Genome-Wide Association Study for Abscission Failure of Fruit Pericarps (Stick-Tights) in Wild Macadamia Germplasm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Trial Design
2.2. Trait Assessment
2.3. Genotyping and Association Analysis
3. Results
3.1. Phenotypic Variation
3.2. Principal Components Analysis
3.3. Heritabilities
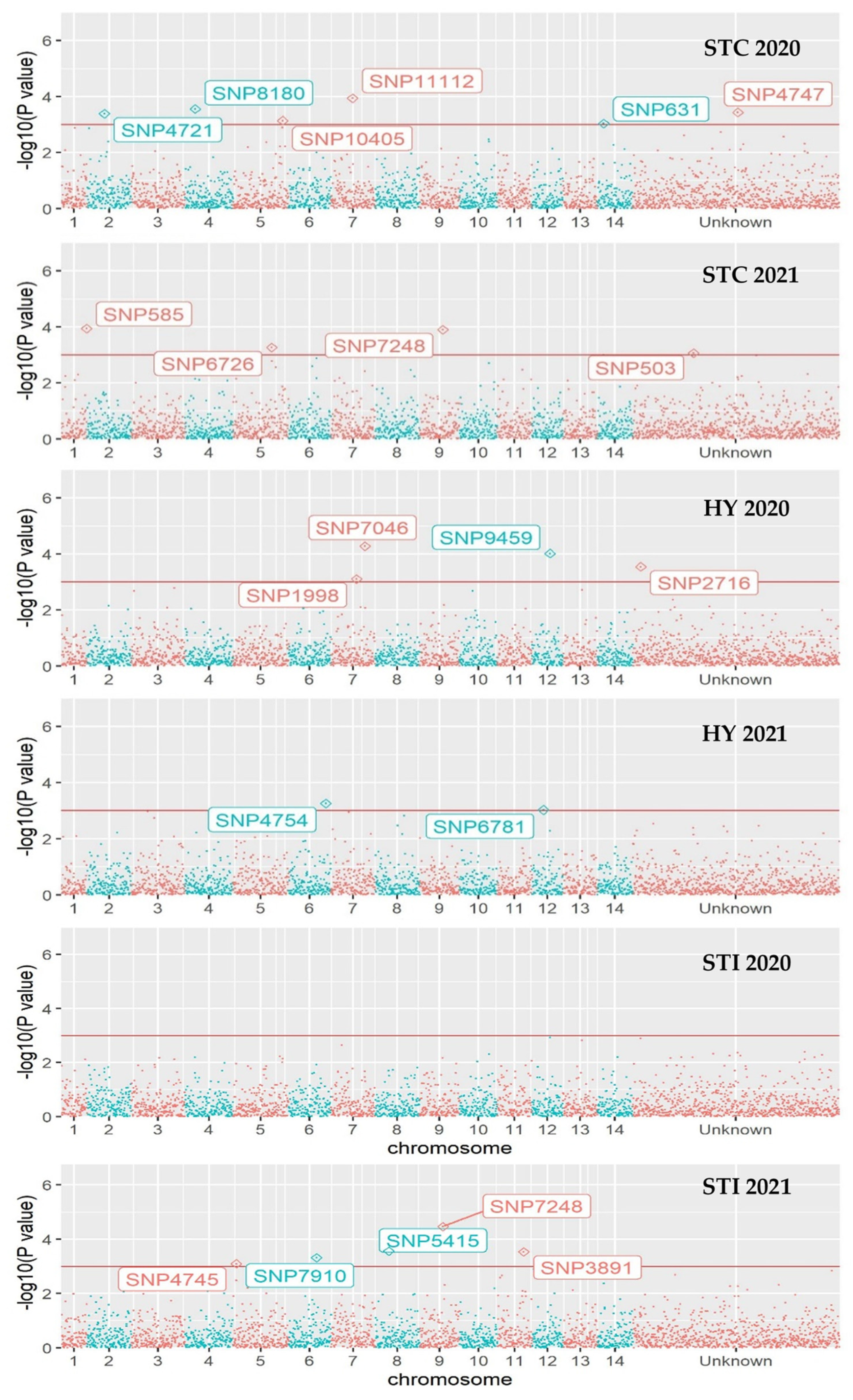
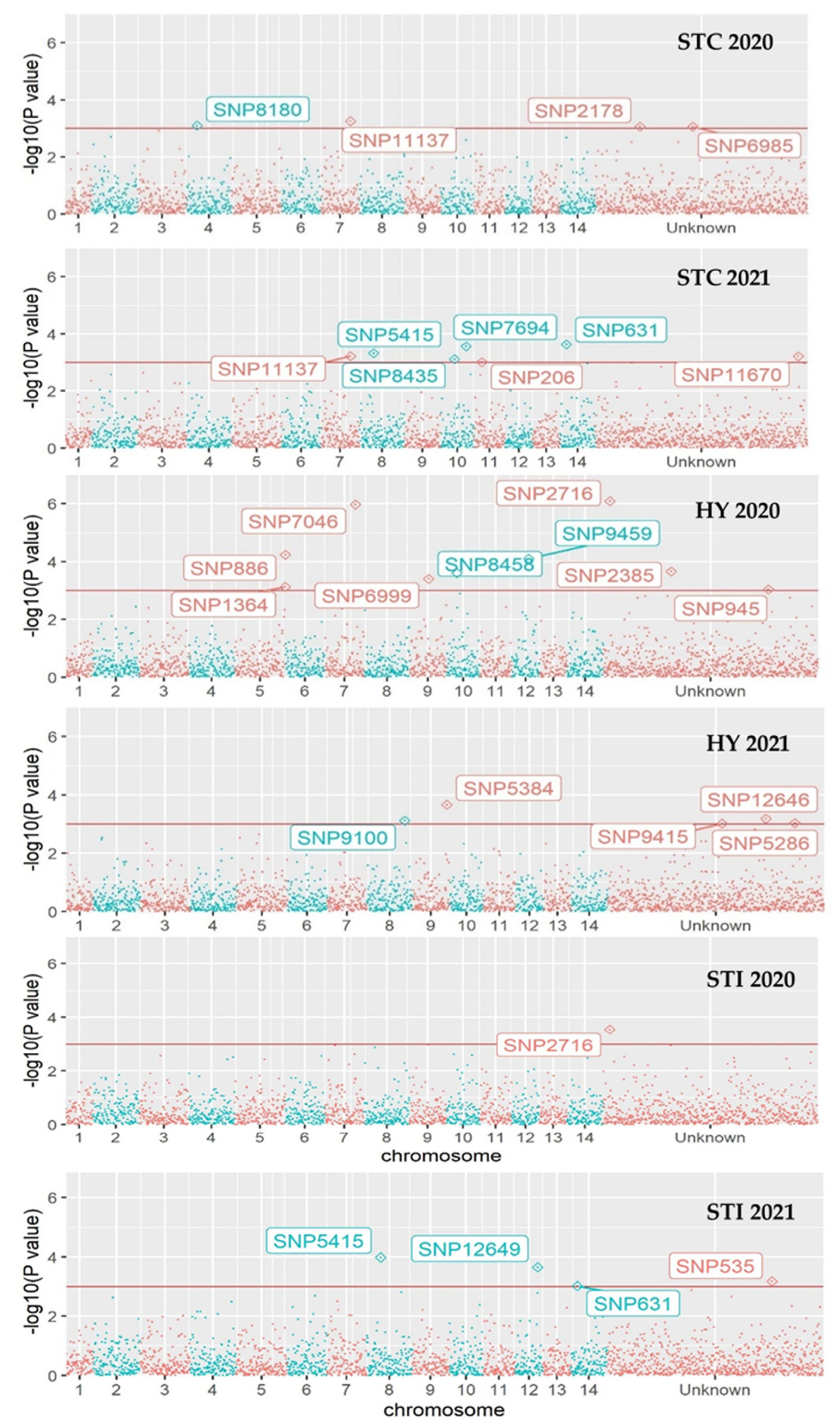
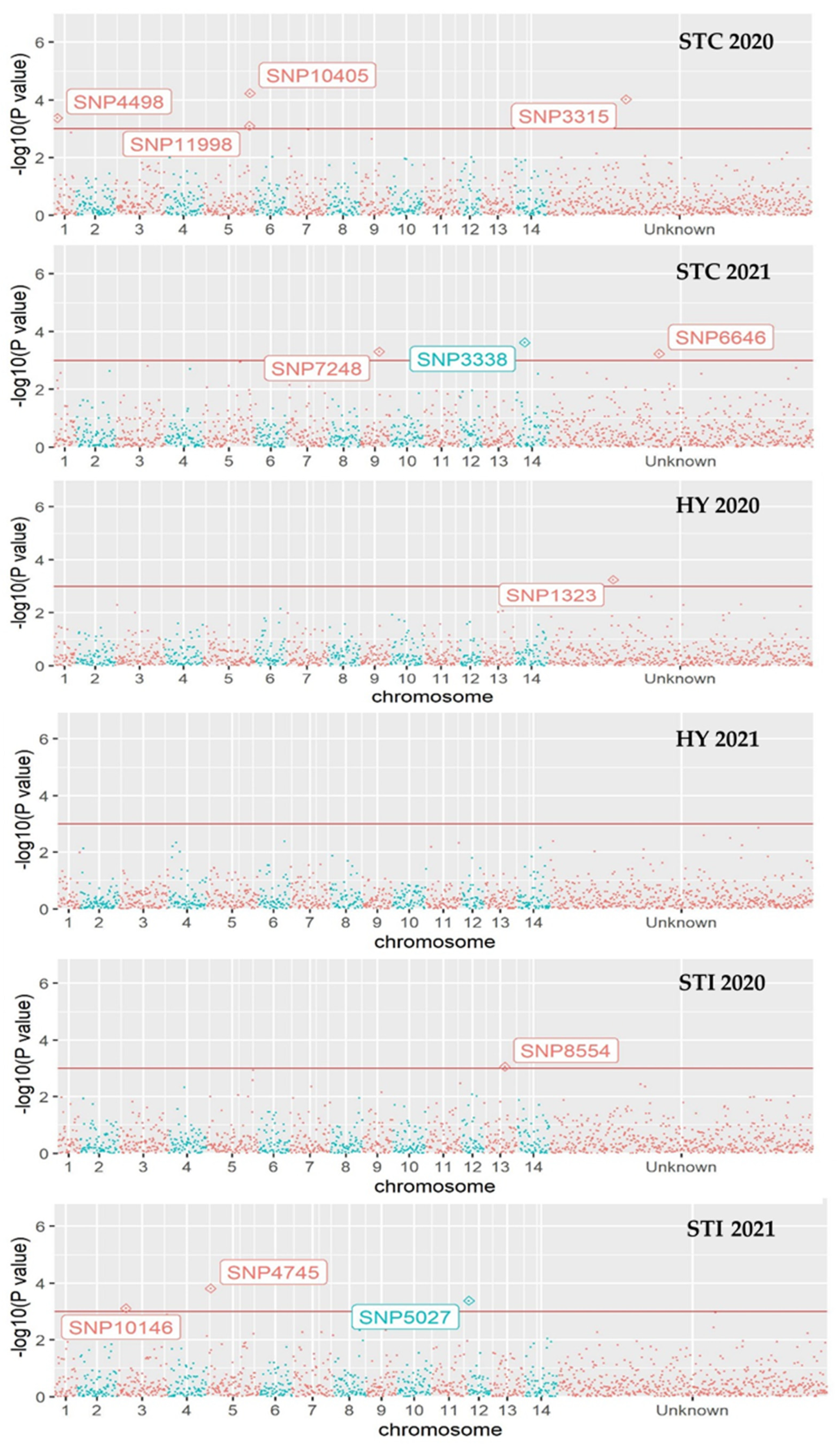
3.4. Genome-Wide Associations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- INC Foundation. Nuts & Dried Fruits Statistical Yearbook 2020/2021; INC: New York, NY, USA, 2021; pp. 1–80. [Google Scholar]
- Hardner, C.; Peace, C.; Lowe, A.; Neal, J.; Pisanu, P.; Powell, M.; Schmidt, A.; Spain, C.; Williams, K. Genetic Resources and Domestication of Macadamia. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2009; Volume 35, pp. 1–125. [Google Scholar]
- Topp, B.L.; Nock, C.J.; Hardner, C.M.; Alam, M.; O’Connor, K.M. Macadamia (Macadamia spp.) breeding. In Advances in Plant Breeding Strategies: Nut and Beverage Crops; Jain, J., Johnson, S.D., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 221–251. [Google Scholar]
- Peace, C. Genomics of Macadamia, a recently domesticated tree nut crop. In Genomics of Tropical Crop Plants; Moore, P.H., Ming, R., Eds.; Springer: New York, NY, USA, 2008; Volume 1, pp. 313–332. [Google Scholar]
- Dahler, J.; McConchie, C.; Turnbull, C. Quantification of cyanogenic glycosides in seedlings of three Macadamia (Proteaceae) species. Aust. J. Bot. 1995, 43, 619–628. [Google Scholar] [CrossRef]
- Gross, C.L. Macadamia. In Flora of Australia; McCarthy, P., Ed.; Australian Nature Conservation Agency: Melbourne, Australia, 1995; Volume 1, pp. 419–425. [Google Scholar]
- Boyton, S.; Hardner, C. Phenology of flowering and nut production in macadamia. In Proceedings of the International Symposium on Tropical and Subtropical Fruits, Cairns, Australia, 30 April 2002; pp. 381–387. [Google Scholar]
- Trueman, S.; Richards, S.; McConchie, C.; Turnbull, C. Relationships between kernel oil content, fruit removal force and abscission in macadamia. Aust. J. Exp. Agric. 2000, 40, 859–866. [Google Scholar] [CrossRef]
- Miles, A.K.; Akinsanmi, O.A.; Aitken, E.A.B.; Drenth, A. Source of Pseudocercospora macadamiae inoculum in macadamia trees and its use for characterising husk spot susceptibility in the field. Crop Prot. 2010, 29, 1347–1353. [Google Scholar] [CrossRef]
- Beilharz, V.; Mayers, P.; Pascoe, I. Pseudocercospora macadamiae sp. nov., the cause of husk spot of macadamia. Australas. Plant Pathol. 2003, 32, 279–282. [Google Scholar] [CrossRef]
- Department of Agriculture and Fisheries. Macadamia Industry Benchmark Report 2009 to 2020 Seasons; MC18002; Queensland Government: Brisbane, Australia, 2021.
- Akinsanmi, O.; Drenth, A. Economic returns from fungicide application to control husk spot of macadamia in Australia is influenced by spray efficiency, rates and costs of application. Crop Prot. 2012, 41, 35–41. [Google Scholar] [CrossRef]
- Akinsanmi, O.; Miles, A.; Drenth, A. Timing of fungicide applications for control of husk spot caused by Pseudocercospora macadamiae in macadamia. Plant Dis. 2007, 91, 1675–1681. [Google Scholar] [CrossRef]
- Akinsanmi, O.; Topp, B.; Drenth, A. Pericarps retained in the tree canopy and stomatal abundance are components of resistance to husk spot caused by Pseudocercospora macadamiae in macadamia. Euphytica 2012, 185, 313–323. [Google Scholar] [CrossRef]
- Drenth, A.; Akinsanmi, O. Integrated Management of Husk Spot Disease (Pseudocercospora macadamiae) in Macadamias; MC03007; The University of Queensland: Brisbane, Australia, 2007. [Google Scholar]
- McConchie, C.; Turnbull, C.; Trueman, S. Morphology of the abscission layer in macadamia fruit. In Control of Nut Abscission in Macadamia; Turnbull, C.J.N., Trueman, J.D., Wilkie, J.D., McConchie, C.A., Eds.; Horticulture Australia Ltd: Sydney, Australia, 2003; pp. 106–120. [Google Scholar]
- Taylor, J.E.; Whitelaw, C.A. Tansley Review No. 127. Signals in Abscission. New Phytol. 2001, 151, 323–339. [Google Scholar] [CrossRef]
- Salter, B.; Wilkie, J.D.; Wiltshire, N.; Forrester, R.; McConchie, C. Fruit and Leaf Abscission of Five Macadamia Cultivars Following the Application of Ethephon at Two Concentrations and Two Spray Volumes; CSIRO: Canberra, Australia, 2005; pp. 29–42. [Google Scholar]
- Trueman, S.J.; McConchie, C.A.; Turnbull, C.G.N. Ethephon promotion of crop abscission for unshaken and mechanically shaken macadamia. Aust. J. Exp. Agric. 2002, 42, 1001–1008. [Google Scholar] [CrossRef]
- Nichols, J. Macadamia Tree Shaking Replaces Fungicides in Eliminating Spread of Costly Nut Disease. Available online: https://www.abc.net.au/news/rural/2020-08-25/macadamia-disease-husk-spot-tree-shaker/12588842 (accessed on 11 August 2022).
- Trueman, S.J. Yield responses to ethephon for unshaken and mechanically shaken macadamia. Aust. J. Exp. Agric. 2003, 43, 1143–1150. [Google Scholar] [CrossRef]
- Akinsanmi, O.A.; Drenth, A. Sustainable control of husk spot of macadamia by cultural practices. Acta Hortic. 2016, 1109, 231–236. [Google Scholar] [CrossRef]
- Topp, B.; Hardner, C.; Neal, J.; Kelly, A.; Russell, D.; McConchie, C.; O’Hare, P. Overview of the Australian macadamia industry breeding program. Acta Hortic. 2016, 1127, 45–50. [Google Scholar] [CrossRef]
- O’Connor, K.; Hayes, B.; Topp, B. Prospects for increasing yield in macadamia using component traits and genomics. Tree Genet. Genomes 2018, 14, 1–14. [Google Scholar] [CrossRef]
- Mai, T. Genomic-Assisted Exploitation of Wild Germplasm for Improvement of Macadamia. Ph.D. Thesis, The University of Queensland, St Lucia, Australia, 2021. [Google Scholar]
- O’Connor, K.; Hayes, B.; Alam, M.; Topp, B. Variability, heritability, and genetic markers associated with disease harbouring stick tight husks in macadamia. In Proceedings of the TropAg International Agriculture Conference, Brisbane, Australia, 20–22 November 2017. [Google Scholar]
- O’Connor, K.; Hayes, B.; Hardner, C.; Alam, M.; Topp, B. Selecting for nut characteristics in macadamia using a genome-wide association study. HortScience 2019, 54, 629–632. [Google Scholar] [CrossRef]
- O’Connor, K.; Hayes, B.; Hardner, C.; Nock, C.; Baten, A.; Alam, M.; Henry, R.; Topp, B. Genome-wide association studies for yield component traits in a macadamia breeding population. BMC Genom. 2020, 21, 199. [Google Scholar] [CrossRef]
- Mai, T.; Alam, M.; Hardner, C.; Henry, R.; Topp, B. Genetic structure of wild germplasm of macadamia: Species assignment, diversity and phylogeographic relationships. Plants 2020, 9, 714. [Google Scholar] [CrossRef]
- Hardner, C.; Pisanu, P.; Boyton, S. National Macadamia Conservation Program; MC99029; Horticulture Australia: Brisbane, Australia, 2004. [Google Scholar]
- Alam, M.; Neal, J.; O’Connor, K.; Kilian, A.; Topp, B. Ultra-high-throughput DArTseq-based silicoDArT and SNP markers for genomic studies in macadamia. PLoS ONE 2018, 13, e0203465. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Narasimhan, B.; Chu, G. impute: Imputation for Microarray Data, R Package Version 1.66.0. 2021. Available online: https://cran.r-project.org/web/packages/impute/index.html (accessed on 11 August 2022).
- Granato, I.; Firitsche-Neto, R. snpReady: Preparing Datasets in Order to Run Genomic Analysis, R Package Version 0.9.6. 2018. Available online: https://cran.r-project.org/web/packages/snpReady/index.html (accessed on 11 August 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Butler, D. Asreml: Fits the Linear Mixed Model; R Package Version 4.1.0.160; 2021. [Google Scholar]
- VanRaden, P. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Gezan, S.; de Oliveira, A.; Murray, D. ASRgenomics: An R Package with Complementary Genomic Functions; R Package Version 1.0.0; 2021. [Google Scholar]
- Al-Tamimi, N.; Brien, C.; Oakey, H.; Berger, B.; Saade, S.; Ho, Y.; Schmöckel, S.; Tester, M.; Negraõ, S. Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat. Commun. 2016, 7, 13342. [Google Scholar] [CrossRef]
- Müller, B.; de Almeida Filho, J.; Lima, B.; Garcia, C.; Missiaggia, A.; Aguiar, A.; Takahashi, E.; Kirst, M.; Gezan, S.; Silva-Junior, O.; et al. Independent and joint-GWAS for growth traits in Eucalyptus by assembling genome-wide data for 3373 individuals across four breeding populations. New Phytol. 2019, 221, 818–833. [Google Scholar] [CrossRef]
- Cox, D.; Snell, E. Analysis of Binary Data, 2nd ed.; Chapan and Hall: London, UK; New York, NY, USA, 1989. [Google Scholar]
- Wang, J.; Yu, J.; Lipka, A.; Zhang, Z. Interpretation of manhattan plots and other outputs of genome-wide association studies. In Genome-Wide Association Studies; Torkamaneh, D., Belzile, F., Eds.; Methods in Molecular Biology; Springer Nature: Berlin/Heidelberg, Germany, 2022; Volume 2481, pp. 63–82. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Genome Data Viewer: Macadamia integrifolia. Available online: https://www.ncbi.nlm.nih.gov/genome/gdv/browser/genome/?id=GCF_013358625.1 (accessed on 9 August 2022).
- Ru, S.; Main, D.; Evans, K.; Peace, C. Current applications, challenges, and perspectives of marker-assisted seedling selection in Rosaceae tree fruit breeding. Tree Genet. Genomes 2015, 11, 1–12. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Mai, T.; Hardner, C.; Alam, M.; Henry, R.; Topp, B. Phenotypic characterisation for growth and nut characteristics revealed the extent of genetic diversity in wild macadamia germplasm. Agriculture 2021, 11, 680. [Google Scholar] [CrossRef]
- Simmonds, N.W.; Smartt, J. Principles of Crop Improvement, 2nd ed.; Blackwell Science: Oxford, MA, USA, 2000. [Google Scholar]
- Falconer, D. Introduction to Quantitative Genetics; Longman Scientific & Technical: New York, NY, USA, 1960. [Google Scholar]
- Drenth, A.; Akinsanmi, O. Disease Management in Macadamia; MC07003; The University of Queensland: Brisbane, Australia, 2012. [Google Scholar]
- Hea-Young, L.; Jeong-Gu, K.; Byoung-Cheorl, K.; Kihwan, S. Assessment of the genetic diversity of the breeding lines and a genome wide association study of three horticultural traits using worldwide Cucumber (Cucumis spp.) germplasm collection. Agronomy 2020, 10, 1736. [Google Scholar] [CrossRef]
- McClure, K.A.; Gardner, K.M.; Douglas, G.M.; Song, J.; Forney, C.F.; DeLong, J.; Fan, L.; Du, L.; Toivonen, P.M.A.; Somers, D.J.; et al. A genome-wide association study of apple quality and scab resistance. Plant Genome 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gore, M.; Buckler, E.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Cao, K.; Zhou, Z.; Wang, Q.; Guo, J.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, X.; et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016, 7, 13246. [Google Scholar] [CrossRef]
- Hardner, C. Macadamia domestication in Hawai‘i. Genet. Resour. Crop Evol. 2015, 63, 1411–1430. [Google Scholar] [CrossRef]
- O’Connor, K.M.; Hayes, B.J.; Hardner, C.M.; Alam, M.; Henry, R.J.; Topp, B.L. Genomic selection and genetic gain for nut yield in an Australian macadamia breeding population. BMC Genom. 2021, 22, 370. [Google Scholar] [CrossRef]
- Hoang, G.T.; Van Dinh, L.; Nguyen, T.T.; Ta, N.K.; Gathignol, F.; Mai, C.D.; Jouannic, S.; Tran, K.D.; Khuat, T.H.; Do, V.N.; et al. Genome-wide association study of a panel of vietnamese rice landraces reveals new QTLs for tolerance to water deficit during the vegetative phase. Rice 2019, 12, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Phung, N.T.P.; Mai, C.D.; Hoang, G.T.; Truong, H.T.M.; Lavarenne, J.; Gonin, M.; Nguyen, K.L.; Ha, T.T.; Do, V.N.; Gantet, P.; et al. Genome-wide association mapping for root traits in a panel of rice accessions from Vietnam. BMC Plant Biol. 2016, 16, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tung, C.-W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef] [PubMed]
- Gondro, C.; Lee, S.; Lee, H.; Porto-Neto, L. Quality control for genome-wide association studies. In Genome-Wide Association Studies and Genomic Prediction; Gondro, C., van der Werf, J., Hayes, B., Eds.; Springer Science: London, UK, 2013. [Google Scholar]
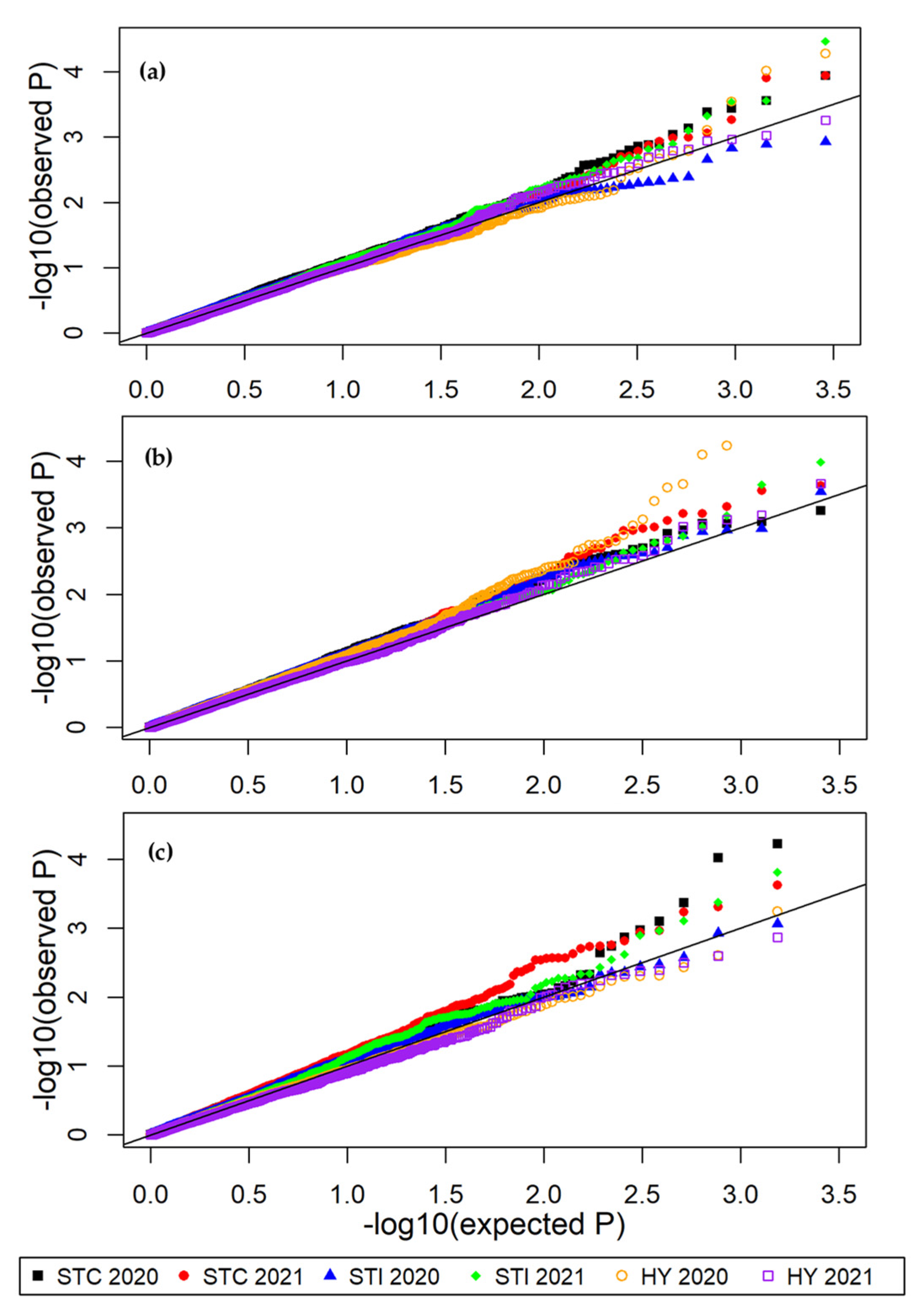
| Panel | Number of Genotypes | Number of Trees | Number of Genotypes with Clonal Replication a |
|---|---|---|---|
| Combined b | 199 in total (102 M. integrifolia + 84 M. tetraphylla + 13 hybrids) | 279 in total (147 M. integrifolia + 115 M. tetraphylla + 17 hybrids) | 66 in total (2–4) (39 M. integrifolia (2–3) + 23 M. tetraphylla (2–4) + 4 hybrids (2)) |
| M. integrifolia | 102 | 147 | 39 (2–3) |
| M. tetraphylla | 84 | 115 | 23 (2–4) |
| Traits a | Combined b (n = 199 Accessions, 279 Trees) | M. integrifolia (n = 102 Accessions, 147 Trees) | M. tetraphylla (n = 84 Accessions, 115 Trees) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | |
| STC 2020 | 0 | 305 | 42.2 | 0 | 305 | 70.3 | 0 | 105 | 8.7 |
| STC 2021 | 0 | 800 | 79.8 | 0 | 700 | 120.0 | 0 | 300 | 28.9 |
| HY 2020 | 0 | 310 | 80.1 | 0 | 310 | 98.3 | 0 | 290 | 57.3 |
| HY 2021 | 0 | 600 | 148.8 | 10 | 600 | 205.1 | 0 | 440 | 70.3 |
| STI 2020 | 0 | 1 | 0.29 | 0 | 1 | 0.38 | 0 | 1 | 0.18 |
| STI 2021 | 0 | 1 | 0.33 | 0 | 0.95 | 0.35 | 0 | 1 | 0.32 |
| p-Value a | |||||
|---|---|---|---|---|---|
| STC 2020 | STC 2021 | HY 2020 | HY 2021 | STI 2020 | STI 2021 |
| 2.99 × 10−2 | 1.79 × 10−1 | 2.52 × 10−1 | 6.18 × 10−2 | 2.45 × 10−1 | 8.53 × 10−1 |
| Traits a | Combined b (n = 199) | M. integrifolia (n = 102) | M. tetraphylla (n = 84) |
|---|---|---|---|
| h2 | h2 | h2 | |
| STC 2020 | 0.26 (0.13) | 0.36 (0.18) | 0.26 (0.21) |
| STC 2021 | 0.46 (0.13) | 0.42 (0.18) | 0.51 (0.22) |
| HY 2020 | 0.15 (0.13) | 0.08 (0.14) | 0.26 (0.24) |
| HY 2021 | 0.12 (0.11) | 0.00 (0.00) | 0.12 (0.19) |
| STI 2020 | 0.30 | 0.39 | 0.02 |
| STI 2021 | 0.22 | 0.17 | 0.31 |
| Number of Accessions with Allelic State a | |||||||
|---|---|---|---|---|---|---|---|
| Trait, Year | SNP ID | Alleles | p-Value | Allele Effect b | 0 | 1 | 2 |
| Combined panel | |||||||
| STC 2020 | 631 | G > A | 9.16 × 10−4 | 0.36 | 186 | 13 | 0 |
| 4721 | G > T | 4.11 × 10−4 | 0.30 | 188 | 11 | 0 | |
| 4747 | T > G | 3.67 × 10−4 | 0.44 | 188 | 7 | 4 | |
| 8180 | C > T | 2.76 × 10−4 | 0.52 | 151 | 44 | 4 | |
| 10405 | C > A | 7.30 × 10−4 | 1.71 | 164 | 25 | 10 | |
| 11112 | C > A | 1.14 × 10−4 | 1.84 | 159 | 33 | 7 | |
| STC 2021 | 503 | T > C | 8.66 × 10−4 | 1.55 | 57 | 85 | 57 |
| 585 | G > A | 1.15 × 10−4 | 0.58 | 156 | 22 | 21 | |
| 631 | G > A | 1.01 × 10−3 | 0.33 | 186 | 13 | 0 | |
| 4747 | T > G | 6.16 × 10−3 | 0.49 | 188 | 7 | 4 | |
| 6726 | T > C | 5.43 × 10−4 | 0.37 | 106 | 23 | 70 | |
| 7248 | T > C | 1.25 × 10−4 | 0.33 | 191 | 5 | 3 | |
| 8180 | C > T | 7.13 × 10−3 | 0.59 | 151 | 44 | 4 | |
| STI 2020 | 8180 | C > T | 6.36 × 10−3 | −0.15 | 151 | 44 | 4 |
| 7248 | T > C | 6.83 × 10−3 | −0.23 | 191 | 5 | 3 | |
| STI 2021 | 8180 | C > T | 5.32 × 10−3 | −0.14 | 151 | 44 | 4 |
| 7248 | T > C | 3.41 × 10−5 | −0.30 | 191 | 5 | 3 | |
| 5415 | T > G | 2.75 × 10−4 | 0.21 | 163 | 35 | 1 | |
| 3891 | A > C | 2.91 × 10−4 | 0.25 | 187 | 9 | 3 | |
| 7910 | C > A | 4.75 × 10−4 | −0.33 | 103 | 10 | 86 | |
| 4745 | A > C | 7.98 × 10−4 | −0.14 | 158 | 27 | 14 | |
| M. integrifolia panel | |||||||
| STC 2020 | 631 | G > A | 2.12 × 10−3 | 0.38 | 89 | 13 | 0 |
| 2178 | G > C | 8.62 × 10−4 | 1.78 | 8 | 14 | 80 | |
| 6985 | G > A | 8.64 × 10−4 | 0.63 | 49 | 32 | 21 | |
| 8180 | C > T | 8.03 × 10−4 | 0.53 | 58 | 40 | 4 | |
| 11137 | C > A | 5.54 × 10−4 | 1.66 | 59 | 30 | 13 | |
| STC 2021 | 206 | A > G | 9.81 × 10−4 | 0.54 | 89 | 4 | 9 |
| 631 | G > A | 2.33 × 10−4 | 0.32 | 89 | 13 | 0 | |
| 5415 | T > G | 4.77 × 10−4 | 2.23 | 66 | 35 | 1 | |
| 6985 | G > A | 2.59 × 10−3 | 0.65 | 49 | 32 | 21 | |
| 7694 | G > C | 2.75 × 10−4 | 1.61 | 53 | 27 | 22 | |
| 8435 | C > G | 7.77 × 10−4 | 0.35 | 94 | 7 | 1 | |
| 11137 | C > A | 6.07 × 10−4 | 1.65 | 59 | 30 | 13 | |
| 11670 | T > C | 6.15 × 10−4 | 2.23 | 4 | 13 | 85 | |
| STI 2020 | 2716 | T > C | 2.88 × 10−4 | −0.21 | 7 | 9 | 86 |
| STI 2021 | 5415 | T > G | 1.04 × 10−4 | 0.21 | 66 | 35 | 1 |
| 12649 | C > T | 2.24 × 10−4 | 0.11 | 48 | 14 | 40 | |
| 535 | G > A | 6.60 × 10−4 | −0.20 | 90 | 10 | 2 | |
| 631 | G > A | 9.54 × 10−4 | −0.24 | 89 | 13 | 0 | |
| M. tetraphylla panel | |||||||
| STC 2020 | 3315 | G > C | 9.50 × 10−5 | 0.45 | 57 | 25 | 2 |
| 4498 | C > T | 4.23 × 10−4 | 1.83 | 9 | 25 | 50 | |
| 10405 | C > A | 5.89 × 10−5 | 1.93 | 53 | 21 | 10 | |
| 11998 | T > A | 7.97 × 10−4 | 1.75 | 49 | 25 | 10 | |
| STC 2021 | 3338 | G > A | 2.38 × 10−4 | 0.34 | 2 | 8 | 74 |
| 4745 | A > C | 8.53 × 10−3 | 0.62 | 51 | 22 | 11 | |
| 6646 | G > A | 5.84 × 10−4 | 1.73 | 29 | 30 | 25 | |
| 7248 | T > C | 4.90 × 10−4 | 0.29 | 78 | 4 | 2 | |
| STI 2020 | 8554 | T > G | 8.70 × 10−4 | 0.20 | 63 | 14 | 7 |
| 4745 | A > C | 9.92 × 10−3 | −0.14 | 51 | 22 | 11 | |
| STI 2021 | 4745 | A > C | 1.53 × 10−4 | −0.19 | 51 | 22 | 11 |
| 5027 | G > A | 4.17 × 10−4 | −0.25 | 64 | 18 | 2 | |
| 10146 | G > C | 7.74 × 10−4 | −0.37 | 80 | 3 | 1 | |
| Presence of SNP in Other Panels d | ||||||
|---|---|---|---|---|---|---|
| SNP ID | Chr a | Bp b | MAF c | Combined e | integf | tetrag |
| Combined panel | ||||||
| 11112 | 7 | 24,733,108 | 8.5 | - | y | y |
| 8180 | 4 | 6,321,754 | 7.7 | - | y | n |
| 4747 | - | - | 26.5 | - | y | n |
| 4721 | 2 | 15,858,489 | 36.2 | - | y | n |
| 10405 | 5 | 43,602,889 | 8.8 | - | n | y |
| 631 | 14 | 3,288,694 | 30.6 | - | y | n |
| 585 | 1 | 35,796,996 | 6.2 | - | y | n |
| 7248 | 9 | 17,297,314 | 36.2 | - | n | y |
| 6726 | 5 | 34,504,332 | 2.4 | - | n | y |
| 503 | - | - | 2.0 | - | y | y |
| 5415 | 4 | 6,115,067 | 8.7 | - | n | y |
| 3891 | 7 | 30,363,260 | 33.2 | - | y | n |
| 7910 | 4 | 31,117,569 | 6.3 | - | n | y |
| 4745 | 5 | 2,290,426 | 7.2 | - | n | y |
| M. integrifolia panel | ||||||
| 11137 | 7 | 31,108,153 | 3.6 | y | - | n |
| 8180 | 4 | 6,321,754 | 4.3 | y | - | n |
| 2178 | - | - | 1.2 | y | - | n |
| 6985 | - | - | 2.8 | y | - | y |
| 631 | 14 | 3,288,694 | 15.7 | y | - | n |
| 7694 | 10 | 27,246,724 | 2.9 | y | - | n |
| 5415 | 8 | 6,115,067 | 5.5 | Y | - | n |
| 11670 | - | - | 1.1 | n | - | n |
| 8435 | 10 | 19,629,398 | 22.7 | n | - | n |
| 206 | 11 | 10,745,223 | 9.3 | n | - | n |
| 2716 | - | - | 1.1 | y | - | n |
| 12649 | 12 | 24,083,560 | 2.2 | y | - | y |
| 535 | - | - | 14.6 | y | - | y |
| M. tetraphylla panel | ||||||
| 10405 | 5 | 43,602,889 | 4.1 | y | n | - |
| 3315 | - | - | 5.8 | y | n | - |
| 4498 | 1 | 2,946,127 | 1.3 | y | y | - |
| 11998 | 5 | 43,602,823 | 3.7 | y | n | - |
| 3338 | 14 | 4,231,774 | 1.1 | y | y | - |
| 7248 | 9 | 17,297,314 | 21.0 | y | n | - |
| 6646 | - | - | 2.1 | y | y | - |
| 4745 | 5 | 2,290,426 | 3.8 | y | n | - |
| 10146 | 3 | 6,830,111 | 33.6 | n | n | - |
| 5027 | 12 | 2,625,752 | 7.6 | y | n | - |
| 8554 | 13 | 13,051,846 | 6.0 | y | n | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunn, J.; De Faveri, J.; O’Connor, K.; Alam, M.; Hardner, C.; Akinsanmi, O.; Topp, B. Genome-Wide Association Study for Abscission Failure of Fruit Pericarps (Stick-Tights) in Wild Macadamia Germplasm. Agronomy 2022, 12, 1913. https://doi.org/10.3390/agronomy12081913
Nunn J, De Faveri J, O’Connor K, Alam M, Hardner C, Akinsanmi O, Topp B. Genome-Wide Association Study for Abscission Failure of Fruit Pericarps (Stick-Tights) in Wild Macadamia Germplasm. Agronomy. 2022; 12(8):1913. https://doi.org/10.3390/agronomy12081913
Chicago/Turabian StyleNunn, Jasmine, Joanne De Faveri, Katie O’Connor, Mobashwer Alam, Craig Hardner, Olufemi Akinsanmi, and Bruce Topp. 2022. "Genome-Wide Association Study for Abscission Failure of Fruit Pericarps (Stick-Tights) in Wild Macadamia Germplasm" Agronomy 12, no. 8: 1913. https://doi.org/10.3390/agronomy12081913
APA StyleNunn, J., De Faveri, J., O’Connor, K., Alam, M., Hardner, C., Akinsanmi, O., & Topp, B. (2022). Genome-Wide Association Study for Abscission Failure of Fruit Pericarps (Stick-Tights) in Wild Macadamia Germplasm. Agronomy, 12(8), 1913. https://doi.org/10.3390/agronomy12081913







