Choi, C.-H. Tribological Properties of Micropored Poly(2-hydroxyethyl methacrylate) Hydrogels in a Biomimetic Aqueous Environment. ACS Appl. Mater. Interfaces 2021, 13, 41473–41484. [Google Scholar] [CrossRef]
- Hitmi, L.; Bouter, D.; Degrange, M. Influence of drying and HEMA treatment on dentin wettability. Dent. Mater. 2002, 18, 503–511. [Google Scholar] [CrossRef]
- Reichl, F.-X.; Löhle, J.; Seiss, M.; Furche, S.; Shehata, M.M.; Hickel, R.; Müller, M.; Dränert, M.; Durner, J. Elution of TEGDMA and HEMA from polymerized resin-based bonding systems. Dent. Mater. 2012, 28, 1120–1125. [Google Scholar] [CrossRef]
- Altunsoy, M.; Botsali, M.S.; Ulker, H.E. Evaluation of HEMA released from four different adhesive systems by HPLC. J. Appl. Biomater. Funct. Mater. 2015, 13, e100–e105. [Google Scholar] [CrossRef]
- Díaz-Álvarez, M.; Martín-Esteban, A.; Turiel, E. Evaluation of 2-hydroxyethyl methacrylate as comonomer in the preparation of water-compatible molecularly imprinted polymers for triazinic herbicides. J. Sep. Sci. 2022, 45, 2356–2365. [Google Scholar] [CrossRef]
- Aalto-Korte, K.; Suuronen, K. Ten years of contact allergy from acrylic compounds in an occupational dermatology clinic. Contact Dermat. 2020, 84, 240–246. [Google Scholar] [CrossRef]
- Symanzik, C.; Weinert, P.; Babić, Ž.; Hallmann, S.; Havmose, M.S.; Johansen, J.D.; Kezic, S.; Macan, M.; Macan, J.; Strahwald, J.; et al. Allergic contact dermatitis caused by 2-hydroxyethyl methacrylate and ethyl cyanoacrylate contained in cosmetic glues among hairdressers and beauticians who perform nail treatments and eyelash extension as well as hair extension applications: A systematic review. Contact Dermat. 2022, 86, 480–492. [Google Scholar] [CrossRef]
- Paranjpe, A.; Bordador, L.; Wang, M.-Y.; Hume, W.; Jewett, A. Resin Monomer 2-Hydroxyethyl Methacrylate (HEMA) is a Potent Inducer of Apoptotic Cell Death in Human and Mouse Cells. J. Dent. Res. 2005, 84, 172–177. [Google Scholar] [CrossRef]
- Buchmueller, J.; Enge, A.-M.; Peters, A.; Ebmeyer, J.; Küpper, J.-H.; Schäfer, B.; Braeuning, A.; Hessel-Pras, S. The chemical structure impairs the intensity of genotoxic effects promoted by 1,2-unsaturated pyrrolizidine alkaloids in vitro. Food Chem. Toxicol. 2022, 164, 113049. [Google Scholar] [CrossRef]
- Tchounwou, P.B. Genotoxic Stress. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 313–317. [Google Scholar]
- Ginzkey, C.; Zinnitsch, S.; Steussloff, G.; Koehler, C.; Hackenberg, S.; Hagen, R.; Kleinsasser, N.H.; Froelich, K. Assessment of HEMA and TEGDMA induced DNA damage by multiple genotoxicological endpoints in human lymphocytes. Dent. Mater. 2015, 31, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Szczepanska, J.; Poplawski, T.; Synowiec, E.; Pawlowska, E.; Chojnacki, C.J.; Chojnacki, J.; Blasiak, J. 2-hydroxylethyl methacrylate (HEMA), a tooth restoration component, exerts its genotoxic effects in human gingival fibroblasts trough meth-acrylic acid, an immediate product of its degradation. Mol. Biol. Rep. 2012, 39, 1561–1574. [Google Scholar] [CrossRef] [PubMed]
- Ansteinsson, V.; Solhaug, A.; Samuelsen, J.; Holme, J.; Dahl, J. DNA-damage, cell-cycle arrest and apoptosis induced in BEAS-2B cells by 2-hydroxyethyl methacrylate (HEMA). Mutat. Res. Toxicol. Environ. Mutagen. 2011, 723, 158–164. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, D.R.; Berger, T.; Mak, T.W. Caspase Functions in Cell Death and Disease. Cold Spring Harb. Perspect. Biol. 2015, 7, a026716. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Jiao, Y.; Ma, S.; Wang, Y.; Li, H.; Shan, L.; Liu, Q.; Liu, Y.; Song, Q.; Yu, F.; Yu, H.; et al. N-Acetyl Cysteine Depletes Reactive Oxygen Species and Prevents Dental Monomer-Induced Intrinsic Mitochondrial Apoptosis In Vitro in Human Dental Pulp Cells. PLoS ONE 2016, 11, e0147858. [Google Scholar] [CrossRef]
- Teti, G.; Salvatore, V.; Mazzotti, M.C.; Orlandini, B.; Focaroli, S.; Durante, S.; Paternostro, F. Ultrastructural changes in human gin-gival fibroblasts after exposure to 2-hydroxyethyl methacrylate. Ital. J. Anat. Embryol. 2014, 119, 130–140. [Google Scholar]
- Samuelsen, J.T.; Dahl, J.E.; Karlsson, S.; Morisbak, E.; Becher, R. Apoptosis induced by the monomers HEMA and TEGDMA involves formation of ROS and differential activation of the MAP-kinases p38, JNK and ERK. Dent. Mater. 2007, 23, 34–39. [Google Scholar] [CrossRef]
- Covarrubias, L.; Hernández-García, D.; Schnabel, D.; Salas-Vidal, E.; Castro-Obregón, S. Function of reactive oxygen species during animal development: Passive or active? Dev. Biol. 2008, 320, 1–11. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef]
- Snezhkina, A.V.; Kudryavtseva, A.V.; Kardymon, O.L.; Savvateeva, M.V.; Melnikova, N.V.; Krasnov, G.S.; Dmitriev, A.A. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. Oxidative Med. Cell. Longev. 2019, 2019, 6175804. [Google Scholar] [CrossRef]
- Zhao, R.Z.; Jiang, S.; Zhang, L.; Yu, Z.B. Mitochondrial electron transport chain, ROS generation and uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef]
- Di Nisio, C.; Zara, S.; Cataldi, A.; Di Giacomo, V. 2-Hydroxyethyl methacrylate inflammatory effects in human gingival fibroblasts. Int. Endod. J. 2012, 46, 466–476. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).