Genome-Wide Characterization of MADS-box Genes Identifies Candidates Associated with Flower and Fruit Development in Loquat (Eriobotrya japonica Lindl.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genome-Wide Identification
2.3. Phylogenic Analysis
2.4. Conserved Motif, Functional Domains, and Gene Structure Analysis
2.5. Synteny Analysis and Chromosome Location
2.6. RNA-seq Analysis
2.7. RNA Extraction and qRT-PCR
3. Results
3.1. MADS-box Genes in 12 Rosaceae Species
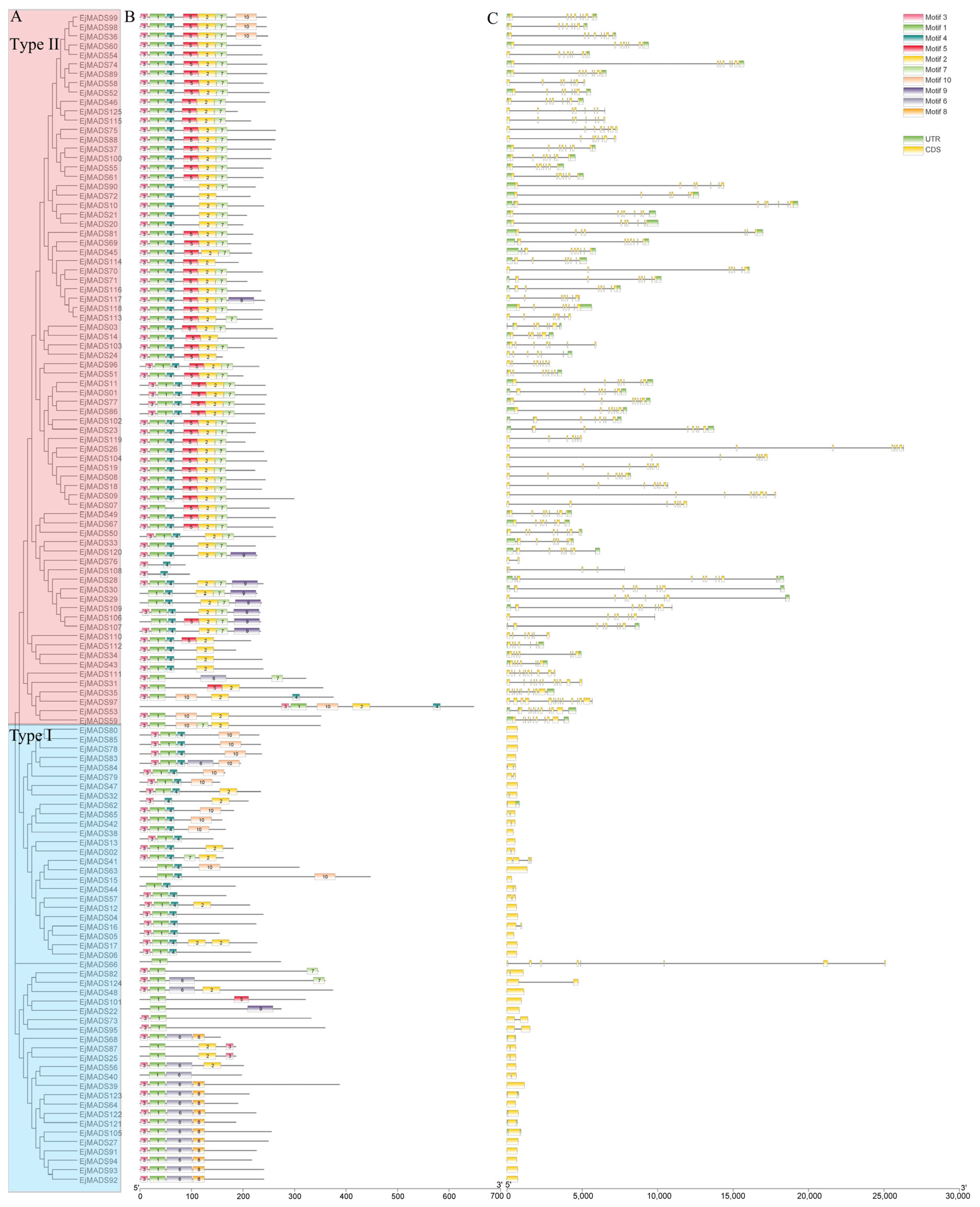
3.2. Phylogenetic Analysis and Gene Characterization
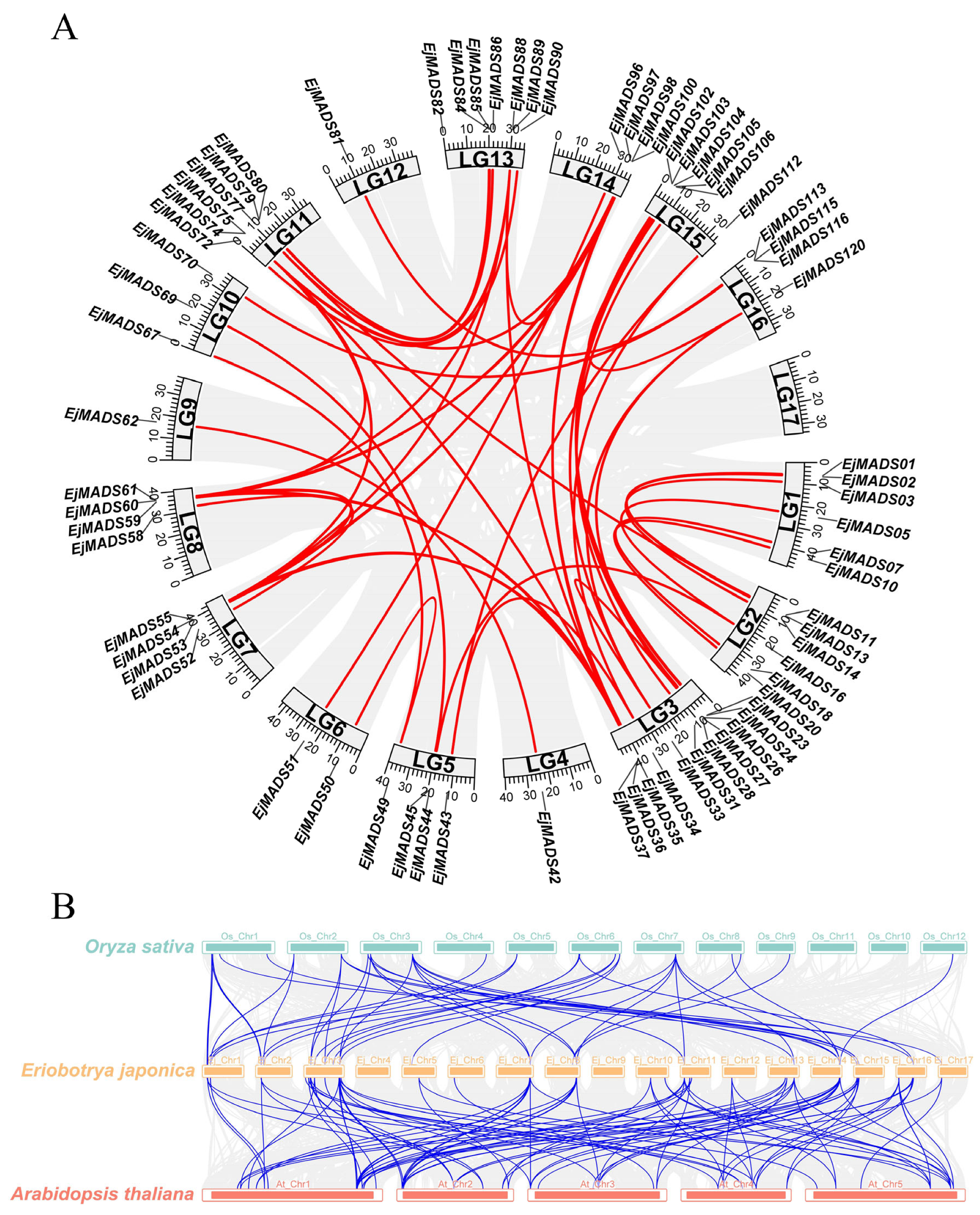
3.3. Chromosomal Locations and Annotation
3.4. Gene Structures and Conserved Motifs
3.5. Gene Duplication and Synteny Analysis
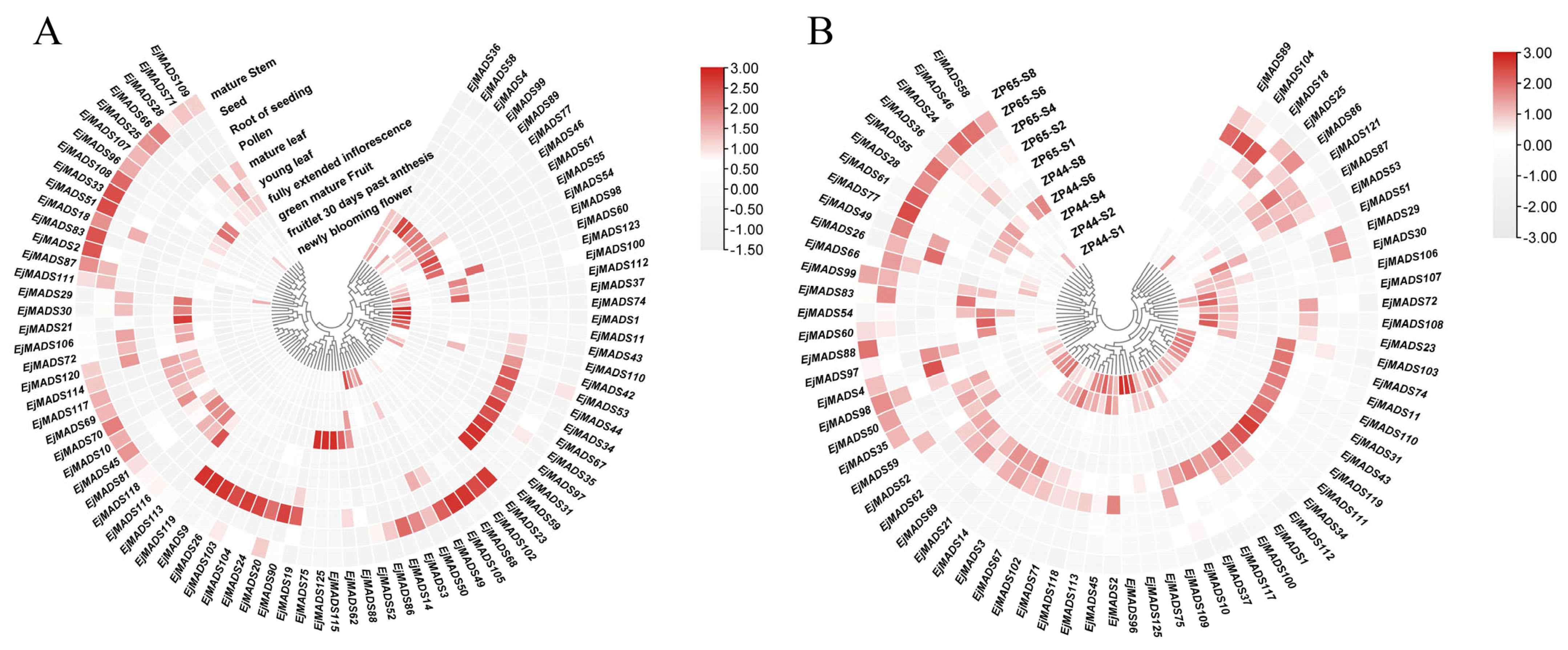
3.6. Expression Profiles in Different Tissues
3.7. Expression Patterns in Flower Buds at Different Stages and in Different Tissues
4. Discussion
4.1. Features of MADS-box Genes
4.2. Expression of EjMADSs in Different Tissues of Loquat
4.3. Functional Conservation of the ABCDE Model Genes in Loquat
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kurokura, T.; Mimida, N.; Battey, N.H.; Hytönen, T. The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 2013, 64, 4131–4141. [Google Scholar] [CrossRef] [PubMed]
- Ó’Maoiléidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014, 201, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Vachon, G.; Engelhorn, J.; Carles, C.C. Interactions between transcription factors and chromatin regulators in the control of flower development. J. Exp. Bot. 2018, 69, 2461–2471. [Google Scholar] [CrossRef]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-Box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef] [PubMed]
- Thakare, D.; Tang, W.; Hill, K.; Perry, S.E. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 Promotes somatic embryo development in arabidopsis and soybean. Plant Physiol. 2008, 146, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Parcy, F. Flowering: A time for integration. Int. J. Dev. Biol. 2005, 49, 585–593. [Google Scholar] [CrossRef]
- Hartmann, U.; Höhmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef]
- Mandel, M.A.; Yanofsky, M.F. A gene triggering flower formation in Arabidopsis. Nature 1995, 377, 522–524. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Theissen, G.; Saedler, H. Plant Biology. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Colombo, L.; Franken, J.; Koetje, E.; van Went, J.; Dons, H.J.; Angenent, G.C.; van Tunen, A.J. The petunia MADS Box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Zhu, Y.; Su, W.; Zhang, L.; Jing, Y.; Lin, S.; Gao, Y. The role of EjSOC1s in flower initiation in Eriobotrya japonica. Front. Plant Sci. 2019, 10, 253. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Zhang, Z.; Lin, S.; Lin, S.; Yang, X. The role of EjSVPs in flower initiation in Eriobotrya japonica. Int. J. Mol. Sci. 2019, 20, 5933. [Google Scholar] [CrossRef] [PubMed]
- Jing, D.; Chen, W.; Xia, Y.; Shi, M.; Wang, P.; Wang, S.; Wu, D.; He, Q.; Liang, G.; Guo, Q. Homeotic transformation from stamen to petal in Eriobotrya japonica is associated with hormone signal transduction and reduction of the transcriptional activity of EjAG. Physiol. Plant 2020, 168, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shi, M.; Chen, W.; Hu, R.; Jing, D.; Wu, D.; Wang, S.; Li, Q.; Deng, H.; Guo, Q.; et al. Expression pattern and functional characterization of PISTILLATA ortholog associated with the formation of petaloid sepals in double-flower Eriobotrya japonica (Rosaceae). Front. Plant Sci. 2019, 10, 1685. [Google Scholar] [CrossRef]
- Liu, Y.; Song, H.; Liu, Z.; Hu, G.; Lin, S. Molecular characterization of loquat EjAP1 gene in relation to flowering. Plant Growth Regul. 2013, 70, 287–296. [Google Scholar] [CrossRef]
- Xia, Y.; Xue, B.; Shi, M.; Zhan, F.; Wu, D.; Jing, D.; Wang, S.; Guo, Q.; Liang, G.; He, Q. Comparative transcriptome analysis of flower bud transition and functional characterization of EjAGL17 involved in regulating floral initiation in loquat. PLoS ONE 2020, 15, e0239382. [Google Scholar] [CrossRef]
- Xu, H.X.; Meng, D.; Yang, Q.; Chen, T.; Qi, M.; Li, X.Y.; Ge, H.; Chen, J.W. Sorbitol induces flower bud formation via the MADS-box transcription factor EjCAL in loquat. J. Integr. Plant Biol. 2023, 65, 1241–1261. [Google Scholar] [CrossRef]
- Díaz-Riquelme, J.; Lijavetzky, D.; Martínez-Zapater, J.M.; Carmona, M.J. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 2009, 149, 354–369. [Google Scholar] [CrossRef]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Jiang, S.; An, H.; Xu, F.; Zhang, X. Chromosome-level genome assembly and annotation of the loquat (Eriobotrya japonica) genome. Gigascience 2020, 9, giaa015. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Jing, D.; Liu, X.; He, Q.; Dang, J.; Hu, R.; Xia, Y.; Wu, D.; Wang, S.; Zhang, Y.; Xia, Q.; et al. Genome assembly of wild loquat (Eriobotrya Japonica) and resequencing provide new insights into the genomic evolution and fruit domestication in loquat. Hortic. Res. 2023, 10, uhac265. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Zhao, C.; Li, S.; Guo, Y.; Xu, H.; Hu, G.; Liu, Z.; Chen, X.; Chen, J.; Lin, S.; et al. Integration of genomics, transcriptomics and metabolomics identifies candidate loci underlying fruit weight in loquat. Hortic. Res. 2022, 9, uhac037. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Jing, Y.; Lin, S.; Yue, Z.; Yang, X.; Xu, J.; Wu, J.; Zhang, Z.; Xia, R.; Zhu, J.; et al. Polyploidy underlies co-option and diversification of biosynthetic triterpene pathways in the apple tribe. Proc. Natl. Acad. Sci. USA 2021, 118, e2101767118. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef]
- Su, W.; Shao, Z.; Wang, M.; Gan, X.; Yang, X.; Lin, S. EjBZR1 represses fruit enlargement by binding to the EjCYP90 promoter in loquat. Hortic. Res. 2021, 8, 152. [Google Scholar] [CrossRef]
- Su, W.; Yuan, Y.; Zhang, L.; Jiang, Y.; Gan, X.; Bai, Y.; Peng, J.; Wu, J.; Liu, Y.; Lin, S. Selection of the optimal reference genes for expression analyses in different materials of Eriobotrya japonica. Plant Methods 2019, 15, 7. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Arya, P.; Gupta, K.; Randhawa, V.; Acharya, V.; Singh, A.K. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 2016, 6, 20695. [Google Scholar] [CrossRef]
- Wang, R.; Ming, M.; Li, J.; Shi, D.; Qiao, X.; Li, L.; Zhang, S.; Wu, J. Genome-wide identification of the MADS-box transcription factor family in pear (Pyrus bretschneideri) reveals evolution and functional divergence. PeerJ 2017, 5, e3776. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 2007, 8, 242. [Google Scholar] [CrossRef]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef]
- Wang, B.; Hu, W.; Fang, Y.; Feng, X.; Fang, J.; Zou, T.; Zheng, S.; Ming, R.; Zhang, J. Comparative analysis of the MADS-box genes revealed their potential functions for flower and fruit development in longan (Dimocarpus longan). Front. Plant Sci. 2021, 12, 813798. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, W.; Xu, Z.; Chen, M.; Yu, D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chang, X.; Zhang, Y.; Yu, X.; Qin, Y.; Sun, Y.; Zhang, L. The pineapple MADS-box gene family and the evolution of early monocot flower. Sci. Rep. 2021, 11, 849. [Google Scholar] [CrossRef]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef]
- Ubi, B.E.; Saito, T.; Bai, S.; Nishitani, C.; Ban, Y.; Ikeda, K.; Ito, A.; Moriguchi, T. Characterization of 10 MADS-box genes from Pyrus pyrifolia and their differential expression during fruit development and ripening. Gene 2013, 528, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Vrebalov, J.; Pan, I.L.; Arroyo, A.J.M.; McQuinn, R.; Chung, M.; Poole, M.; Rose, J.; Seymour, G.; Grandillo, S.; Giovannoni, J. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF Gene TAGL1. Plant Cell 2009, 21, 3041–3062. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.A.; Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meri-stem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Winter, C.M.; Austin, R.S.; Blanvillain-Baufumé, S.; Reback, M.A.; Monniaux, M.; Wu, M.-F.; Sang, Y.; Yamaguchi, A.; Yamaguchi, N.; Parker, J.E. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 2011, 20, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Goto, K.; Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes. Dev. 1994, 8, 1548–1560. [Google Scholar] [CrossRef]
- Wagner, D.; Sablowski, R.W.; Meyerowitz, E.M. Transcriptional activation of APETALA1 by LEAFY. Science 1999, 285, 582–584. [Google Scholar] [CrossRef]
- Ng, M.; Yanofsky, M.F. Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 2001, 13, 739–753. [Google Scholar] [CrossRef]
- Hu, T.; Li, X.; Du, L.; Manuela, D.; Xu, M. LEAFY and APETALA1 down-regulate ZINC FINGER PROTEIN 1 and 8 to release their repression on class B and C floral homeotic genes. Proc. Natl. Acad. Sci. USA 2023, 120, e2221181120. [Google Scholar] [CrossRef]
- Litt, A.; Irish, V.F. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 2003, 165, 821–833. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Liu, C.; Xi, W.; Shen, L.; Tan, C.; Yu, H. Regulation of floral patterning by flowering time genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef]
- Moon, J.; Lee, H.; Kim, M.; Lee, I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 2005, 46, 292–299. [Google Scholar] [CrossRef]
- Amasino, R. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Liu, X.; Zhao, C.; Wu, W.; Jiang, Y.; Su, W.; Lin, S.; Yang, X.; Peng, Z. Genome-Wide Characterization of MADS-box Genes Identifies Candidates Associated with Flower and Fruit Development in Loquat (Eriobotrya japonica Lindl.). Agronomy 2023, 13, 2709. https://doi.org/10.3390/agronomy13112709
Li W, Liu X, Zhao C, Wu W, Jiang Y, Su W, Lin S, Yang X, Peng Z. Genome-Wide Characterization of MADS-box Genes Identifies Candidates Associated with Flower and Fruit Development in Loquat (Eriobotrya japonica Lindl.). Agronomy. 2023; 13(11):2709. https://doi.org/10.3390/agronomy13112709
Chicago/Turabian StyleLi, Wenxiang, Xiaopei Liu, Chongbin Zhao, Wendong Wu, Yuanyuan Jiang, Wenbing Su, Shunquan Lin, Xianghui Yang, and Ze Peng. 2023. "Genome-Wide Characterization of MADS-box Genes Identifies Candidates Associated with Flower and Fruit Development in Loquat (Eriobotrya japonica Lindl.)" Agronomy 13, no. 11: 2709. https://doi.org/10.3390/agronomy13112709
APA StyleLi, W., Liu, X., Zhao, C., Wu, W., Jiang, Y., Su, W., Lin, S., Yang, X., & Peng, Z. (2023). Genome-Wide Characterization of MADS-box Genes Identifies Candidates Associated with Flower and Fruit Development in Loquat (Eriobotrya japonica Lindl.). Agronomy, 13(11), 2709. https://doi.org/10.3390/agronomy13112709






