Biological and Molecular Characterization of Clover Yellow Vein Virus Infecting Trifolium repens in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral Source
2.2. Electron Microscopy
2.3. Sap Transmission Experiments
2.4. RNA Sequencing and RT-PCR Validation
2.5. Determination of ClYVV-IM Genome Sequence
2.6. Genetic Variation, Phylogenetic and Recombination Analysis
3. Results
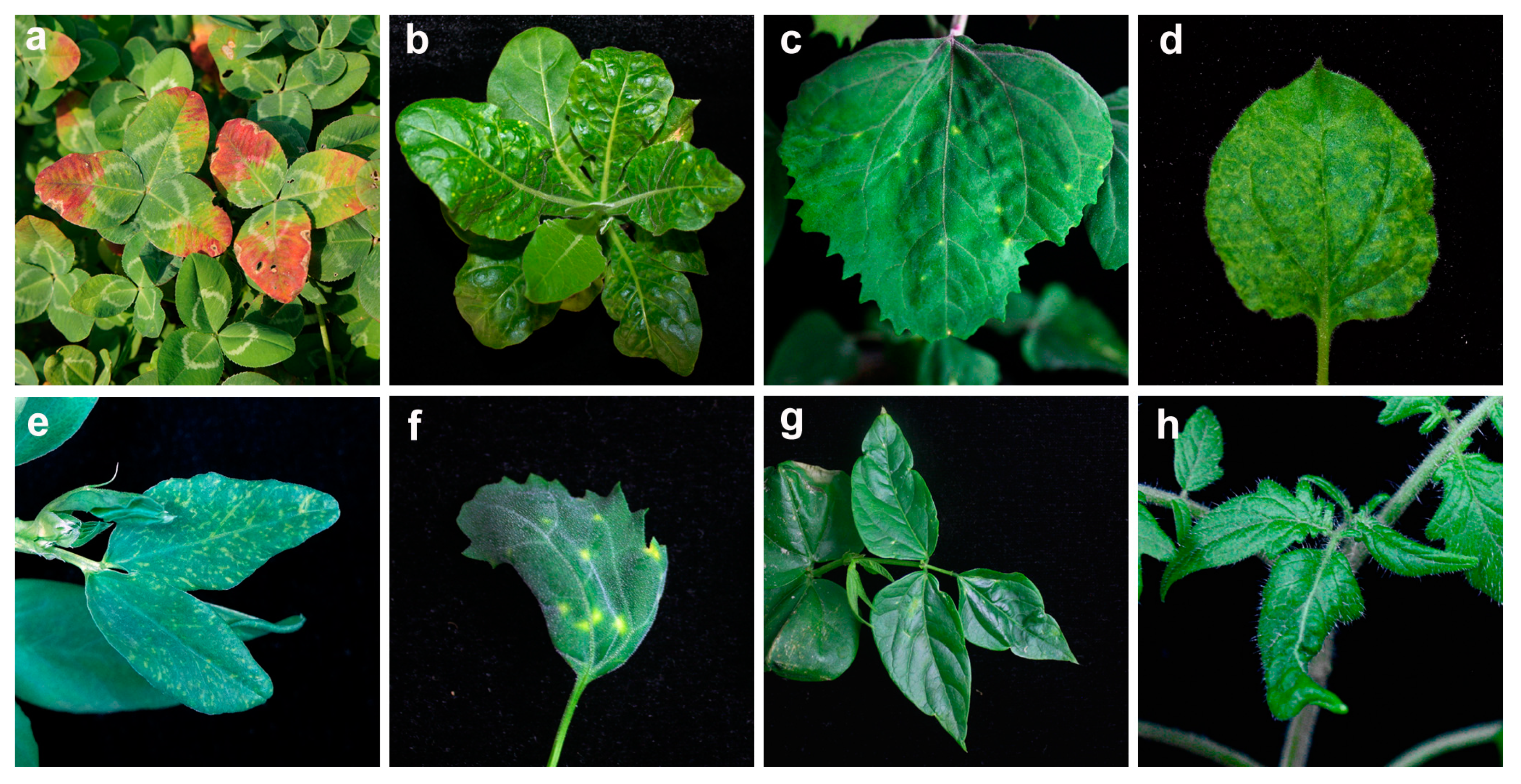
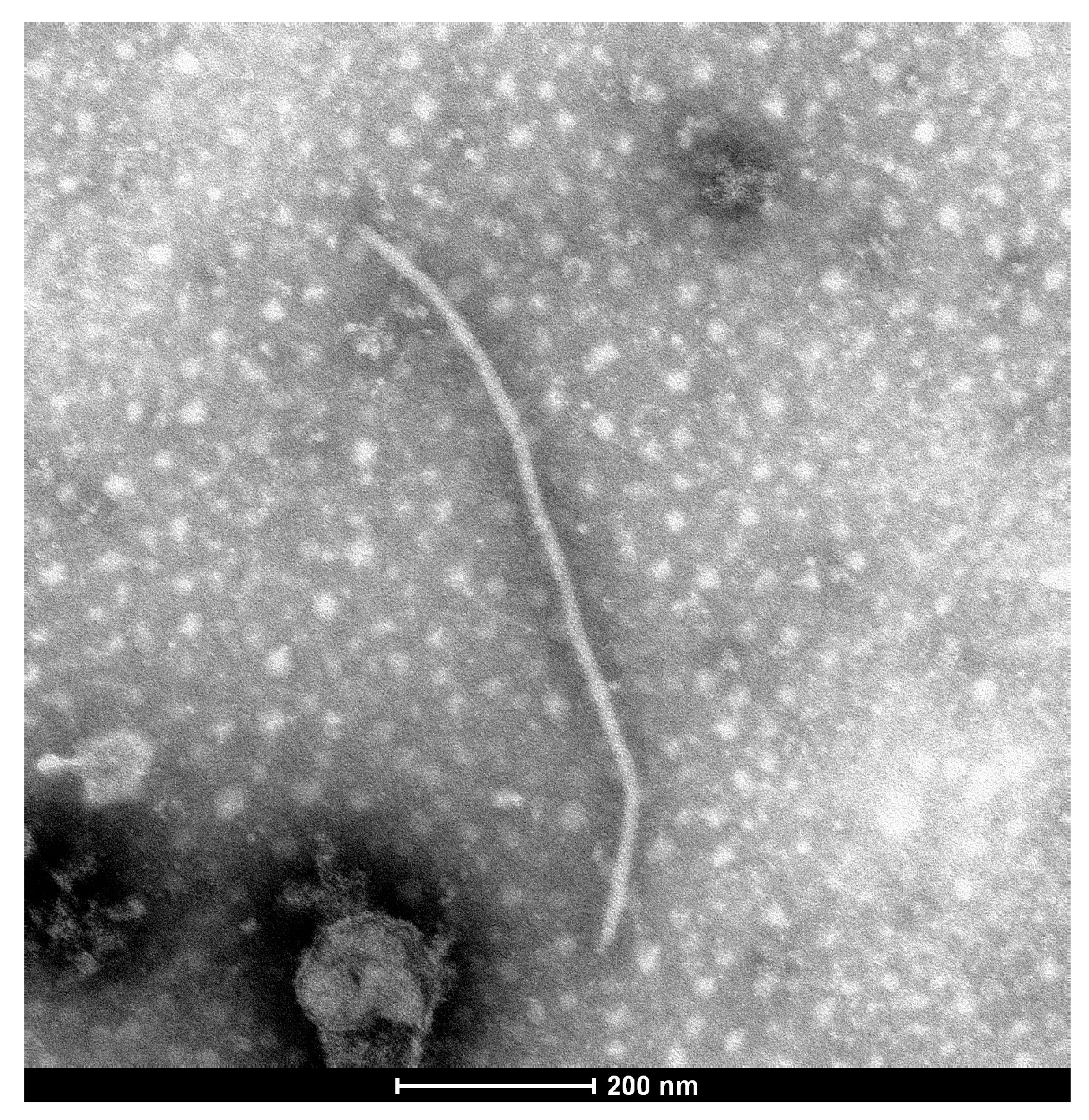
3.1. Electron Microscopy
3.2. Sap Transmission Experiments
3.3. Small RNA Sequencing and RT-PCR Validation
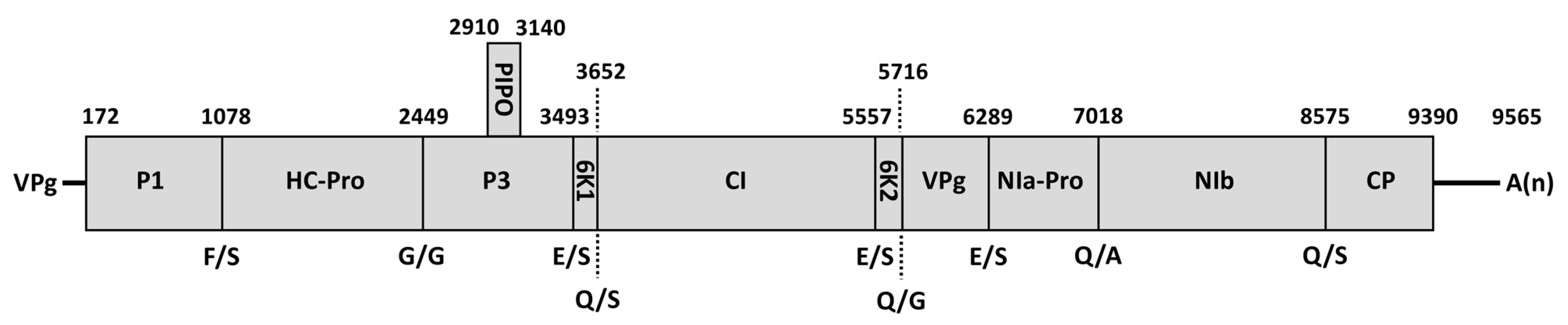
3.4. Characterization of ClYVV-IM Genome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Isobe, S.N.; Hisano, H.; Sato, S.; Hirakawa, H.; Okumura, K.; Shirasawa, K.; Sasamoto, S.; Watanabe, A.; Wada, T.; Kishida, Y. Comparative genetic mapping and discovery of linkage disequilibrium across linkage groups in white clover (Trifolium repens L.). G3-Genes Genom. Genet. 2012, 2, 607–617. [Google Scholar] [CrossRef]
- Wu, F.; Ma, S.; Zhou, J.; Han, C.; Zhang, X. Genetic diversity and population structure analysis in a large collection of white clover (Trifolium repens L.) germplasm worldwide. PeerJ 2021, 9, e11325. [Google Scholar] [CrossRef] [PubMed]
- Abberton, M.T.; Fothergill, M.; Collins, R.P.; Marshall, A.H. Breeding forage legume for sustainable and profitable farming systems. Asp. Appl. Biol. 2006, 80, 81–87. [Google Scholar]
- Griffiths, A.G.; Barrett, B.A.; Simon, D.; Khan, A.K.; Bickerstaff, P.; Anderson, C.B.; Franzmayr, B.K.; Hancock, K.R.; Jones, C.S. An integrated genetic linkage map for white clove (Trifolium repens L.) with alignment to Medicago. BMC Genom. 2013, 14, 388. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Wei, L.; Xu, B.; Calderón-Urrea, A.; Xiang, D. Study of viruses co-infecting white clover (Trifolium repens) in China. J. Integr. Agric. 2017, 16, 1990–1998. [Google Scholar] [CrossRef]
- Panter, S.; Chu, P.G.; Ludlow, E.; Garrett, R.; Kalla, R.; Jahufer, M.Z.Z.; de Lucas Arbiza, A.; Rochfort, S.; Mouradow, A.; Smith, K.F.; et al. Molecular breeding of transgenic white clover (Trifolium repens L.) with field resistance to Alfalfa mosaic virus through the expression of its coat protein gene. Transgenic Res. 2012, 21, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Hart, J.P.; Griffiths, P.D. Resistance to Clover yellow vein virus in common bean germplasm. Crop Sci. 2014, 54, 2609–2617. [Google Scholar] [CrossRef]
- Hisa, Y.; Suzuki, H.; Atsumi, G.; Choi, S.H.; Nakahara, K.S.; Uyeda, I. P3N-PIPO of Clover yellow vein virus exacerbates symptoms in pea infected with White clover mosaic virus and is implicated in viral synergism. Virology 2014, 449, 200–206. [Google Scholar] [CrossRef]
- Chuang, B.Y.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef]
- Hollings, M.; Nariani, T.K. Some properties of clover yellow vein, a virus from Trifolium repens L. Ann. Appl. Biol. 1965, 56, 99–109. [Google Scholar] [CrossRef]
- Hart, J.P.; Griffiths, P.D. A serious of eIF4E alleles at the Bc-3 locus are associated with recessive resistance to Clover yellow vein virus in common bean. Theor. Appl. Genet. 2013, 126, 2849–28663. [Google Scholar] [CrossRef] [PubMed]
- Li, C.S.; Zhu, H.C. Identification of Clover yellow vein virus infecting Phaseolus vulgaris. J. Shandong Agric. Univ. 1989, 4, 223–226. [Google Scholar]
- Li, C.S. Identification of Clover yellow vein virus infecting Dolichos lablab. Virol. Sin. 1991, 6, 223–226. [Google Scholar]
- Zhang, C.; Zheng, H.; Yan, D.; Han, K.; Peng, J.; Lu, Y. Identification of Clover yellow vein virus infecting broad bean in China by deep sequencing and characterization of virus-derived small interfering RNAs. Acta Agric. Univ. Zhejiangensis 2018, 30, 406–412. [Google Scholar]
- Song, S.; Cui, J.; Lei, G.; Chen, Y.; Yang, M.; Li, Z.; Zhang, J. Occurrence, infectivity and molecular characterization of hosta virus X in North-east China. Can. J. Plant Pathol. 2020, 42, 595–603. [Google Scholar] [CrossRef]
- Li, Z.; Sun, P.; Zhang, L.; Song, S. First report of basella rugose mosaic virus infecting Anredera cordifolia in mainland of China. Crop Prot. 2021, 139, 105350. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Palese, P. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patters in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Nury, S.; Hosseini, A.; Gibbs, A.J.; Mohammadi, M. Poisono hemlock virus (PHVY), a novel potyvirus from Iranian Conium maculatum (Apiaceae). Australas Plant Pathol. 2020, 49, 119–216. [Google Scholar] [CrossRef]
- Denny, B.L.; Guy, P.L. Incidence and spread of viruses in white-clover pastures of the South Island, New Zealand. Australas Plant Path. 2009, 38, 270–276. [Google Scholar] [CrossRef]
- Pratt, M.J. Studies on clover yellow mosaic and white clover mosaic viruses. Can. J. Bot. 2011, 39, 655–665. [Google Scholar] [CrossRef]
- Fletcher, J.; Tang, J.; Blouin, A.; Ward, L.; MacDiarmid, R.; Ziebell, H. Red clover mosaic virus- A novel virus to New Zealand that is widespread in Legumes. Plant Dis. 2016, 100, 890–895. [Google Scholar] [CrossRef]
- Larsen, R.C.; Miklas, P.N.; Eastwell, K.C.; Grau, C.R. A strain of Clover yellow vein virus that causes severe pod necrosis disease in snap bean. Plant Dis. 2008, 92, 1026–1032. [Google Scholar] [CrossRef]
- Liang, P.; Navarro, B.; Zhang, Z.; Wang, H.; Lu, M. Identification and characterization of a novel geminivirus with a monopartite genome infecting apple tree. J. Gen. Virol. 2015, 96, 2411–2420. [Google Scholar] [CrossRef]
- Roossinck, M.J. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 1997, 35, 191–209. [Google Scholar] [CrossRef]
- Boulila, M. Molecular evidence for recombination in Prunus necrotic ringspot virus. Plant Mol. Biol. Rep. 2009, 27, 189–198. [Google Scholar] [CrossRef]
- Li, X.; Zhu, T.; Yin, X.; Zhang, C.; Chen, J.; Tian, Y.; Liu, J. The genetic structure of Turnip mosaic virus population reveals the rapid expansion of a new emergent lineage in China. Virol. J. 2017, 14, 165. [Google Scholar] [CrossRef] [PubMed]
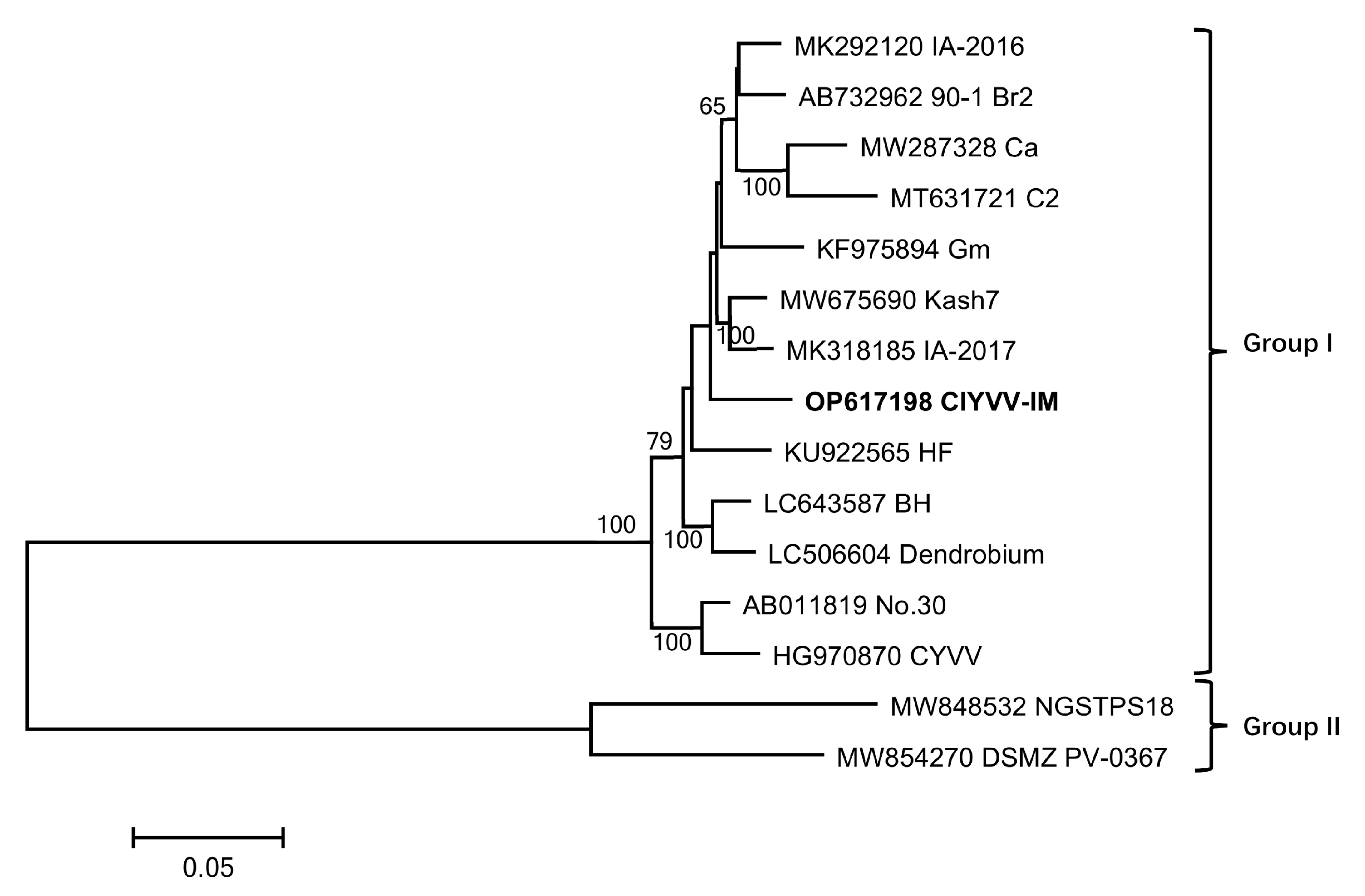
| Isolate | GenBank No. | Host | Country | Genome Identities (%) | Polyprotein Identities (%) | |
|---|---|---|---|---|---|---|
| Nucleotide | Nucleotide | Amino Acid | ||||
| Kash7 a | MW675690 | Phaseolus vulgaris | India | 96.30 | 96.32 | 98.60 |
| IA-2017 a | MK318185 | Glycine max | USA | 96.10 | 96.10 | 98.50 |
| IA-2016 a | MK292120 | Glycine max | USA | 96.02 | 96.02 | 98.80 |
| HF a | KU922565 | Vicia faba | China | 95.62 | 95.63 | 98.40 |
| BH a | LC643587 | Aquilegia buergeriana | South Korea | 95.61 | 95.50 | 98.50 |
| Gm a | KF975894 | Glycine max | South Korea | 95.38 | 95.37 | 98.60 |
| Dendrobium a | LC506604 | Dendrobium spp. | South Korea | 95.33 | 95.31 | 98.60 |
| No. 30 a | AB011819 | - | - | 95.16 | 95.02 | 98.70 |
| Ca a | MW287328 | Centella asiatica | USA | 94.57 | 94.52 | 97.90 |
| NGSTPS18 a | MW848532 | Vicia faba | Germany | 83.05 | 82.91 | 92.80 |
| CYVV b | HG970870 | Trifolium repens | Australia | 94.34 | 98.50 | |
| C2 b | MT631721 | Crotalaria micans | USA | 93.94 | 97.70 | |
| DSMZ-PV-0367 b | MW854270 | Phaseolus vulgaris | Germany | 83.15 | 93.10 | |
| 90–1 Br2 b | AB732962 | Pisum sativum | Japan | 95.85 | 98.70 | |
| Event No. | Recombinant | Parental Isolate | Region (nt) | p-Value a | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Major | Minor | R | G | B | M | C | S | 3S | |||
| 1 | IM | Gm | No. 30 | 5153–5694 | 1.0 × 10−11 | 3.33 × 10−11 | 1.62 × 10−12 | 3.80 × 10−8 | 9.77 × 10−9 | 8.62 × 10−13 | 3.59 × 10−10 |
| Kash7 | |||||||||||
| IA-2017 | |||||||||||
| IA-2016 | |||||||||||
| HF | |||||||||||
| BH | |||||||||||
| Dendrobium | |||||||||||
| 2 | IM | Dendrobium | HF | 914–2970 | 6.6 × 10−5 | - | 2.97 × 10−3 | 7.53 × 10−8 | 3.30 × 10−7 | 9.70 × 10−10 | 1.77 × 10−23 |
| 3 | IA-2016 | Kash7 | Ca | 1999–4539 | 5.0 × 10−7 | - | 1.62 × 10−4 | 2.42 × 10−9 | 5.20 × 10−10 | 3.55 × 10−5 | 3.58 × 10−4 |
| 4 | HF | IA-2017 | No. 30 | 3069–4579 | 8.8 × 10−11 | 3.47 × 10−11 | 9.25 × 10−12 | 2.15 × 10−12 | 1.54 × 10−13 | 4.17 × 10−14 | 9.64 × 10−17 |
| 5 | BH | IA-2016 | No. 30 | 3676–4181 | 1.41 × 10−4 | 4.29 × 10−4 | 1.67 × 10−4 | 5.43 × 10−10 | 3.31 × 10−9 | 2.74 × 10−10 | 7.76 × 10−9 |
| 6 | BH | No. 30 | IA-2017 | 3676–4190 | 8.86 × 10−11 | 3.47 × 10−11 | 9.25 × 10−12 | 2.15 × 10−12 | 1.54 × 10−13 | 4.17 × 10−14 | 9.64 × 10−17 |
| 7 | BH | IA-2017 | IA-2016 | 2971–4190 | 2.41 ×10−4 | 3.40 × 10−4 | 1.15 × 10−4 | 4.11 × 10−4 | 2.13 × 10−4 | 9.96 × 10−7 | 1.12 × 10−5 |
| 8 | BH | Dendrobium | IM | 69–406 | 3.55 × 10−5 | 9.55 × 10−4 | 3.59 × 10−5 | 6.21 × 10−5 | 1.17 × 10−6 | 3.80 × 10−7 | 6.90 × 10−9 |
| 9 | BH | Dendrobium | No. 30 | 835–2702 | 2.39 × 10−21 | 4.95 × 10−10 | 4.16 × 10−12 | 9.15 × 10−21 | 3.79 × 10−18 | 4.15 × 10−15 | 4.49 × 10−33 |
| 10 | Dendrobium | Gm | Ca | 867–1662 | 3.43 × 10−11 | 4.18 × 10−4 | 2.57 × 10−10 | 1.59 × 10−7 | 1.26 × 10−6 | 6.02 × 10−10 | 3.13 × 10−4 |
| 11 | Dendrobium | IA-2017 | IA-2016 | 2842–3675 | 2.41 × 10−4 | 3.40 × 10−4 | 1.15 × 10−4 | 4.11 × 10−4 | 2.13 × 10−4 | 9.96 × 10−7 | 1.12 × 10−5 |
| 12 | Dendrobium | IA-2016 | No. 30 | 3676–4181 | 1.41 × 10−4 | 4.29 × 10−4 | 1.67 × 10−4 | 5.43 × 10−10 | 3.31 × 10−9 | 2.74 × 10−10 | 7.76 × 10−9 |
| 13 | No. 30 | Dendrobium | HF | 948–1599 | 6.69 × 10−5 | - | 2.97 × 10−3 | 7.53 × 10−8 | 3.30 × 10−7 | 9.70 × 10−10 | 1.77 × 10−23 |
| 14 | No. 30 | IM | BH | 2703–5694 | 5.53 × 10−11 | 3.66 × 10−7 | 2.20 × 10−12 | 3.78 × 10−6 | 3.93 × 10−3 | 1.14 × 10−16 | 1.44 × 10−50 |
| 15 | Ca | IA-2016 | No. 30 | 4540–5694 | 3.00 × 10−14 | - | 2.97 × 10−12 | 2.72 × 10−15 | 1.44 × 10−16 | - | 4.76 × 10−26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Xu, L.; Sun, P.; Zhu, M.; Zhang, L.; Zhang, B.; Song, S. Biological and Molecular Characterization of Clover Yellow Vein Virus Infecting Trifolium repens in China. Agronomy 2023, 13, 1193. https://doi.org/10.3390/agronomy13051193
Li Z, Xu L, Sun P, Zhu M, Zhang L, Zhang B, Song S. Biological and Molecular Characterization of Clover Yellow Vein Virus Infecting Trifolium repens in China. Agronomy. 2023; 13(5):1193. https://doi.org/10.3390/agronomy13051193
Chicago/Turabian StyleLi, Zhengnan, Lei Xu, Pingping Sun, Mo Zhu, Lei Zhang, Bin Zhang, and Shuang Song. 2023. "Biological and Molecular Characterization of Clover Yellow Vein Virus Infecting Trifolium repens in China" Agronomy 13, no. 5: 1193. https://doi.org/10.3390/agronomy13051193
APA StyleLi, Z., Xu, L., Sun, P., Zhu, M., Zhang, L., Zhang, B., & Song, S. (2023). Biological and Molecular Characterization of Clover Yellow Vein Virus Infecting Trifolium repens in China. Agronomy, 13(5), 1193. https://doi.org/10.3390/agronomy13051193








