Genome-Wide Identification and Evolutionary Analysis of AOMT Gene Family in Pomegranate (Punica granatum)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Data Sources and Gene Identification of AOMTs
2.3. Phylogenetic Analysis
2.4. Exon-Intron Structure
2.5. Motif Analysis
2.6. RNA Extraction, cDNA Synthesis and RT-PCR Analysis
2.7. Expression Analyses of PgAOMT Gene Family Members
3. Results
3.1. Identification of the AOMT Genes in Pomegranate
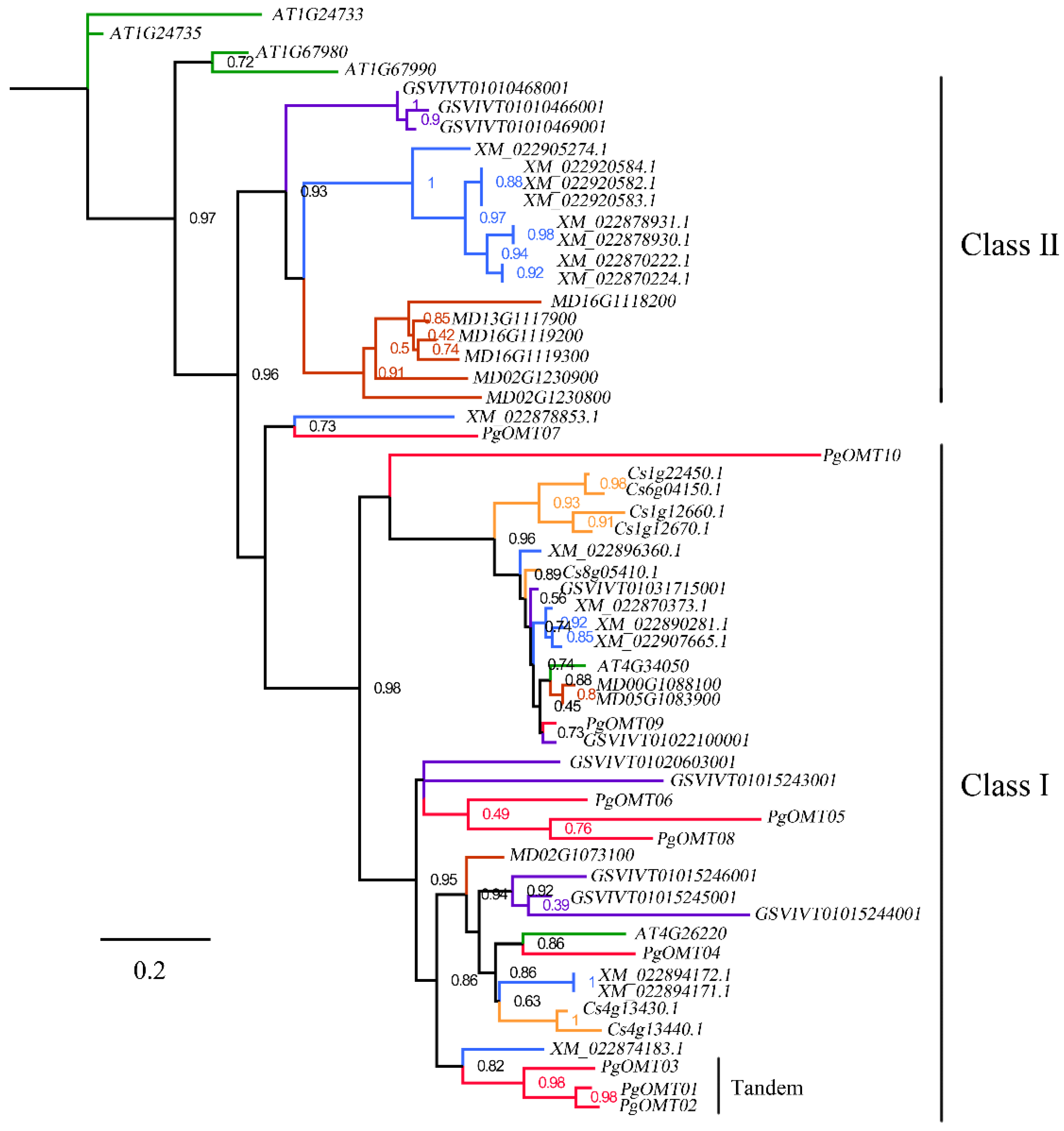
3.2. Phylogenetic Analysis of PgAOMT Gene Family
3.3. Gene Structure and Protein Conserved Motifs of PgAOMT Genes Family
3.4. RT-PCR Analysis of PgAOMT Candidate Gene Sequences
3.5. Expression Patterns of PgAOMT in Peel and Aril during Different Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- DeMuth, J.P.; Hahn, M.W. The life and death of gene families. BioEssays 2009, 31, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elisabeth, K.; Leng, S.; Alexander, H. The effects of repeated whole genome duplication events on the evolution of cytokinin signaling pathway. BMC Evol. Biol. 2018, 18, 1–19. [Google Scholar]
- Van De Peer, Y.; Mizrachi, Y.V.D.P.E.; Marchal, Y.V.D.P.K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, M.; Espley, R.V.; Stevenson, D.; Cooney, J.; Datson, P.M.; Saiz, A.; Atkinson, R.G.; Hellens, R.P.; Allan, A.C. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 2010, 65, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-N.; Yao, G.-F.; Zheng, D.; Zhang, S.; Wang, C.; Zhang, M.-Y.; Wu, J. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2014, 34, 189–198. [Google Scholar] [CrossRef]
- Chiu, L.-W.; Zhou, X.; Burke, S.; Wu, X.; Prior, R.L.; Li, L. The Purple Cauliflower Arises from Activation of a MYB Transcription Factor. Plant Physiol. 2010, 154, 1470–1480. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Wu, J.; Ji, K.-X.; Zeng, Q.-Y.; Bhuiya, M.-W.; Su, S.; Shu, Q.-Y.; Ren, H.-X.; Liu, Z.-A.; Wang, L.-S. Methylation mediated by an anthocyanin, O-methyltransferase, is involved in purple flower coloration in Paeonia. J. Exp. Bot. 2015, 66, 6563–6577. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhu, N.; Zhu, X.; Wu, M.; Jiang, C.-Z.; Grierson, D.; Luo, Y.; Shen, W.; Zhong, S.; Fu, D.-Q.; et al. Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic. Res. 2019, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Giusti, M.M. Anthocyanins: Natural Colorants with Health-Promoting Properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef]
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Otegui, M.S.; Grotewold, E. Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal. Behav. 2015, 10, e1027850. [Google Scholar] [CrossRef] [Green Version]
- Offen, W.; Martinez-Fleites, C.; Yang, M.; Kiat-Lim, E.; Davis, B.G.; Tarling, C.A.; Ford, C.M.; Bowles, D.J.; Davies, G. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006, 25, 1396–1405. [Google Scholar] [CrossRef] [Green Version]
- Noda, N.; Yoshioka, S.; Kishimoto, S.; Nakayama, M.; Douzono, M.; Tanaka, Y.; Aida, R. Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci. Adv. 2017, 3, e1602785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Nishiyama, Y.; Fuell, C.; Taguchi, G.; Elliott, K.; Hill, L.; Tanaka, Y.; Kitayama, M.; Yamazaki, M.; Bailey, P.; et al. Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 2007, 50, 678–695. [Google Scholar] [CrossRef]
- Noel, J.P.; Dixon, R.A.; Pichersky, E.; Zubieta, C.; Ferrer, J.-L. Chapter two Structural, functional, and evolutionary basis for methylation of plant small molecules. In The Chemistry and Biochemistry of Plant Hormones—Recent Advances in Phytochemistry; Elsevier BV: Amsterdam, The Netherlands, 2003; Volume 37, pp. 37–58. [Google Scholar]
- Lücker, J.; Martens, S.; Lund, S.T. Characterization of a Vitis vinifera cv. Cabernet Sauvignon 3′,5′-O-methyltransferase showing strong preference for anthocyanins and glycosylated flavonols. Phytochemistry 2010, 71, 1474–1484. [Google Scholar] [CrossRef] [PubMed]
- Hugueney, P.; Provenzano, S.; Verriès, C.; Ferrandino, A.; Meudec, E.; Batelli, G.; Merdinoglu, D.; Cheynier, V.; Schubert, A.; Ageorges, A. A Novel Cation-Dependent O-Methyltransferase Involved in Anthocyanin Methylation in Grapevine. Plant Physiol. 2009, 150, 2057–2070. [Google Scholar] [CrossRef] [PubMed]
- Roldan, M.V.G.; Outchkourov, N.; Van Houwelingen, A.; Lammers, M.; De La Fuente, I.R.; Ziklo, N.; Aharoni, A.; Hall, R.D.; Beekwilder, J. AnO-methyltransferase modifies accumulation of methylated anthocyanins in seedlings of tomato. Plant J. 2014, 80, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Barathikannan, K.; Venkatadri, B.; Khusro, A.; Al-Dhabi, N.A.; Agastian, P.; Arasu, M.V.; Choi, H.S.; Kim, Y.O. Chemical analysis of Punica granatum fruit peel and its in vitro and in vivo biological properties. BMC Complement. Altern. Med. 2016, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, Y.; Ren, Y.; Wang, Y.; Yuan, Z. Fruit Breeding in Regard to Color and Seed Hardness: A Genomic View from Pomegranate. Agronomy 2020, 10, 991. [Google Scholar] [CrossRef]
- Costa, A.M.; Moretti, L.K.; Simões, G.; Silva, K.A.; Calado, V.; Tonon, R.V.; Torres, A.G. Microencapsulation of pomegranate (Punica granatum L.) seed oil by complex coacervation: Development of a potential functional ingredient for food application. LWT 2020, 131, 109519. [Google Scholar] [CrossRef]
- De Vries, J.; Ischebeck, T. Ties between Stress and Lipid Droplets Pre-date Seeds. Trends Plant Sci. 2020, 25, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Kalaycıoğlu, Z.; Erim, F.B. Total phenolic contents, antioxidant activities, and bioactive ingredients of juices from pomegranate cultivars worldwide. Food Chem. 2017, 221, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Bowen-Forbes, C.S.; Zhang, Y.; Nair, M.G. Anthocyanin content, antioxidant, anti-inflammatory and anticancer properties of blackberry and raspberry fruits. J. Food Compos. Anal. 2010, 23, 554–560. [Google Scholar] [CrossRef]
- Albert, N.W.; Thrimawithana, A.H.; McGhie, T.K.; Clayton, W.A.; Deroles, S.C.; Schwinn, K.E.; Bowman, J.L.; Jordan, B.R.; Davies, K.M. Genetic analysis of the liverwort Marchantia polymorpha reveals that R2R3MYB activation of flavonoid production in response to abiotic stress is an ancient character in land plants. New Phytol. 2018, 218, 554–566. [Google Scholar] [CrossRef] [Green Version]
- Fürst-Jansen, J.M.R.; De Vries, S.; De Vries, J. Evo-physio: On stress responses and the earliest land plants. J. Exp. Bot. 2020, 71, 3254–3269. [Google Scholar] [CrossRef] [Green Version]
- De Vries, J.; De Vries, S.; Slamovits, C.H.; Rose, L.E.; Archibald, J.M. How Embryophytic is the Biosynthesis of Phenylpropanoids and their Derivatives in Streptophyte Algae? Plant Cell Physiol. 2017, 58, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef] [Green Version]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. Smart, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [Green Version]
- Kazutaka, K.; Standley, D.M. Mafft Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772. [Google Scholar]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, C.; Huang, X.; Zhang, H.; Yuan, Z. Land-plant Phylogenomic and Pomegranate Transcriptomic Analyses Reveal an Evolutionary Scenario of CYP75 Genes Subsequent to Whole Genome Duplications. J. Plant Biol. 2019, 62, 48–60. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Liu, X.; Zhang, Z.; Sang, M.; Sun, X.; He, C.; Xin, P.; Zhang, H. Functional Analysis of the FZF1 Genes of Saccharomyces uvarum. Front. Microbiol. 2018, 9, 96. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Panu, A.; Manohar, J.; Konstantin, A.; Delphine, B.; Gabor, C.; Edouard, D.C.; Séverine, D.; Volker, F.; Arnaud, F.; Elisabeth, G. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar]
- Lee, H.-L.; Irish, V.F. Gene Duplication and Loss in a MADS Box Gene Transcription Factor Circuit. Mol. Biol. Evol. 2011, 28, 3367–3380. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Leebens-Mack, J.; Ayyampalayam, S.; Bowers, J.E.; McKain, M.R.; McNeal, J.; Rolf, M.; Ruzicka, D.R.; Wafula, E.; Wickett, N.J.; et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012, 13, R3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaltenegger, E.; Eich, E.; Ober, D. Evolution of Homospermidine Synthase in the Convolvulaceae: A Story of Gene Duplication, Gene Loss, and Periods of Various Selection Pressures. Plant Cell 2013, 25, 1213–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kondo, E.; Nakayama, M.; Kameari, N.; Tanikawa, N.; Morita, Y.; Akita, Y.; Hase, Y.; Tanaka, A.; Ishizaka, H. Red-purple flower due to delphinidin 3,5-diglucoside, a novel pigment for Cyclamen spp., generated by ion-beam irradiation. Plant Biotechnol. 2009, 26, 565–569. [Google Scholar] [CrossRef]
- Provenzano, S.; Spelt, C.; Hosokawa, S.; Nakamura, N.; Brugliera, F.; Demelis, L.; Geerke, D.P.; Schubert, A.; Tanaka, Y.; Quattrocchio, F.; et al. Genetic Control and Evolution of Anthocyanin Methylation. Plant Physiol. 2014, 165, 962–977. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.; Li, K.; Poudel, K.; Zhao, D.; et al. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft- and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef] [Green Version]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef] [Green Version]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Trainin, T.; Harel-Beja, R.; Bar-Ya’Akov, I.; Borochov-Neori, H.; Holland, D. A “White” Anthocyanin-less Pomegranate (Punica granatum L.) Caused by an Insertion in the Coding Region of the Leucoanthocyanidin Dioxygenase (LDOX; ANS) Gene. PLoS ONE 2015, 10, e0142777. [Google Scholar] [CrossRef]
- Carocha, V.; Soler, M.; Hefer, C.; Cassan-Wang, H.; Fevereiro, P.; Myburg, A.A.; Paiva, J.A.P.; Grima-Pettenati, J. Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytol. 2015, 206, 1297–1313. [Google Scholar] [CrossRef] [Green Version]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar]
- Xu, Q.; Chen, L.-L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.-B.; Hao, B.-H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, A.-M.; Tesch, D.; Nothwang, H.G.; Bininda-Emonds, O.R. Evolution of the Cation Chloride Cotransporter Family: Ancient Origins, Gene Losses, and Subfunctionalization through Duplication. Mol. Biol. Evol. 2013, 31, 434–447. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremblay-Savard, O.; Bertrand, D.; El-Mabrouk, N. Evolution of orthologous tandemly arrayed gene clusters. BMC Bioinform. 2011, 12, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, C.; Lehti-Shiu, M.D.; Thibaud-Nissen, F.; Prakash, T.; Buell, C.R.; Shiu, S.-H. Evolutionary and Expression Signatures of Pseudogenes in Arabidopsis and Rice. Plant Physiol. 2009, 151, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Nei, M.; Rooney, A.P. Concerted and Birth-and-Death Evolution of Multigene Families*. Annu. Rev. Genet. 2005, 39, 121–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Wang, M. Ultraviolet A-specific induction of anthocyanin biosynthesis and PAL expression in tomato (Solanum lycopersicum L.). Plant Growth Regul. 2010, 62, 1–8. [Google Scholar] [CrossRef]
- Yang, R.-Z.; Wei, X.-L.; Gao, F.-F.; Wang, L.-S.; Zhang, H.-J.; Xu, Y.-J.; Li, C.-H.; Ge, Y.-X.; Zhang, J.-J.; Zhang, J. Simultaneous analysis of anthocyanins and flavonols in petals of lotus (Nelumbo) cultivars by high-performance liquid chromatography-photodiode array detection/electrospray ionization mass spectrometry. J. Chromatogr. A 2009, 1216, 106–112. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, Z.; Yin, Y.; Feng, L. Patterns of Pigment Changes in Pomegranate (Punica granatum L.) Peel During Fruit Ripening. Acta Hortic. 2015, 1089, 83–89. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Yuan, Z.; Zhao, X.; Yin, Y.; Feng, L. Composition and Contents of Anthocyanins in Different Pomegranate Cultivars. Acta Hortic. 2015, 35–41. [Google Scholar] [CrossRef]


| Species | Gene ID | Gene Name | Protein Length/aa | Molecular Weight/ku | PI | Instability Index | GRAVY |
|---|---|---|---|---|---|---|---|
| Pomegranate | Pg002344.1 | PgOMT01 | 250 | 27,320.50 | 5.45 | 46.31 | −0.088 |
| Pg002346.1 | PgOMT02 | 250 | 27,519.67 | 5.05 | 45.46 | −0.052 | |
| Pg002348.1 | PgOMT03 | 239 | 26,378.53 | 5.35 | 31.20 | −0.079 | |
| Pg002351.1 | PgOMT04 | 242 | 27,172.27 | 5.44 | 36.49 | −0.160 | |
| Pg002849.1 | PgOMT05 | 183 | 20,421.47 | 6.21 | 56.67 | −0.152 | |
| Pg003086.1 | PgOMT06 | 109 | 11,927.73 | 5.89 | 23.63 | −0.110 | |
| Pg006183.1 | PgOMT07 | 286 | 31,785.38 | 4.79 | 39.20 | 0.009 | |
| Pg017894.1 | PgOMT08 | 93 | 10,153.52 | 5.18 | 45.87 | −0.200 | |
| Pg021629.1 | PgOMT09 | 258 | 28,709.93 | 5.72 | 37.09 | −0.246 | |
| Pg026019.1 | PgOMT10 | 414 | 45,306.63 | 9.26 | 34.14 | 0.093 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Yuan, W.; Zhao, Y.; Ren, Y.; Zhao, X.; Yuan, Z. Genome-Wide Identification and Evolutionary Analysis of AOMT Gene Family in Pomegranate (Punica granatum). Agronomy 2021, 11, 318. https://doi.org/10.3390/agronomy11020318
Zhang X, Yuan W, Zhao Y, Ren Y, Zhao X, Yuan Z. Genome-Wide Identification and Evolutionary Analysis of AOMT Gene Family in Pomegranate (Punica granatum). Agronomy. 2021; 11(2):318. https://doi.org/10.3390/agronomy11020318
Chicago/Turabian StyleZhang, Xinhui, Weicheng Yuan, Yujie Zhao, Yuan Ren, Xueqing Zhao, and Zhaohe Yuan. 2021. "Genome-Wide Identification and Evolutionary Analysis of AOMT Gene Family in Pomegranate (Punica granatum)" Agronomy 11, no. 2: 318. https://doi.org/10.3390/agronomy11020318
APA StyleZhang, X., Yuan, W., Zhao, Y., Ren, Y., Zhao, X., & Yuan, Z. (2021). Genome-Wide Identification and Evolutionary Analysis of AOMT Gene Family in Pomegranate (Punica granatum). Agronomy, 11(2), 318. https://doi.org/10.3390/agronomy11020318







