Genome-Wide Analysis and Expression of MYC Family Genes in Tomato and the Functional Identification of slmyc1 in Response to Salt and Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification and Characterization of the Tomato MYC Transcription Factors
2.3. Protein Sequence, Phylogenesis, Gene Structure, Mapping MYC Genes, and Subcellular Localization of SlMYC Analyses
2.4. Analysis of cis-Regulatory Elements of the SlMYC
2.5. Gene Expression Analysis
2.6. Plasmid Construction, Plant Transformation, and Mutant Analysis
2.7. Salt and Mannitol Treatments to slmyc1 Seeds
2.8. Statistical Analysis
3. Results
3.1. Identification and Characterization of SlMYC Genes
3.2. Chromosomal Localization and Synteny Relationship of SlMYC Genes
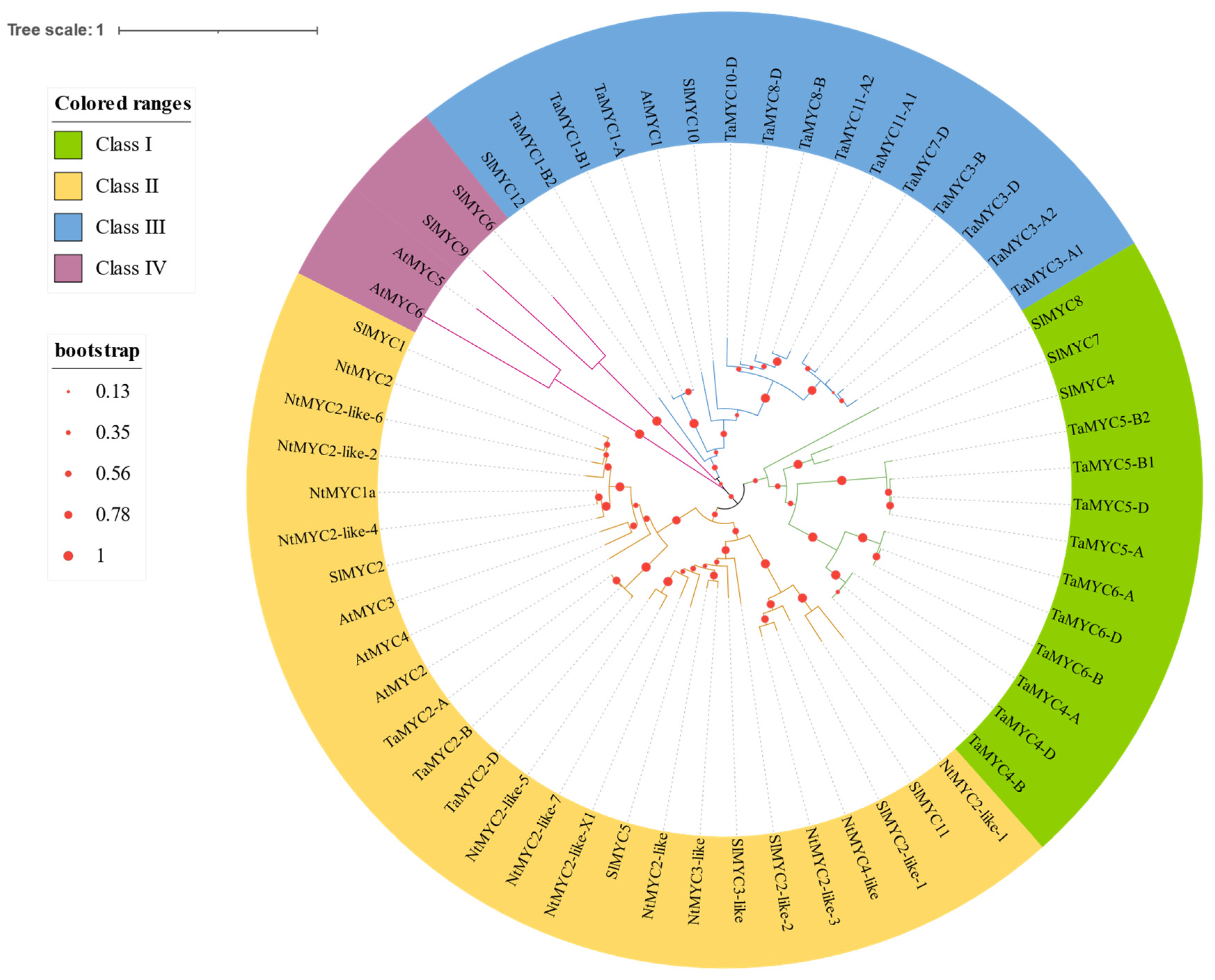
3.3. Phylogenetic Analysis of SlMYC Proteins
3.4. Conserved Motifs and SlMYC Protein Sequence Alignment
3.5. Analysis of Exon–Intron Structure and cis-Regulatory Elements of SlMYC Genes
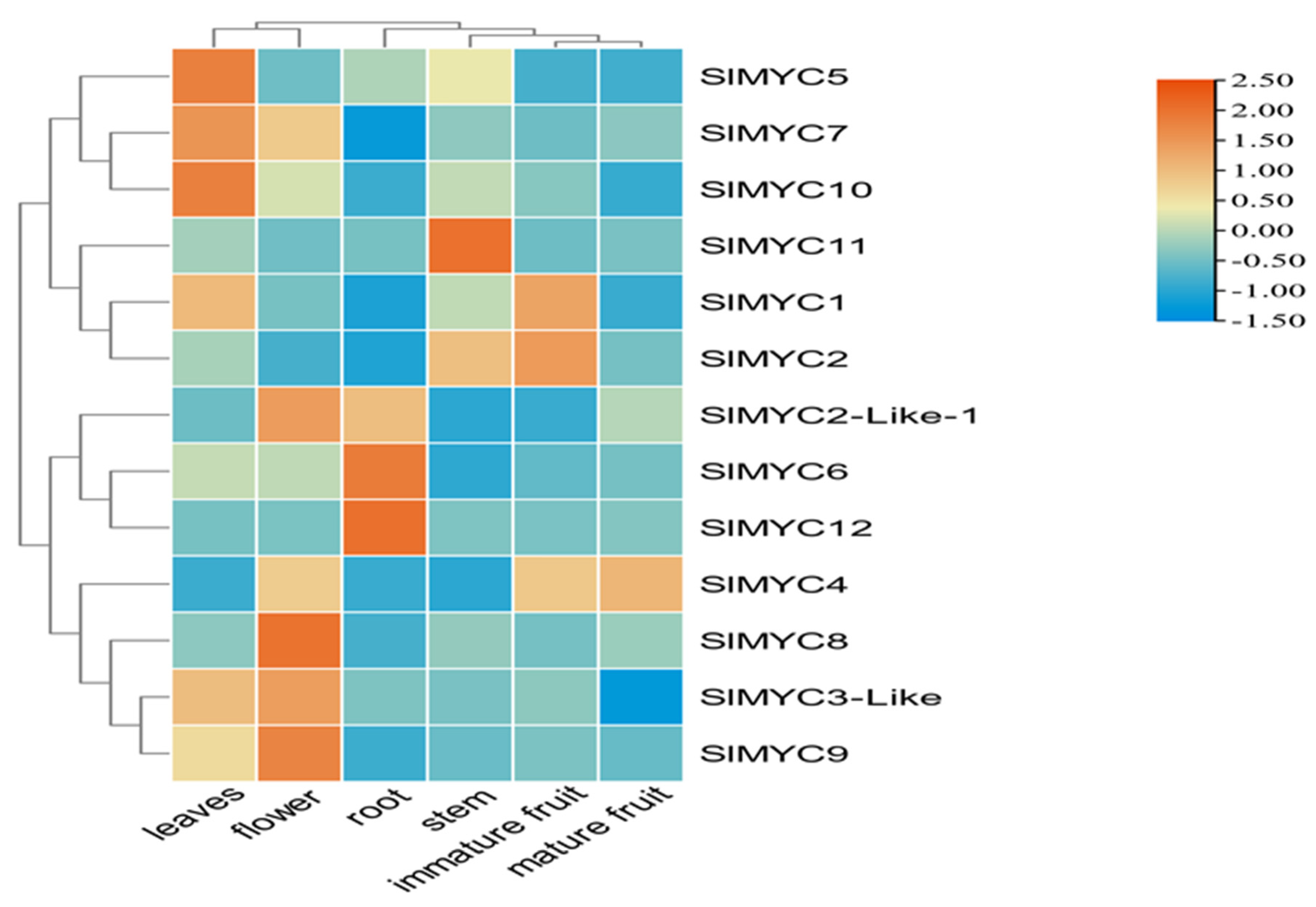
3.6. Expression Profiles of SlMYC in Various Organs of Tomato Plants
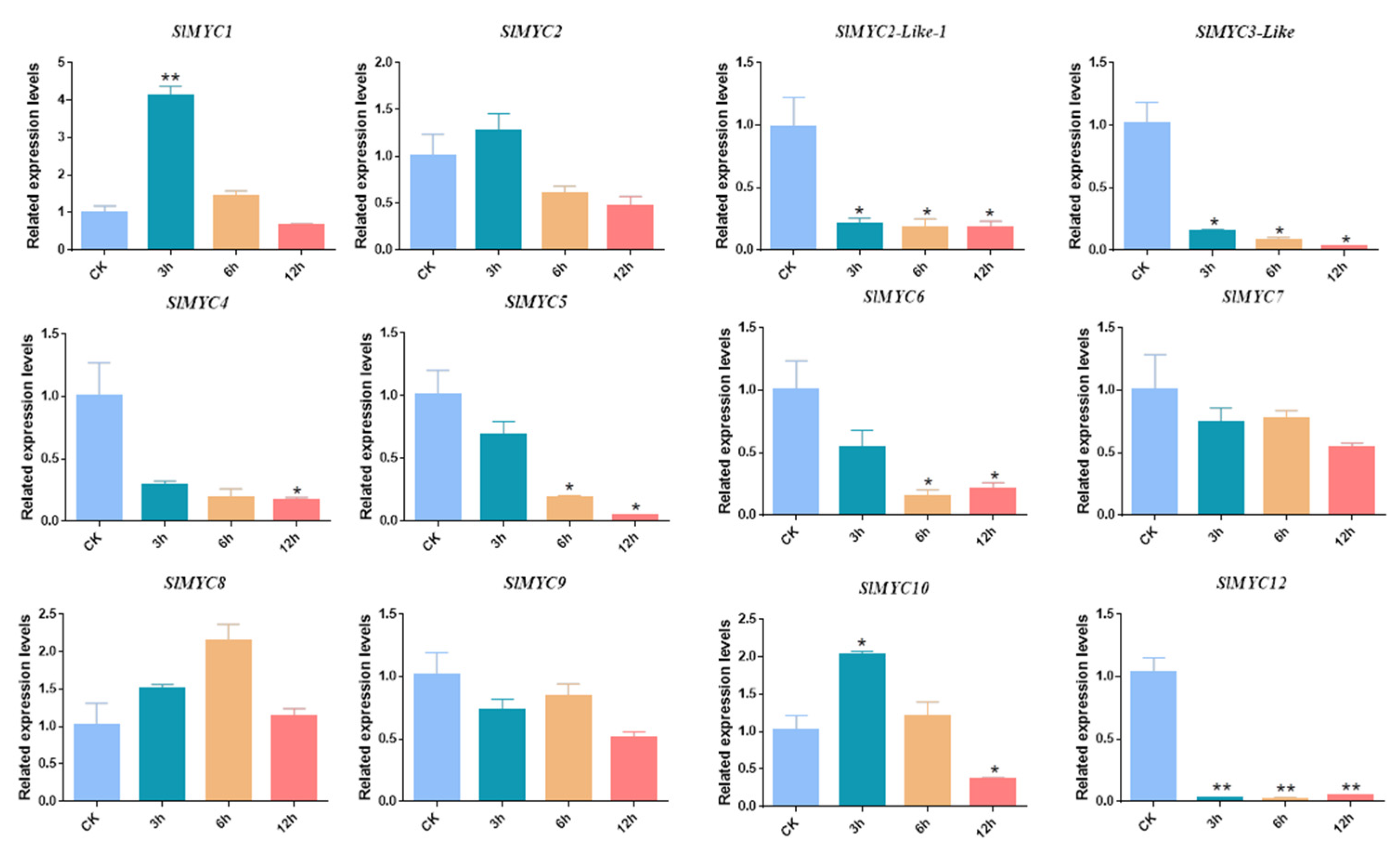
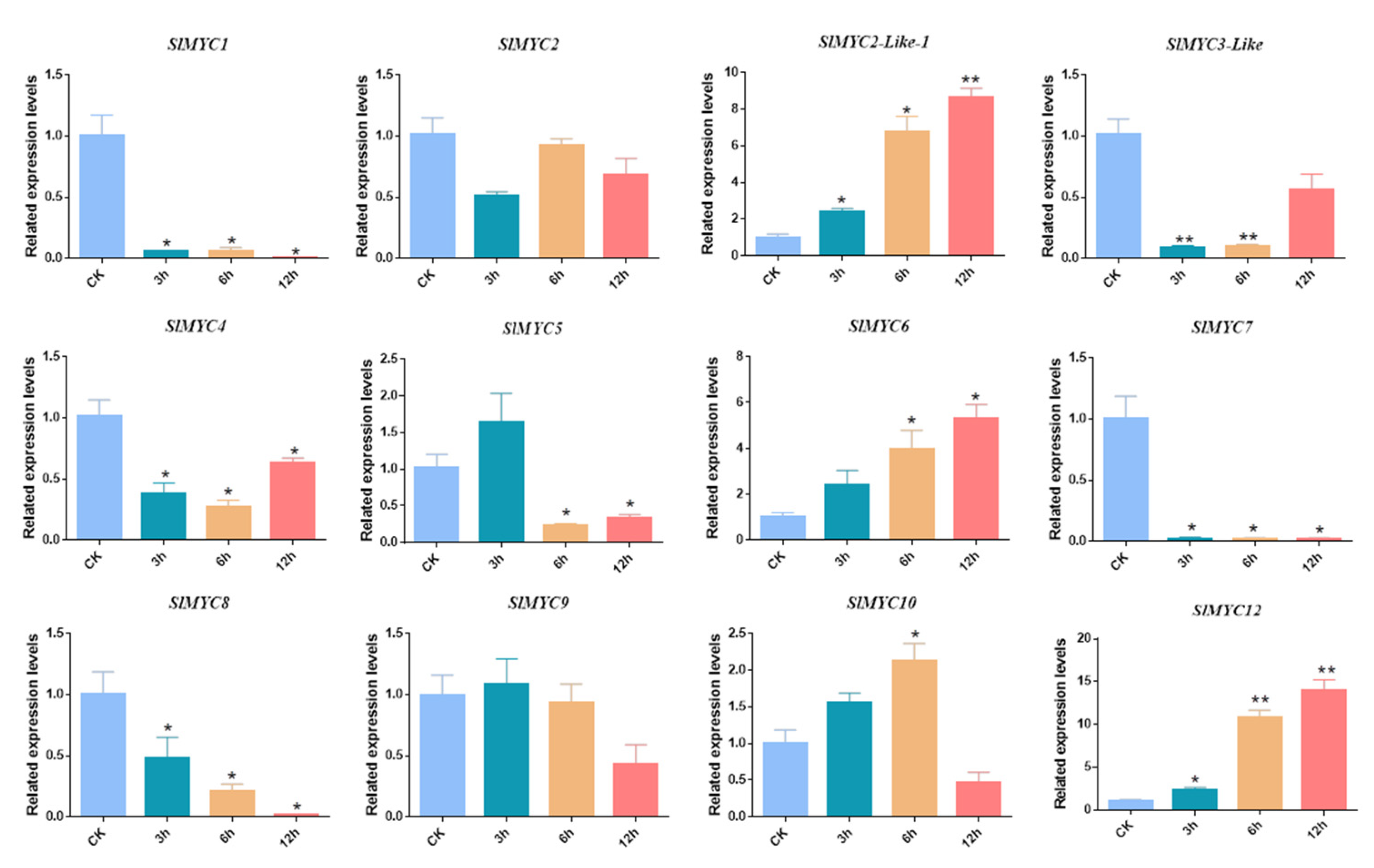
3.7. Expression Patterns of SlMYC under MeJA, NaCl, and Mannitol Stress
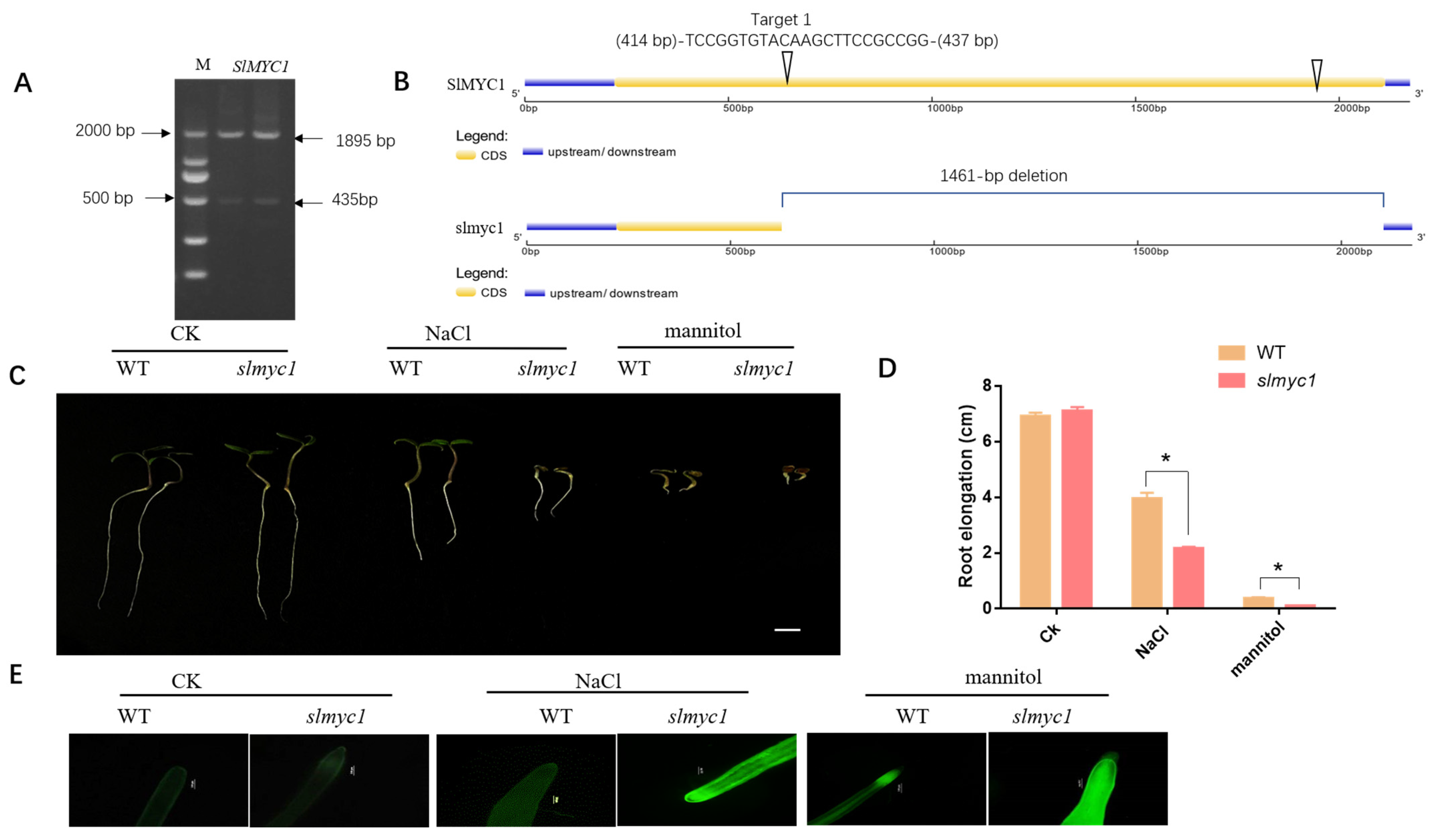
3.8. CRISPR/Cas9-Mediated Mutagenesis in slmyc1
3.9. slmyc1 Had Decreased Tolerance to NaCl and Mannitol Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ferré-D’Amaré, A.R.; Pognonec, P.; Roeder, R.G.; Burley, S.K.J.E.J. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994, 13, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Massari, M.E.; Murre, C. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional Dependence, Cliques, and Predictive Motifs in the bHLH Protein Domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Penuelas, M.; Monte, I.; Schweizer, F.; Vallat, A.; Reymond, P.; Garcia-Casado, G.; Franco-Zorrilla, J.M.; Solano, R. Jasmonate-Related MYC Transcription Factors Are Functionally Conserved in Marchantia polymorpha. Plant Cell 2019, 31, 2491–2509. [Google Scholar] [CrossRef]
- Reinbothe, C.; Springer, A.; Samol, I.; Reinbothe, S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009, 276, 4666–4681. [Google Scholar] [CrossRef]
- Zheng, Y. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, H.; Luo, T.; Liu, Y.; Li, H. Characteristics and Expression Pattern of MYC Genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon. Plants 2019, 8, 274. [Google Scholar] [CrossRef]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D.J.N.C. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef]
- Uji, Y.; Taniguchi, S.; Tamaoki, D.; Shishido, H.; Akimitsu, K.; Gomi, K.J.P.; Physiology, C. Overexpression ofOsMYC2Results in the Up-Regulation of Early JA-Rresponsive Genes and Bacterial Blight Resistance in Rice. Plant Cell Physiol. 2016, 57, 1814–1827. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Xie, D.J.P.C. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef] [PubMed]
- Debora, G.; Aurore, C.; Acosta, I.F.; Jonas, G.; Laurens, P.; Alain, G.; René, D.; Esteban, A.; Farmer, E.E.; Hao, Y.J.P.G. Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth. PLoS Genet. 2015, 11, e1005300. [Google Scholar]
- Qi, T.; Huang, H.; Song, S.; Xie, D.J.P.C. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Qi, S.; Liu, K.; Li, D.; Jin, C.; Li, Z.; Huang, G.; Hai, J.; Zhang, M.; Chen, M.; et al. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 63–70. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef]
- Schweizer, F.; Fernández-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Reymond, S.; Reymond, P. Arabidopsis Basic Helix-Loop-Helix Transcription Factors MYC2, MYC3, and MYC4 Regulate Glucosinolate Biosynthesis, Insect Performance, and Feeding Behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef]
- Yuan, Z.; Xinyuan, X.; Shiming, L.; Wenjie, C.; Bo, Z.; Dengcai, L.; Baolong, L.; Daowen, W.; Huaigang, Z. Allelic Variation and Transcriptional Isoforms of Wheat TaMYC1 Gene Regulating Anthocyanin Synthesis in Pericarp. Front. Plant Sci. 2017, 8, 1645. [Google Scholar]
- Yu, S.O.; Hans-Peter, M.; Kukoeva, T.V.; Andreas, B.; Khlestkina, E.K. Regulation of the Flavonoid Biosynthesis Pathway Genes in Purple and Black Grains of Hordeum vulgare. PLoS ONE 2016, 11, e0163782. [Google Scholar]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB Homologs in Drought and Abscisic Acid-Regulated Gene Expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Yamaguchi-Shinozaki, S. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221. [Google Scholar] [CrossRef] [PubMed]
- Shyu, C.; Figueroa, P.; Depew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.E.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Ma, H.; Gu, X.; Thelen, A.; Brunzelle, J.S.; Li, J.; Xu, H.E.; Melcher, K.J.S.A. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 2015, 1, e1500107. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Chen, Q.; Zhai, Q.; Fang, M.; Li, C.; Shen, J.; Lu, Y.; Deng, L.; Wang, Q.; Liu, Y. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Boter, M.; Ruiz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Gene Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef]
- Panda, S.; Jozwiak, A.; Sonawane, P.D.; Szymanski, J.; Kazachkova, Y.; Vainer, A.; Kilambi, H.V.; Almekias-Siegl, E.; Dikaya, V.; Bocobza, S.; et al. Steroidal alkaloids defence metabolism and plant growth are modulated by the joint action of gibberellin and jasmonate signalling. New Phytol. 2022, 233, 1220–1237. [Google Scholar] [CrossRef]
- Hua, B.; Chang, J.; Wu, M.L.; Xu, Z.J.; Zhang, F.Y.; Yang, M.N.; Xu, H.M.; Wang, L.J.; Chen, X.Y.; Wu, S. Mediation of JA signalling in glandular trichomes by thewoolly/SlMYC1regulatory module improves pest resistance in tomato. Plant Biotechnol. J. 2021, 19, 375–393. [Google Scholar] [CrossRef]
- Xu, J.S.; van Herwijnen, Z.O.; Drager, D.B.; Sui, C.; Haring, M.A.; Schuurink, R.C. SlMYC1 Regulates Type VI Glandular Trichome Formation and Terpene Biosynthesis in Tomato Glandular Cells. Plant Cell 2018, 30, 2988–3005. [Google Scholar] [CrossRef]
- Swinnen, G.; De Meyer, M.; Pollier, J.; Molina-Hidalgo, F.J.; Ceulemans, E.; Venegas-Molina, J.; De Milde, L.; Fernandez-Calvo, P.; Ron, M.; Pauwels, L.; et al. The basic helix-loop-helix transcription factors MYC1 and MYC2 have a dual role in the regulation of constitutive and stress-inducible specialized metabolism in tomato. New Phytol. 2022, 236, 911–928. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Rui, X.; Hao, C.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018, 10, 289660. [Google Scholar]
- Kim, W.-N.; Kim, H.-J.; Chung, Y.-S.; Kim, H.-U. Construction of Multiple Guide RNAs in CRISPR/Cas9 Vector Using Stepwise or Simultaneous Golden Gate Cloning: Case Study for Targeting the FAD2 and FATB Multigene in Soybean. Plants 2021, 10, 2542. [Google Scholar] [CrossRef] [PubMed]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Qi, Q.; Yanyan, D.; Yuanlin, L.; Kunzhi, L.; Huini, X.; Xudong, S. Overexpression of SlMDHAR in transgenic tobacco increased salt stress tolerance involving S-nitrosylation regulation. Plant Sci. 2020, 299, 110609. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K.J.P.P. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genom. 2017, 18, 152. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Yamaguchi-Shinozaki, S.K.J.P.C. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Figueroa, B. The Arabidopsis JAZ2 promoter contains a G-box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. In Proceedings of the Society for Advancement of Hispanics/Chicanos & Native Americans in Science National Conference, Kansas, MO, USA, 28–30 October 2021. [Google Scholar]
- Iwamoto, M.; Maekawa, M.; Saito, A.; Higo, H.; Higo, K. Evolutionary relationship of plant catalase genes inferred from exon-intron structures: Isozyme divergence after the separation of monocots and dicots. Theor. Appl. Genet. 1998, 97, 9–19. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Huang, H.; Wang, J.; Liu, B.; Qi, T.; Xie, D.J.P.; Physiology, C. MYC5 Is Involved in Jasmonate-regulated Plant Growth, Leaf Senescence and Defense Responses. Plant Cell Physiol. 2017, 58, 1752. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.F.; Wang, Y.K.; Guo, L.P.; Guo, X.M.; Guo, H.Y.; Yuan, S.H.; Duan, W.J.; Liu, Z.; Zhao, C.P.; Zhang, F.T.; et al. Genomic identification and characterization of MYC family genes in wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 1032. [Google Scholar] [CrossRef] [PubMed]
- Adrián, G.-B.; Figueroa, N.E.; Figueroa, P.M.; Figueroa, C.R.; Blazquez, M.A.J.P.O. Jasmonate signalling pathway in strawberry: Genome-wide identification, molecular characterization and expression of JAZs and MYCs during fruit development and ripening. PLoS ONE 2018, 13, e0197118. [Google Scholar]
- Strygina, K.V.; Khlestkina, E.K. Myc-like transcriptional factors in wheat: Structural and functional organization of the subfamily I members. BMC Plant Biol. 2019, 19, 50. [Google Scholar] [CrossRef]
- Wei, K.; Chen, H. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 2018, 18, 309. [Google Scholar] [CrossRef]
- Yanfang, Y.; Kaikai, Z.; Liying, Y.; Xing, L.; Ying, W.; Hongwei, L.; Qiang, L.; Duanfen, C.; Deyou, Q. Identification and characterization of MYC transcription factors in Taxus sp. Gene 2018, 675, 1–8. [Google Scholar] [CrossRef]




| Gene | Accession Number | Chr. Location | No. of Amino Acids (aa) | Length of Gene (bp) | Length of CDS (bp) | Molecular Weight (Da) | Predicted Isoelectric Point (PI) | Subcellular Localization Prediction | GRAVY |
|---|---|---|---|---|---|---|---|---|---|
| MYC1 | NM_001301178.2 | Chromosome 8: 33,463–35,295 | 630 | 2174 | 1893 | 69,346.51 | 5.78 | Nucleus | −0.564 |
| MYC2 | NM_001324483.1 | Chromosome 8: 60,990,466–60,992,535 | 689 | 2485 | 2070 | 75,041.11 | 5.51 | Nucleus | −0.598 |
| MYC2-like-1 | XM_004244608.3 | Chromosome 8: 2,993,511–2,996,747 | 452 | 1556 | 1359 | 51,296.03 | 6.47 | Nucleus | −0.431 |
| MYC2-like-2 | XM_004248047.4 | Chromosome 10: 3,248,348–3,250,830 | 393 | 1608 | 1182 | 45,112.04 | 5.85 | Nucleus | −0.454 |
| MYC3-like | XM_004248044.4 | Chromosome 10: 3,273,458–3,274,813 | 451 | 1769 | 1356 | 50,463.80 | 5.98 | Nucleus | −0.435 |
| MYC4 | XM_004229943.4 | Chromosome 1: 87,017,635–87,020,664 | 605 | 3030 | 1818 | 67,261.91 | 6.83 | Nucleus | −0.528 |
| MYC5 | XM_004229974.4 | Chromosome 1: 87,325,345–87,327,252 | 450 | 1665 | 1353 | 49,496.80 | 5.44 | Nucleus | −0.348 |
| MYC6 | XM_010322267.3 | Chromosome 5: 240,028–243,178 | 482 | 2143 | 1449 | 54,013.27 | 5.84 | Nucleus | −0.394 |
| MYC7 | XM_010323205.3 | Chromosome 5: 61,560,052–61,562,617 | 579 | 2556 | 1740 | 64,271.00 | 7.59 | Nucleus | −0.425 |
| MYC8 | XM_004242307.4 | Chromosome 6: 49,306,848–49,309,630 | 477 | 2182 | 1434 | 52,944.89 | 6.59 | Nucleus | −0.472 |
| MYC9 | XM_019215077.2 | Chromosome 8: 51,962,518–51,966,949 | 602 | 2432 | 1809 | 68,432.78 | 5.61 | Nucleus | −0.874 |
| MYC10 | NM_001347001.1 | Chromosome 8: 64,341,490–64,346,297 | 626 | 2353 | 1881 | 70,053.83 | 5.49 | Nucleus | −0.546 |
| MYC11 | XM_004246037.4 | Chromosome 8: 65,832,927–65,835,049 | 413 | 2123 | 1242 | 46,922.74 | 8.41 | Nucleus | −0.234 |
| MYC12 | XM_010328066.3 | Chromosome 9: 63,177,650–63,185,525 | 686 | 2425 | 2061 | 75,694.23 | 5.10 | Nucleus | −0.407 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Zeng, S.; Yan, J.; Li, K.; Xu, H. Genome-Wide Analysis and Expression of MYC Family Genes in Tomato and the Functional Identification of slmyc1 in Response to Salt and Drought Stress. Agronomy 2023, 13, 757. https://doi.org/10.3390/agronomy13030757
Feng Y, Zeng S, Yan J, Li K, Xu H. Genome-Wide Analysis and Expression of MYC Family Genes in Tomato and the Functional Identification of slmyc1 in Response to Salt and Drought Stress. Agronomy. 2023; 13(3):757. https://doi.org/10.3390/agronomy13030757
Chicago/Turabian StyleFeng, Yang, Senlin Zeng, Jinping Yan, Kunzhi Li, and Huini Xu. 2023. "Genome-Wide Analysis and Expression of MYC Family Genes in Tomato and the Functional Identification of slmyc1 in Response to Salt and Drought Stress" Agronomy 13, no. 3: 757. https://doi.org/10.3390/agronomy13030757
APA StyleFeng, Y., Zeng, S., Yan, J., Li, K., & Xu, H. (2023). Genome-Wide Analysis and Expression of MYC Family Genes in Tomato and the Functional Identification of slmyc1 in Response to Salt and Drought Stress. Agronomy, 13(3), 757. https://doi.org/10.3390/agronomy13030757







