Impact of Phosphogypsum Application on Fungal Community Structure and Soil Health in Saline–Alkali-Affected Paddy Fields
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites and Environmental Conditions
2.2. Soil Sampling and Physicochemical Properties Analysis
2.3. Microbial Diversity Analysis
2.4. Statistical Analysis
3. Results
3.1. Physicochemical Properties
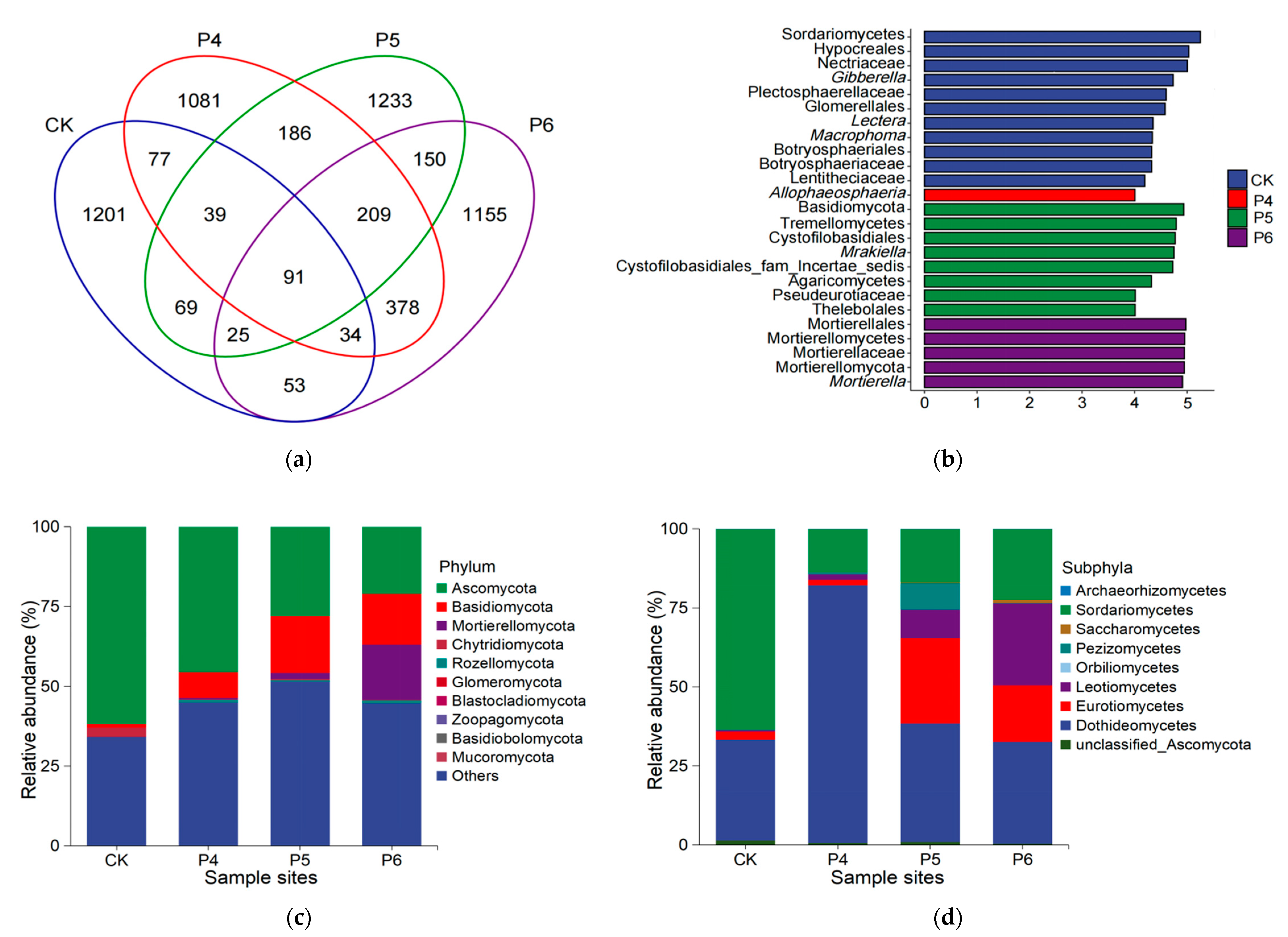
3.2. Fungal Community Composition and Diversity
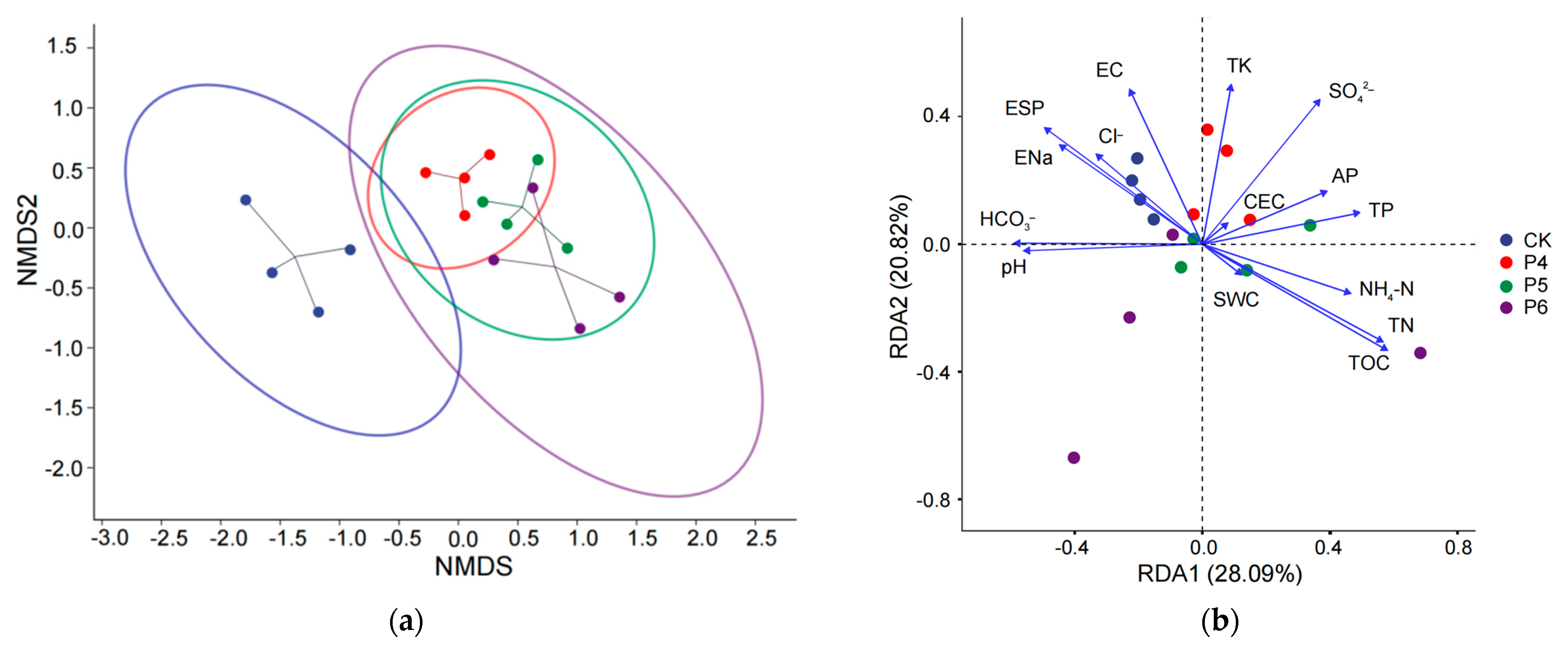
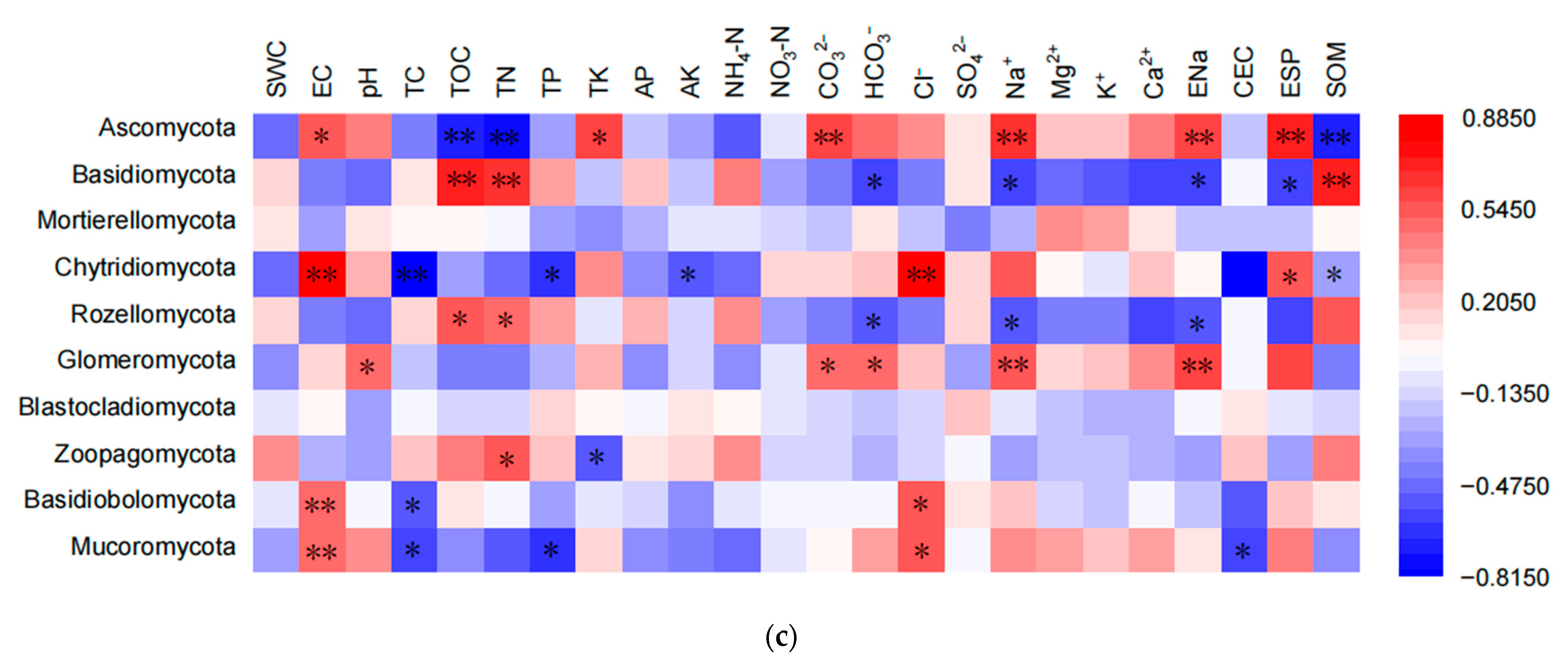
3.3. Influences of Environmental Parameters on Fungal Diversity and Community Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wirsenius, S.; Azar, C.; Berndes, G. How much land is needed for global food production under scenarios of dietary changes and livestock productivity increases in 2030? Agric. Syst. 2010, 103, 621–638. [Google Scholar] [CrossRef]
- Food and Agricultural Organization. FAO Statistical Yearbook 2020, World Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2022; p. 2. [Google Scholar]
- Wicke, B.; Smeets, E.; Dornburg, V.; Vashey, B.; Gaiser, T.; Turkenburg, W.; Faaij, A. The global technical and economic potential of bioenergy from salt-affected soils. Energy Environ. Sci. 2011, 4, 2669–2681. [Google Scholar] [CrossRef]
- Singh, A. Salinization of agricultural lands due to poor drainage: A viewpoint. Ecol. Indic. 2018, 95, 127–130. [Google Scholar] [CrossRef]
- Zaman, F. The Socioeconomic Impact of Climate Change: Assessing the Vulnerability and Adaptive Capacity of the Coastal People in Bangladesh. Master’s Thesis, University of Dhaka, Dhaka, Bangladesh, 2018. [Google Scholar]
- Wang, M.M.; Rengasamy, P.; Wang, Z.C.; Yang, F.; Ma, H.Y.; Huang, L.H.; Liu, M.; Yang, H.Y.; Li, J.P.; An, F.H.; et al. Identification of the most limiting factor for rice yield using soil data collected before planting and during the reproductive stage. Land Degrad. Dev. 2018, 29, 2310–2320. [Google Scholar] [CrossRef]
- Dobermann, A.; Fairhurst, T. Rice: Nutrient Disorders & Nutrient Management; Potash and Phosphate Institute, and Manila, IRRI: Singapore, 2000. [Google Scholar]
- Mahmoodabadi, M.; Yazdanpanah, N.; Rodríguez Sinobas, L.; Pazira, E.; Neshat, A. Reclamation of calcareous saline sodic soil with different amendments (I): Redistribution of soluble cations within the soil profile. Agric. Water Manag. 2013, 120, 30–38. [Google Scholar] [CrossRef]
- Hanay, A.; Büyüksönmez, F.; Kiziloglu, F.M.; Canbolat, M.Y. Reclamation of saline-sodic soils with Gypsum and MSW compost. Compost Sci. Util. 2013, 12, 175–179. [Google Scholar] [CrossRef]
- Sun, W.; Xiao, E.; Pu, Z.; Krumins, V.; Dong, Y.; Li, B.; Hu, M. Paddy soil microbial communities driven by environment- and microbe-microbe interactions: A case study of elevation-resolved microbial communities in a rice terrace. Sci. Total Environ. 2018, 612, 884–893. [Google Scholar] [CrossRef]
- Tian, T.; Zhang, C.L.; Zhu, F.; Yuan, S.X.; Guo, Y.; Xue, S.G. Effect of phosphogypsum on saline-alkalinity and aggregate stability of bauxite residue. T. Nonferr. Metal. Soc. 2021, 31, 1484–1495. [Google Scholar] [CrossRef]
- Miao, L.; Liang, Z.; Ma, H.Y.; Huang, L.H.; Wang, M.M. Responses of rice (Oryza saliva L.) growth and yield to phosphogypsumamendment in saline-sodic soils of North-East China. J. Food Agric. Environ. 2010, 8, 827–833. [Google Scholar]
- Zhou, S.M.; Zhang, M.; Zhang, K.K.; Yang, X.W.; He, D.X.; Yin, J.; Wang, C.Y. Effects of reduced nitrogen and suitable soil moisture on wheat (Triticum aestivum L.) rhizosphere soil microbiological, biochemical properties and yield in the Huanghuai Plain, China. J. Integ. Agric. 2020, 19, 234–250. [Google Scholar] [CrossRef]
- Aon, M.A.; Cabello, M.N.; Sarena, D.E.; Colaneri, A.C.; Franco, M.G.; Burgos, J.L.; Cortassa, S.I. Spatio-temporal patterns of soil microbial and enzymatic activities in an agricultural soil. Appl. Soil Ecol. 2001, 18, 239–254. [Google Scholar] [CrossRef]
- Goberna, M.; Sánchez, J.; Pascual, J.A.; Garciá, C. Surface and subsurface organic carbon, microbial biomass and activity in a forest soil sequence. Soil. Biol. Biochem. 2006, 38, 2233–2243. [Google Scholar] [CrossRef]
- Khan, M.N.; Mobin, M.; Abbas, Z.K.; Alamri, S.A. Fertilizers and their contaminants in soils, surface and groundwater. Encycl. Anthr. 2018, 5, 225–240. [Google Scholar] [CrossRef]
- Gomes, S.I.F.; Merckx, V.S.F.T.; Kehl, J.; Gebauer, G. Mycoheterotrophic plants living on arbuscular mycorrhizal fungi are generally enriched in 13C, 15N and 2H isotopes. J. Ecol. 2020, 108, 1250–1261. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2010, 32, 666–681. [Google Scholar] [CrossRef]
- Broeckling, C.D.; Broz, A.K.; Bergelson, J.; Manter, D.K.; Vivanco, J.M. Root exudates regulate soil fungal community composition and diversity. Appl. Environ. Microbiol. 2008, 74, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Bringhurst, R.M.; Cardon, Z.G.; Gage, D.J. Galactosides in the rhizosphere: Utilization by Sinorhizobium meliloti and development of a biosensor. Proc. Natl. Acad. Sci. USA 2001, 98, 4540–4545. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Spor, A.; Hénault, C.; Bru, D.; Bizouard, F.; Jones, C.M.; Sarr, A.; Maron, P.A. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 2013, 7, 1609–1619. [Google Scholar] [CrossRef]
- Bridge, P.; Spooner, B. Soil fungi: Diversity and detection. Plant Soil 2001, 232, 147–154. [Google Scholar] [CrossRef]
- Augé, R. Arbuscular mycorrhizae and soil/plant water relations. Can. J. Soil Sci. 2004, 84, 373–381. [Google Scholar] [CrossRef]
- Bashan, Y.; Salazar, B.; Puente, M.E. Responses of native legume desert trees used for reforestation in the Sonoran Desert to plant growth-promoting microorganisms in screen house. Biol. Fert. Soils 2009, 45, 655–662. [Google Scholar] [CrossRef]
- Burke, R.A.; Molina, M.; Cox, J.E.; Osher, L.J.; Piccolo, M.C. Stable carbon isotope ratio and composition of microbial fatty acids in tropical soils. J. Environ. Qual. 2003, 32, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Sparling, G.P. Biological indicators of soil health. In Soil Microbial Biomass, Activity and Nutrient Cycling as Indicators of Soil Health; Pankhurst, C., Doube, B.M., Gupta, V.V.S.R., Eds.; CAB International: Wallingford, UK, 1997; Volume 12, pp. 97–120. [Google Scholar]
- Saggar, S.; Yeates, G.W.; Shepherd, T.G. Cultivation effects on soil biological properties, microfauna and organic matter dynamics in Eutric Gleysol and Gleyic Luvisol soils in New Zealand. Soil Till. Res. 2001, 58, 55–68. [Google Scholar] [CrossRef]
- Ji, L.; Xin, Y.; Guo, D. Soil Fungal Community Structure and Its Effect on CO2 Emissions in the Yellow River Delta. Int. J. Environ. Res. Public Health 2023, 20, 4190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, B.; Yin, R.; Xing, S.; Fu, W.; Wu, H.; Hao, Z.; Ma, Y.; Zhang, X. Long-term nickel contamination increased soil fungal diversity and altered fungal community structure and co-occurrence patterns in agricultural soils. J. Hazard Mater. 2022, 436, 129113. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 3, 1722–1736. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Veresoglou, S.D.; Tedersoo, L.; Xu, T.; Ge, T.; Liu, L.; Chen, Y.; Hao, Z.; Su, Y.; Rillig, M.C.; et al. Contrasting latitudinal diversity and co-occurrence patterns of soil fungi and plants in forest ecosystems. Soil Biol. Biochem. 2019, 131, 100–110. [Google Scholar] [CrossRef]
- Bender, S.F.; Wagg, C.; van der Heijden, M.G.A. An Underground Revolution: Biodiversity and Soil Ecological Engineering for Agricultural Sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 2018, 6, e27295v2. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DAD2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Boylen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Li, Y.; Li, S. Effects of the interaction between biochar and nutrients on soil organic carbon sequestration in soda saline-alkali grassland: A review. Glob. Ecol. Conserv. 2021, 26, e01449. [Google Scholar] [CrossRef]
- Brzostek, E.R.; Greco, A.; Drake, J.E.; Finzi, A.C. Root carbon inputs to the rhizosphere stimulate extracellular enzyme activity and increase nitrogen availability in temperate forest soils. Biogeochemistry 2013, 115, 65–76. [Google Scholar] [CrossRef]
- Crowther, T.W.; Van Den Hoogen, J.; Wan, J.; Mayes, M.A.; Keiser, A.D.; Mo, L.; Averill, C.; Maynard, D.S. The global soil community and its influence on biogeochemistry. Science 2019, 365, eaav0550. [Google Scholar] [CrossRef]
- Huang, L.; Bai, J.; Wen, X.; Zhang, G.; Zhang, C.; Cui, B.; Liu, X. Microbial resistance and resilience in response to environmental changes under the higher intensity of human activities than global average level. Glob. Chang. Biol. 2020, 26, 2377–2389. [Google Scholar] [CrossRef]
- Maroua, J.; Labidi, S.; Bahri, B.A.; Laurelle, F.; Tisserant, B.; Jeddi, F.B.; Lounès-Hadj Sahraoui, A. Soil properties and climate affect arbuscular mycorrhizal fungi and soil microbial communities in Mediterranean rainfed cereal cropping systems. Pedobiologia 2021, 87–88, 150748. [Google Scholar] [CrossRef]
- Lui, D.; Liu, G.; Chen, L.; Wang, J.; Zhang, L. Soil pH determines fungal diversity along an elevation gradient in Southwestern China. Sci. China Life Sci. 2018, 61, 718–726. [Google Scholar] [CrossRef]
- Tao, Z.; Wang, N.F.; Liu, H.Y.; Zhang, Y.Q.; Yu, L.Y. Soil pH is a key determinant of soil fungal community composition in the Ny-Ålesund region, Svalbard (High Arctic). Front. Microbiol. 2016, 7, 227. [Google Scholar] [CrossRef]
- Sun, R.; Dsouza, M.; Gilbert, J.A.; Guo, X.; Wang, D.; Guo, Z.; Ni, Y.; Chu, H. Fungal community composition in soils subjected to long-term chemical fertilization is most influenced by the type of organic matter. Environ. Microbiol. 2016, 18, 5137–5150. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. Soil carbon content drives the biogeographical distribution of fungal communities in the black soil zone of northeast China. Soil Biol. Biochem. 2015, 83, 29–39. [Google Scholar] [CrossRef]
- Liu, K.; Liu, Z.; Zhou, N.; Shi, X.; Lock, T.R.; Kallenbach, R.L.; Yuan, Z. Diversity-stability relationships in temperate grasslands as a function of soil pH. Land Degrad. Dev. 2022, 33, 1704–1717. [Google Scholar] [CrossRef]
- Williams, M.A.; Rice, C.W. Seven years of enhanced water availability influences the physiological, structural, and functional attributes of a soil microbial community. Appl. Soil Ecol. 2007, 35, 535–545. [Google Scholar] [CrossRef]
- Sterkenburg, E.; Bahr, A.; Durling, M.B.; Clemmensen, K.E.; Lindahl, B.D. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol. 2015, 207, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gao, Y.; Wang, S.; Xu, D.; Yu, H.; Wu, L.; Lin, Q.; Hu, Y.; Li, X.; He, Z.; et al. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 2014, 8, 430–440. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Chen, Y.; Dai, Q.; Hu, J. Mudflat reclamation causes change in the composition of fungal communities under long-term rice cultivation. Can. J. Microbiol. 2019, 65, 530–537. [Google Scholar] [CrossRef]
- Letuma, P.; Arafat, Y.; Waqa, M.; Lin, F.; Lin, W.; Zhang, Y.; Masita, M.; Fan, K.; Li, Z.; Lin, W. Gene mutation associated with esl mediates shifts on fungal community composition in rhizosphere soil of rice at grain-filling stage. Sci. Rep. 2018, 8, 17521. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Lundell, T.K.; Mäkelä, M.R.; Hildén, K. Lignin-modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J. Basic Microbiol. 2010, 50, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Marian, I.M.; Vonk, P.J.; Valdes, I.D.; Barry, K.; Bostock, B.; Carver, A.; Daum, C.; Lerner, H.; Lipzen, A.; Park, H.; et al. The Transcription Factor Roc1 Is a Key Regulator of Cellulose Degradation in the Wood-Decaying Mushroom Schizophyllum commune. mBio 2022, 13, e0062822. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Qiu, L.; Guo, L.; Man, J.; Shang, B.; Pu, R.; Ou, X.; Dai, C.; Liu, P.; Yang, Y.; et al. Fertilizers Reduce the Accumulation of Cd in Panax notoginseng (Burk.) F.H. by Improving the Quality of the Microbial Community. Front. Plant Sci. 2020, 11, 888. [Google Scholar] [CrossRef] [PubMed]
- Osorio, N.W.; Habte, M. Soil phosphate desorption induced by a phosphate-solubilizing fungus. Commun. Soil Sci. Plant 2014, 45, 451–460. [Google Scholar] [CrossRef]
- Cao, J.; Wang, H.; Holden, N.M.; Adamowski, J.F.; Biswas, A.; Zhang, X.; Feng, Q. Soil properties and microbiome of annual and perennial cultivated grasslands on the Qinghai-Tibetan Plateau. Land Degrad. Dev. 2021, 32, 5306–5321. [Google Scholar] [CrossRef]

| CK | P4 | P5 | P6 | |
|---|---|---|---|---|
| SWC (%) | 15.05 ± 0.53 a | 21.78 ± 1.03 b | 19.77 ± 1.41 b | 19.48 ± 1.38 b |
| EC (μS·cm−1) | 1319.50 ± 377.81 a | 643.50 ± 75.53 b | 538.75 ± 124.52 b | 348.25 ± 17.88 b |
| pH | 10.03 ± 0.19 a | 8.39 ± 0.15 b | 9.21 ± 0.30 c | 9.28 ± 0.18 c |
| TC (%) | 1.15 ± 0.14 a | 1.23 ± 0.05 a | 1.34 ± 0.05 a | 1.27 ± 0.05 a |
| TOC (%) | 0.28 ± 0.02 a | 0.53 ± 0.04 b | 0.53 ± 0.08 b | 0.57 ± 0.07 b |
| SOM (%) | 0.49 ± 0.04 a | 0.92 ± 0.07 b | 0.92 ± 0.14 b | 0.99 ± 0.11 b |
| TN (mg·kg−1) | 321.24 ± 34.02 a | 561.82 ± 39.11 b | 559.34 ± 86.66 b | 556.15 ± 54.04 b |
| TP (mg·kg−1) | 356.10 ± 36.71 a | 522.14 ± 28.29 b | 443.56 ± 21.79 bc | 410.79 ± 18.23 ac |
| TK (mg·kg−1) | 22,370.98 ± 381.40 a | 22,227.74 ± 160.32 ab | 20,854.42 ± 794.73 ab | 20,633.82 ± 572.73 b |
| AP (mg·kg−1) | 9.57 ± 1.93 a | 45.90 ± 7.94 b | 23.58 ± 3.20 c | 18.24 ± 2.21 ac |
| AK (mg·kg−1) | 118.28 ± 7.94 a | 120.17 ± 11.64 a | 124.70 ± 6.35 a | 100.77 ± 8.57 a |
| NH4-N (mg·kg−1) | 2.54 ± 0.43 a | 4.06 ± 0.53 b | 3.53 ± 0.52 ab | 3.60 ± 0.29 ab |
| NO3-N (mg·kg−1) | 57.71 ± 43.14 a | 3.94 ± 2.27 a | 1.86 ± 0.23 a | 10.41 ± 1.63 a |
| ENa (cmol·kg−1) | 6.36 ± 1.09 a | 1.54 ± 0.59 b | 2.46 ± 0.74 b | 1.52 ± 0.42 b |
| CEC (cmol·kg−1) | 16.58 ± 3.05 a | 17.86 ± 0.65 a | 18.87 ± 0.46 a | 15.70 ± 0.98 a |
| ESP (%) | 39.75 ± 3.75 a | 8.39 ± 3.10 b | 13.33 ± 4.06 b | 9.58 ± 2.42 b |
| CK | P4 | P5 | P6 | |
|---|---|---|---|---|
| CO32− (mg·kg−1) | 372.60 ± 154.89 | - | - | - |
| HCO3− (mg·kg−1) | 9274.81 ± 1195.90 a | 543.51 ± 188.02 b | 1963.59 ± 625.71 b | 2685.53 ± 1728.99 b |
| Cl− (mg·kg−1) | 812.95 ± 293.27 a | 108.28 ± 7.46 b | 118.93 ± 9.67 b | 146.44 ± 15.90 b |
| SO42− (mg·kg−1) | 445.65 ± 284.79 ac | 1662.78 ± 165.00 b | 914.52 ± 164.85 a | 331.67 ± 54.04 c |
| Na+ (mg·kg−1) | 2268.71 ± 125.65 a | 664.20 ± 181.23 b | 748.15 ± 178.08 b | 560.57 ± 94.56 b |
| Mg2+ (mg·kg−1) | 373.62 ± 118.29 a | 35.52 ± 13.65 b | 75.64 ± 24.72 b | 169.37 ± 141.28 ab |
| K+ (mg·kg−1) | 46.74 ± 12.45 a | 9.51 ± 1.18 b | 14.10 ± 3.48 b | 22.45 ± 13.92 ab |
| Ca2+ (mg·kg−1) | 2117.52 ± 293.94 a | 302.37 ± 156.33 b | 452.01 ± 132.43 b | 593.93 ± 445.62 b |
| Chao1 | Observed_Species | Shannon | Simpson | |
|---|---|---|---|---|
| SWC (%) | 0.816 ** | 0.812 ** | 0.593 * | 0.320 |
| EC (μS·cm−1) | −0.455 | −0.454 | −0.460 | −0.130 |
| pH | −0.642 ** | −0.637 ** | −0.238 | 0.041 |
| TC (%) | 0.267 | 0.269 | 0.386 | 0.270 |
| TOC (%) | 0.592 * | 0.588 * | 0.523 * | 0.057 |
| SOM (%) | 0.592 * | 0.588 * | 0.523 * | 0.057 |
| TN (mg·kg−1) | 0.660 ** | 0.654 ** | 0.517 * | 0.104 |
| TP (mg·kg−1) | 0.676 ** | 0.668 ** | 0.243 | 0.068 |
| TK (mg·kg−1) | −0.361 | −0.361 | −0.598 * | −0.310 |
| AP (mg·kg−1) | 0.612 * | 0.607 * | 0.184 | 0.612 * |
| AK (mg·kg−1) | 0.001 | 0.004 | 0.064 | 0.270 |
| NH4-N (mg·kg−1) | 0.554 * | 0.542 * | 0.221 | −0.047 * |
| NO3-N (mg·kg−1) | −0.470 | −0.466 | −0.272 | 0.058 |
| ENa (cmol·kg−1) | −0.503 * | −0.499 * | −0.430 | −0.007 |
| CEC (cmol·kg−1) | 0.276 | 0.276 | 0.156 | 0.193 |
| ESP (%) | −0.579 * | −0.575 * | −0.467 | −0.055 |
| CO32− (mg·kg−1) | −0.221 | −0.219 | −0.305 | −0.092 |
| HCO3− (mg·kg−1) | −0.541 * | −0.538 * | −0.461 | −0.132 |
| Cl− (mg·kg−1) | −0.508 * | −0.506 * | −0.372 | −0.074 |
| SO42− (mg·kg−1) | 0.413 | 0.409 | −0.012 | −0.065 |
| Na+ (mg·kg−1) | −0.573 * | −0.570 * | −0.502 * | −0.069 |
| Mg2+ (mg·kg−1) | −0.408 | −0.404 | −0.387 | −0.211 |
| K+ (mg·kg−1) | −0.416 | −0.412 | −0.370 | −0.153 |
| Ca2+ (mg·kg−1) | −0.477 | −0.475 | −0.475 | −0.159 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, G.; Feng, Z.; Xu, Y.; Jin, Y.; Zhang, G.; Hu, J.; Yu, T.; Wang, M.; Liu, M.; Yang, H.; et al. Impact of Phosphogypsum Application on Fungal Community Structure and Soil Health in Saline–Alkali-Affected Paddy Fields. Agronomy 2023, 13, 2726. https://doi.org/10.3390/agronomy13112726
Lu G, Feng Z, Xu Y, Jin Y, Zhang G, Hu J, Yu T, Wang M, Liu M, Yang H, et al. Impact of Phosphogypsum Application on Fungal Community Structure and Soil Health in Saline–Alkali-Affected Paddy Fields. Agronomy. 2023; 13(11):2726. https://doi.org/10.3390/agronomy13112726
Chicago/Turabian StyleLu, Guanru, Zhonghui Feng, Yang Xu, Yangyang Jin, Guohui Zhang, Jiafeng Hu, Tianhe Yu, Mingming Wang, Miao Liu, Haoyu Yang, and et al. 2023. "Impact of Phosphogypsum Application on Fungal Community Structure and Soil Health in Saline–Alkali-Affected Paddy Fields" Agronomy 13, no. 11: 2726. https://doi.org/10.3390/agronomy13112726





