Molecular Characterization of Genetic Diversity in Apricot Cultivars: Current Situation and Future Perspectives
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Amplification
2.3. Data Analysis
3. Results and discussion
3.1. Microsatellite Polymorphism and Genetic Diversity
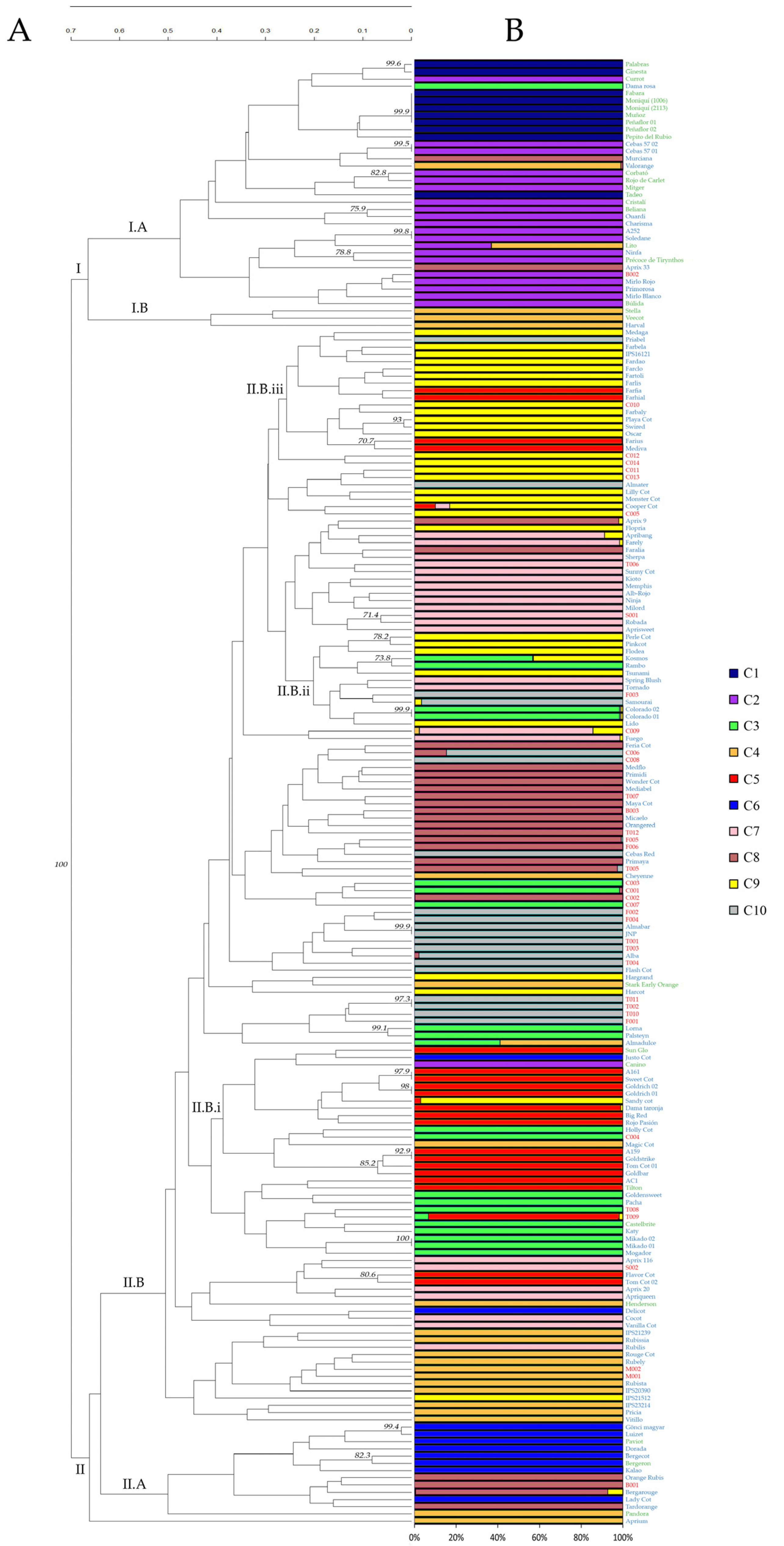
3.2. Identification of the Different Genotypes
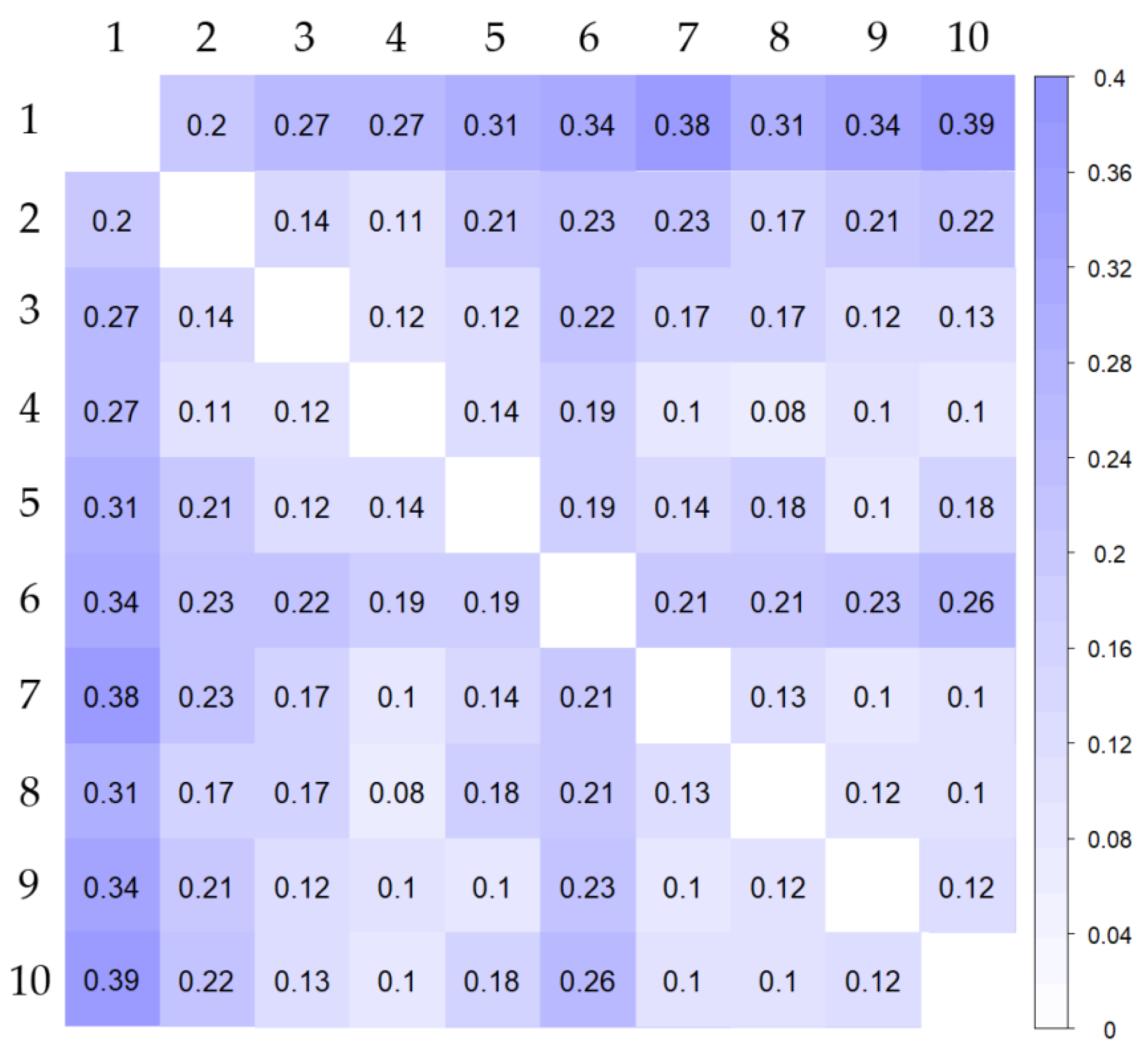
3.3. Genetic Relationships and Similarities among Genotypes
3.4. Population Structure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faust, M.; Surányi, D.; Nyujtó, F. Origin and dissemination of Apricot. Hortic. Rev. 1998, 22, 225–266. [Google Scholar] [CrossRef]
- Janick, J. The origins of fruits, fruit growing, and fruit breeding. Plant Breed. Rev. 2005, 25, 255–320. [Google Scholar] [CrossRef]
- Bourguiba, H.; Scotti, I.; Sauvage, C.; Zhebentyayeva, T.; Ledbetter, C.; Krška, B.; Remay, A.; D’Onofrio, C.; Iketani, H.; Christen, D.; et al. Genetic structure of a worldwide germplasm collection of Prunus armeniaca L. reveals three major diffusion routes for varieties coming from the species’ center of origin. Front. Plant Sci. 2020, 11, 638. [Google Scholar] [CrossRef]
- Bourguiba, H.; Audergon, J.M.; Krichen, L.; Trifi-Farah, N.; Mamouni, A.; Trabelsi, S.; D’Onofrio, C.; Asma, B.M.; Santoni, S.; Khadari, B. Loss of genetic diversity as a signature of apricot domestication and diffusion into the Mediterranean Basin. BMC Plant Biol. 2012, 12, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groppi, A.; Liu, S.; Cornille, A.; Decroocq, S.; Bui, Q.T.; Tricon, D.; Cruaud, C.; Arribat, S.; Belser, C.; Marande, W.; et al. Population genomics of apricots unravels domestication history and adaptive events. Nat. Commun. 2021, 12, 3956. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Available online: http://www.fao.org/faostat/es/ (accessed on 26 July 2021).
- Layne, R.E.C.; Bailey, C.; Hough, L.F. Apricots. In Fruit Breeding, Volume I: Tree and Tropical Fruits; Janick, J., Moore, J.N., Eds.; JohnWiley & Sons, Inc.: New York, NY, USA, 1996; pp. 79–111. [Google Scholar]
- Herrera, S.; Lora, J.; Hormaza, J.I.; Rodrigo, J. Pollination management in stone fruit crops. In Production Technology of Stone Fruits; Mir, M.M., Iqbal, M., Mir, S.A., Eds.; Springer: Singapore, 2021; pp. 75–102. [Google Scholar] [CrossRef]
- Fadón, E.; Herrera, S.; Guerrero, B.I.; Guerra, M.E.; Rodrigo, J. Chilling and heat requirements of temperate stone fruit trees (Prunus sp.). Agronomy 2020, 10, 409. [Google Scholar] [CrossRef] [Green Version]
- Mehlenbacher, S.A.; Cociu, V.; Hough, L.F. Apricot (Prunus). Acta Hortic. 1991, 290, 65–110. [Google Scholar] [CrossRef]
- Hormaza, J.I.; Yamane, H.; Rodrigo, J. Apricot. In Fruits and Nuts. Genome Mapping and Molecular Breeding in Plants; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4, pp. 171–187. [Google Scholar] [CrossRef]
- Herrera, S.; Rodrigo, J.; Hormaza, J.I.; Lora, J. Identification of self-incompatibility alleles by specific PCR analysis and S-RNase sequencing in Apricot. Int. J. Mol. Sci. 2018, 19, 3612. [Google Scholar] [CrossRef] [Green Version]
- Herrera, S.; Lora, J.; Hormaza, J.I.; Herrero, M.; Rodrigo, J. Optimizing production in the new generation of apricot cultivars: Self-incompatibility, S-RNase allele identification, and incompatibility group assignment. Front. Plant Sci. 2018, 9, 527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhebentyayeva, T.; Ledbetter, C.; Burgos, L.; Llácer, G. Apricot. In Fruit Breeding; Badenes, M.L., Byrne, D.H., Eds.; Springer: Boston, MA, USA, 2012; ISBN 978-1-4419-0762-2. [Google Scholar]
- Martín, C.; Hormaza, J.I.; Herrero, M. Molecular Characterization of apricot germplasm from an old stone collection. PLoS ONE 2011, 6, e23979. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, E.J.; Pádua, J.G.; Zucchi, M.I.; Vencovsky, R.; Vieira, M.L.C. Origin, evolution and genome distribution of microsatellites. Genet. Mol. Biol. 2006, 29, 294–307. [Google Scholar] [CrossRef]
- Vieira, M.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Badenes, M.L.; Fernandez i Marti, A.; Ríos, G.; Rubio-Cabetas, M.J. Application of genomic technologies to the breeding of trees. Front. Genet 2016, 7, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maghuly, F.; Fernandez, E.B.; Ruthner, S.; Pedryc, A.; Laimer, M. Microsatellite variability in apricots (Prunus armeniaca L.) reflects their geographic origin and breeding history. Tree Genet. Genomes 2005, 1, 151–165. [Google Scholar] [CrossRef]
- Decroocq, S.; Cornille, A.; Tricon, D.; Babayeva, S.; Chague, A.; Eyquard, J.P.; Karychev, R.; Dolgikh, S.; Kostritsyna, T.; Liu, S.; et al. New insights into the history of domesticated and wild apricots and its contribution to Plum pox virus resistance. Mol. Ecol. 2016, 25, 4712–4729. [Google Scholar] [CrossRef]
- Liu, S.; Cornille, A.; Decroocq, S.; Tricon, D.; Chague, A.; Eyquard, J.P.; Liu, W.S.; Giraud, T.; Decroocq, V. The complex evolutionary history of apricots: Species divergence, gene flow and multiple domestication events. Mol. Ecol. 2019, 28, 5299–5314. [Google Scholar] [CrossRef]
- Hormaza, J.I. Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theor. Appl. Genet. 2002, 104, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Mora, C.; Rodríguez, J.; Cenis, J.L. Genetic variability among local apricots (Prunus armeniaca L.) from the Southeast of Spain. Span. J. Agric. Res. 2009, 7, 855–868. [Google Scholar] [CrossRef] [Green Version]
- Murathan, Z.T.; Kafkas, S.; Asma, B.M.; Topçu, H. S_allele identification and genetic diversity analysis of apricot cultivars. J. Hortic. Sci. Biotechnol. 2017, 92, 251–260. [Google Scholar] [CrossRef]
- Bourguiba, H.; Khadari, B.; Krichen, L.; Trifi-Farah, N.; Mamouni, A.; Trabelsi, S.; Audergon, J.M. Genetic relationships between local North African apricot (Prunus armeniaca L.) germplasm and recently introduced varieties. Sci. Hortic. 2013, 152, 61–69. [Google Scholar] [CrossRef]
- Batnini, M.A.; Krichen, L.; Bourguiba, H.; Trifi-Farah, N.; Ruiz, D.; Martínez-Gómez, P.; Rubio, M. Comparative analysis of traditional and modern apricot breeding programs: A case of study with Spanish and Tunisian apricot breeding germplasm. Span. J. Agric. Res. 2016, 14, e0706. [Google Scholar] [CrossRef] [Green Version]
- Raji, R.; Jannatizadeh, A.; Fattahi, R.; Esfahlani, M.A. Investigation of variability of apricot (Prunus armeniaca L.) using morphological traits and microsatellite markers. Sci. Hortic. 2014, 176, 225–231. [Google Scholar] [CrossRef]
- CPVO. Community Plant Variety Office. Available online: https://cpvo.europa.eu/en (accessed on 26 July 2021).
- Pérez, V.; Larrañaga, N.; Abdallah, D.; Wünsch, A.; Hormaza, J.I. Genetic diversity of local peach (Prunus persica) accessions from La Palma island (Canary Islands, Spain). Agronomy 2020, 10, 457. [Google Scholar] [CrossRef] [Green Version]
- Herrera, S.; Lora, J.; Hormaza, J.I.; Rodrigo, J. Determination of Self- and Inter-(in)compatibility Relationships in Apricot Combining Hand-Pollination, Microscopy and Genetic Analyses. J. Vis. Exp. 2020, 160, e60241. [Google Scholar] [CrossRef] [PubMed]
- Sosinski, B.; Gannavarapu, M.; Hager, L.D.; Beck, L.E.; King, G.J.; Ryder, C.D.; Rajapakse, S.; Baird, W.V.; Ballard, R.E.; Abbott, A.G. Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch]. Theor. Appl. Genet. 2000, 101, 421–428. [Google Scholar] [CrossRef]
- Cipriani, G.; Lot, G.; Huang, W.G.; Marrazzo, M.T.; Peterlunger, E.; Testolin, R. AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: Isolation, characterisation and cross-species amplification in Prunus. Theor. Appl. Genet. 1999, 99, 65–72. [Google Scholar] [CrossRef]
- Lopes, M.S.; Sefc, K.M.; Laimer, M.; Machado, A.D.C. Characterization of microsatellite loci in Apricot. Mol. Ecol. Notes 2002, 2, 24–26. [Google Scholar] [CrossRef]
- Messina, R.; Lain, O.; Marrazzo, M.T.; Cipriani, G.; Testolin, R. New set of microsatellite loci isolated in apricot. Mol. Ecol. Notes 2004, 4, 432–434. [Google Scholar] [CrossRef]
- Testolin, R.; Marrazzo, T.; Cipriani, G.; Quarta, R.; Verde, I.; Dettori, M.T.; Pancaldi, M.; Sansavini, S. Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 2000, 43, 512–520. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 26 July 2021).
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [Green Version]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E. Pegas: An R package for population genetics with an integrated–modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamack, A.T.; Gruber, B. PopGenReport: Simplifying basic population genetic analyses in R. Methods Ecol. Evol. 2014, 5, 384–387. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V. R Package corrplot: Visualization of a Correlation Matrix (Version 0.90). 2021. Available online: https://github.com/taiyun/corrplot (accessed on 26 July 2021).
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dray, S.; Dufour, A.-B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; Available online: https://ggplot2.tidyverse.org (accessed on 26 July 2021).
- Khadivi-Khub, A.; Yarahmadi, M.; Jannatizadeh, A.; Ebrahimi, A. Genetic relationships and diversity of common apricot (Prunus armeniaca L.) based on simple sequence repeat (SSR) markers. Genet. Resour. Crop Evol. 2013, 61, 357–368. [Google Scholar] [CrossRef]
- Tian-Ming, H.; Xue-Sen, C.; Zheng, X.; Jiang-Sheng, G.; Pei-Jun, L.; Wen, L.; Qing, L.; Yan, W. Using SSR markers to determine the population genetic structure of wild apricot (Prunus armeniaca L.) in the Ily Valley of West China. Genet. Resour. Crop Evol. 2007, 54, 563–572. [Google Scholar] [CrossRef]
- Gatti, E.; Defilippi, B.G.; Predieri, S.; Infante, R. Apricot (Prunus armeniaca L.) quality and breeding perspectives. J. Food Agric. Environ. 2009, 7, 573–580. [Google Scholar]
- Krška, B. Genetic Apricot resources and their utilisation in breeding. In Breeding and Health Benefits of Fruit and Nut Crops; Soneji, J.R., Nageswara-Rao, M., Eds.; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Romero, C.; Pedryc, A.; Muñoz, V.; Llácer, G.; Badenes, M.L. Genetic diversity of different apricot geographical groups determined by SSR markers. Genome 2003, 46, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Zhebentyayeva, T.N.; Reighard, G.L.; Gorina, V.M.; Abbott, A.G. Simple sequence repeat (SSR) analysis for assessment of genetic variability in apricot germplasm. Theor. Appl. Genet. 2003, 106, 435–444. [Google Scholar] [CrossRef]
- Ruthner, S.; Pedryc, A.; Kriska, B.; Romero, C.; Badenes, M.L. Molecular characterization of apricot (Prunus armeniaca L.) cultivars using cross species SSR amplification with peach primers. Int. J. Hortic. Sci. 2006, 12, 53–57. [Google Scholar] [CrossRef]
- Akpinar, A.E.; Koçal, H.; Ergül, A.; Kazan, K.; Şelli, M.E.; Bakir, M.; Aslantaş, Ş.; Kaymak, S.; Saribaş, R. SSR-based molecular analysis of economically important Turkish apricot cultivars. Genet. Mol. Res. 2010, 9, 324–332. [Google Scholar] [CrossRef]
- Gürcan, K.; Önal, N.; Yilmaz, K.U.; Ullah, S.; Erdoğan, A.; Zengin, Y. Evaluation of Turkish apricot germplasm using SSR markers: Genetic diversity assessment and search for Plum pox virus resistance alleles. Sci. Hortic. 2015, 193, 155–164. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Errea, P.; Miranda, C.; Santesteban, L.G.; Pina, A. Genetic diversity of Spanish Prunus domestica L. germplasm reveals a complex genetic structure underlying. PLoS ONE 2018, 13, e0195591. [Google Scholar] [CrossRef] [Green Version]
- Halász, J.; Kodad, O.; Galiba, G.M.; Skola, I.; Ercisli, S.; Ledbetter, C.A.; Hegedűs, A. Genetic variability is preserved among strongly differentiated and geographically diverse almond germplasm: An assessment by simple sequence repeat markers. Tree Genet. Genomes 2019, 15, 12. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Pérez, R.; Ruiz, D.; Dicenta, F.; Egea, J.; Martínez-Gómez, P. Application of simple sequence repeat (SSR) markers in apricot breeding: Molecular characterization, protection, and genetic relationships. Sci. Hortic. 2005, 103, 305–315. [Google Scholar] [CrossRef]
- Pedryc, A.; Ruthner, S.; Hermán, R.; Krska, B.; Hegedus, A.; Halász, J. Genetic diversity of apricot revealed by a set of SSR markers from linkage group G1. Sci. Hortic. 2009, 121, 19–26. [Google Scholar] [CrossRef]
- Milatović, D.; Nikolić, D.; Krška, B. Testing of self-(in)compatibility in apricot cultivars from European breeding programmes. Hortic. Sci. 2013, 40, 65–71. [Google Scholar] [CrossRef] [Green Version]


| Accession | Country | Accession | Country | Accession | Country | Accession | Country |
|---|---|---|---|---|---|---|---|
| Beliana | France a | F003 | Spain b,9 | Faralia | France c,18 | Mikado 01 | Spain c,23 |
| Bergeron | France a | F004 | Spain b,9 | Farbaly | France c,18 | Mikado 02 | Spain c,28 |
| Búlida | Spain a | F005 | Spain b,9 | Farbela | France c,18 | Milord | Spain c,23 |
| Canino | Spain a | F006 | Spain b,9 | Farclo | France c,18 | Mirlo Blanco | Spain c,4 |
| Castelbrite | Italy a | M001 | France b,21 | Fardao | France c,18 | Mirlo Rojo | Spain c,4 |
| Corbató | Spain a | M002 | France b,21 | Farely | France c,18 | Mogador | Spain c,23 |
| Cristalí | Spain a | T001 | Spain b,28 | Farfia | France c,18 | Monster Cot | USA c,27 |
| Currot | Spain a | T002 | Spain b,28 | Farhial | France c,18 | Murciana | Spain c,4 |
| Fabara | Spain a | T003 | Spain b,28 | Farius | France c,18 | Ninfa | Italy c,6 |
| Ginesta | Spain a | T004 | Spain b,28 | Farlis | France c,18 | Ninja | France c,7 |
| Henderson | USA a | T005 | Spain b,28 | Fartoli | France c,18 | Orange Rubis | France c,8 |
| Lito | Greece a | T006 | Spain b,28 | Feria Cot | France c,5 | Orangered | USA c,20 |
| Mitger | Spain a | T007 | Spain b,28 | Flash Cot | USA c,27 | Oscar | Spain c,23 |
| Moniquí (1006) | Spain a | T008 | Spain b,28 | Flavor Cot | USA c,31 | Ouardi | Tunisia c,15 |
| Moniquí (2113) | Spain a | T009 | Spain b,28 | Flodea | Spain c,23 | Pacha | Spain c,23 |
| Muñoz | Spain a | T010 | Spain b,28 | Flopria | Spain c,23 | Palsteyn | South Africa c,2 |
| Palabras | Spain a | T011 | Spain b,28 | Fuego | Spain c,23 | Perle Cot | USA c,26 |
| Pandora | Greece a | T012 | Spain b,28 | Goldbar | USA c,31 | Pinkcot | France c,7 |
| Paviot | France a | A159 | Unknown c,33 | Goldensweet | USA c,17 | Playa Cot | France c,5 |
| Peñaflor 01 | Spain a | A161 | Unknown c,33 | Goldrich 01 | USA c,29 | Priabel | France c,18 |
| Peñaflor 02 | Spain a | A252 | Unknown c,33 | Goldrich 02 | USA c,29 | Pricia | France c,18 |
| Pepito del Rubio | Spain a | AC1 | USA c,26 | Goldstrike | USA c,31 | Primaya | France c,18 |
| P. de Tirynthos | Greece a | Alba | Spain c,33 | Gönci magyar | Hungary c,3 | Primidi | France c,21 |
| Rojo de Carlet | Spain a | Alb-Rojo | Spain c,28 | Harcot | Canada c,12 | Primorosa | Spain c,4 |
| Stark E. Orange | USA a | Almabar | Spain c,9 | Hargrand | Canada c,12 | Rambo | Spain c,23 |
| Stella | USA a | Almadulce | Spain c,9 | Harval | Canada c,12 | Robada | USA c,30 |
| Sun Glo | USA a | Almater | Spain c,9 | Holly Cot | France c,5 | Rojo Pasión | Spain c,4 |
| Tadeo | Spain a | Apribang | France c,1 | IPS16121 | France c,18 | Rouge Cot | France c,5 |
| Tilton | USA a | Apriqueen | France c,1 | IPS20390 | France c,21 | Rubely | France c,21 |
| Veecot | Canada a | Aprisweet | France c,1 | IPS21239 | France c,21 | Rubilis | France c,21 |
| S001 | France b,1 | Aprium | Spain c,28 | IPS21512 | France c,21 | Rubissia | France c,21 |
| S002 | France b,1 | Aprix 116 | Spain c,24 | IPS23214 | France c,21 | Rubista | France c,18 |
| C001 | France b,5 | Aprix 20 | Spain c,24 | JNP | Unknown c,33 | Samourai | France c,7 |
| C002 | France b,5 | Aprix 33 | Spain c,24 | Justo Cot | France c,5 | Sandy cot | France c,5 |
| C003 | France b,5 | Aprix 9 | Spain c,24 | Kalao | Spain c,23 | Sherpa | Spain c,23 |
| C004 | France b,5 | Bergarouge | France c,13 | Katy | USA c,32 | Soledane | France c,14 |
| C005 | France b,5 | Bergecot | France c,22 | Kioto | France c,7 | Spring Blush | France c,7 |
| C006 | France b,5 | Big Red | France c,7 | Kosmos | Spain c,23 | Sunny Cot | USA c,26 |
| C007 | France b,5 | Cebas 57 01 | Spain c,4 | Lady Cot | France c,19 | Sweet Cot | USA c,31 |
| C008 | France b,5 | Cebas 57 02 | Spain c,4 | Lido | Spain c,23 | Swired | Switzerland c,25 |
| C009 | France b,5 | Cebas Red | Spain c,4 | Lilly Cot | USA c,26 | Tardorange | Spain c,4 |
| C010 | France b,5 | Charisma | South Africa c,2 | Lorna | USA c,30 | Tom Cot 01 | USA c,31 |
| C011 | France b,5 | Cheyenne | Spain c,23 | Luizet | France c,11 | Tom Cot 02 | USA c,31 |
| C012 | France b,5 | Cocot | France c,5 | Magic Cot | USA c,26 | Tornado | France c,7 |
| C013 | France b,5 | Colorado 01 | Spain c,23 | Maya Cot | France c,5 | Tsunami | France c,7 |
| C014 | France b,5 | Colorado 02 | Spain c,23 | Medaga | France c,18 | Valorange | Spain c,4 |
| B001 | Spain b,4 | Cooper Cot | France c,5 | Medflo | France c,21 | Vanilla Cot | USA c,26 |
| B002 | Spain b,4 | Dama rosa | Spain c,16 | Mediabel | France c,18 | Vitillo | Italy c,10 |
| B003 | Spain b,4 | Dama taronja | Spain c,16 | Mediva | France c,18 | Wonder Cot | USA c,26 |
| F001 | Spain b,9 | Delicot | France c,19 | Memphis | Spain c,23 | ||
| F002 | Spain b,9 | Dorada | Spain c,4 | Micaelo | Spain c,4 |
| Locus | Sequence (5′→ 3′) | Linkage Group | SSR Motive | Predicted Length (bp) | Ta (°C) | Species |
|---|---|---|---|---|---|---|
| pchgms3 1 | ACGGTATGTCCGTACACTCTCCATG | 1 | (CT)19 | 179 | 57 | Peach |
| CAACCTGTGATTGCTCCTATTAAAC | ||||||
| UDP96-001 2 | AGTTTGATTTTCTGATGCATCC | Unknown | (CA)17 | 120 | 57 | Peach |
| TGCCATAAGGACCGGTATGT | ||||||
| ssrPaCITA7 3 | CTTTTGTGCCTCAGCTTCCCAACAC | 1 | (AG)22 | 211 | 51 | Apricot |
| CCTGGCCTGACCCTAAGCAATTCG | ||||||
| ssrPaCITA10 3 | GGTGAGGTCTGTGCTGAATATGCCA | 3 | (CT)26 | 175 | 47 | Apricot |
| CGATTAAAGAAATAAGAAAAAGAGC | ||||||
| ssrPaCITA12 3 | GAGACACCCCAACCACCCATCATGT | 6 | (TC)16 | 151 | 47 | Apricot |
| GGTGTTGGAAATGTGGAAAGAAATG | ||||||
| ssrPaCITA19 3 | GACAAATACAATCAAGAAGTGTCGC | 2 | (TC)16 | 114 | 51 | Apricot |
| GAACAGCTAGCCCCTTTGTCATAC | ||||||
| ssrPaCITA23 3 | GTGAATACAAAATTTTACTACATTG | 3 | (AC)2(AG)18 | 146 | 51 | Apricot |
| CGGTCTCTGACTCTCTGACTTGCGG | ||||||
| ssrPaCITA27 3 | GATCCCTCAACTGAATCTCTC | Unknown | (TC)8(TA)6(TG)17 | 262 | 47 | Apricot |
| CGTCACAACAATAGATGCGAAGG | ||||||
| UDAp-410 4 | TTGTTGACAAGAAGAAAACAAAGC | 1 | (AG)24 | 155 | 56 | Apricot |
| CAACGGGTTGGTTTCAGAAG | ||||||
| UDAp-415 4 | AACTGATGAGAAGGGGCTTG | 1 | (GA)21 | 156 | 56 | Apricot |
| ACTCCCGACATTTGTGCTTC | ||||||
| UDAp-420 4 | TTCCTTGCTTCCCTTCATTG | 6 | (CT)20 | 175 | 56 | Apricot |
| CCCAGAACTTGATTCTGACCA | ||||||
| UDP98-409 2 | GCTGATGGGTTTTATGGTTTTC | 8 | (AG)19 | 129 | 60 | Peach |
| CGGACTCTTATCCTCTATCAACA | ||||||
| UDP98-412 5 | AGGGAAAGTTTCTGCTGCAC | 6 | (AG)28 | 129 | 60 | Peach |
| GCTGAAGACGACGATGATGA |
| Group | Statistic | Na | Ar | Pa | Ho | He | FIS |
|---|---|---|---|---|---|---|---|
| Landraces | 6.50 | 6.20 | 1 | 0.68 | 0.73 | 0.08 | |
| Bred cultivars | 9.40 * | 6.97 | 30 | 0.77 | 0.74 | −0.04 | |
| t-test | ns | ns | ns | ns | |||
| p-value | 1.000 | 0.999 | 0.994 | 0.866 | |||
| Recent releases | 6.30 | 5.84 | 1 | 0.77 | 0.70 | −0.10 | |
| Commercial releases | 9.30 ** | 7.17 | 22 | 0.77 | 0.74 | −0.03 | |
| t-test | ns | ns | ns | ns | |||
| p-value | 1.000 | 1.000 | 0.447 | 0.989 |
| Source of Variation | df | Sum of Squares | Estimated Variance | % of Variation | p-Value |
|---|---|---|---|---|---|
| Among Source | 1 | 87.34 | 0.520 | 6.55 | 0.001 |
| Among Classification within Source | 1 | 18.21 | 0.018 | 0.22 | 0.241 |
| Among Breeding Program within Classification | 37 | 386.14 | 0.490 | 6.18 | 0.001 |
| Among Cultivars within Breeding Program | 162 | 1034.95 | −0.516 | −6.50 | 1.000 |
| Within Cultivars | 202 | 1498.83 | 7.420 | 93.55 | 0.214 |
| Total | 403 | 3025.47 | 7.931 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herrera, S.; Hormaza, J.I.; Lora, J.; Ylla, G.; Rodrigo, J. Molecular Characterization of Genetic Diversity in Apricot Cultivars: Current Situation and Future Perspectives. Agronomy 2021, 11, 1714. https://doi.org/10.3390/agronomy11091714
Herrera S, Hormaza JI, Lora J, Ylla G, Rodrigo J. Molecular Characterization of Genetic Diversity in Apricot Cultivars: Current Situation and Future Perspectives. Agronomy. 2021; 11(9):1714. https://doi.org/10.3390/agronomy11091714
Chicago/Turabian StyleHerrera, Sara, José I. Hormaza, Jorge Lora, Guillem Ylla, and Javier Rodrigo. 2021. "Molecular Characterization of Genetic Diversity in Apricot Cultivars: Current Situation and Future Perspectives" Agronomy 11, no. 9: 1714. https://doi.org/10.3390/agronomy11091714






