Abstract
Castanea sativa Mill. is a valuable species with historical and economic importance in Europe, particularly in the Mediterranean area. In Italy, chestnut cultivation has been developed for centuries, leading to the recognition of more than 300 varieties. Nevertheless, a profusion of local names has been assigned by growers, causing the occurrence of synonyms and homonyms across the country. This research focused on genetic characterization and identification using 21 single sequence repeats (SSRs) for four chestnut varieties (i.e., Pastinese, Nerattino, Carpinese, and Rossola) commonly used for flour production in the Tuscan Apennine Mountains (Pistoia Province). A comprehensive number of 55 accessions identified by local growers as belonging to the four varieties were analyzed, in addition to a few “Marrone” accessions as outgroups. The 21 microsatellites were highly informative, detecting 98 alleles and displaying an average polymorphism information content (PIC) equal to 0.582. In addition, a considerable amount of genetic diversity was revealed, as shown by the heterozygosity levels (He = 0.634 and Ho = 0.475). The STRUCTURE analysis provided clear distinctions among the different varieties, splitting them into four separate groups. This result was also confirmed by UPGMA dendrogram and principal co-ordinates analysis (PCoA). However, one accession (Carp_5), previously identified as Carpinese, showed an allelic profile attributable to Pastinese, suggesting that farmers might have performed mislabeling or grafting propagation errors. Thus, our results confirm the use of SSRs to allocate the accessions of different varieties, uncovering possible synonyms and homonyms. Specifically, in the context of the Pistoiese mountain region, this tool can favor the traceability of processed products, such as flour, enhancing the quality and economic value of the local market. Lastly, our findings have revealed a considerable genetic variability within the Tuscan chestnut varieties whose preservation is mandatory to face climate change challenges through sustainable forest management practices.
1. Introduction
The European or sweet chestnut (Castanea sativa Mill.) is the sole species of the genus Castanea (Fagaceae; x = 12, 2n = 24), which is widespread in Europe, especially in the Mediterranean region from the Iberian Peninsula to the Caspian Sea [1,2]. Chestnut cultivation has an ancient history, being a pre-eminent and essential food source of rural mountain populations in many countries for centuries [3,4]. Once imported into Italy by the Greeks about 5000 years ago [5], the sweet chestnut was expanded into Europe by the Roman Empire, reaching the current areas of cultivation. [6,7,8,9]. Moreover, during the Middle Ages, chestnut production intensified in many mountain areas, such as in the Italian Apennines [7], leading to a diversification of the final product as well as of the adopted varieties [3]. Nowadays, chestnut trees have been cultivated in Italy for over 2000 years. They can be found in many Italian regions, thriving in natural forests, specialized orchards, and, often, in mixed situations due to the partial or periodic abandonment of harsh mountain territories resulting from societal and economic changes that have occurred in the last decades [10].
On the other hand, along with the diffusion and cultivation of chestnut, a wide range of European C. sativa varieties were selected in relation to the suitability of this species to produce both wood and fruit [11]. Indeed, due to the different Italian environmental conditions in many regions, more than 300 varieties have been detected and are widespread in the whole country [3,12]. As a matter of fact, a plethora of local names have been attributed to chestnut populations based on their morphological traits. This led to a varietal misunderstanding with a large number of homonyms and synonyms diffused in all Italian territories [13,14,15], including Tuscany, as reported by Breviglieri [16]. Nowadays, the Italian national and Tuscan regional policies are supporting the recovery of old chestnut orchards, as well as the establishment of new plantations aiming to take advantage of the multipurpose functions of this species related to environmental aspects (e.g., CO2 sequestration and soil conservation), social wellbeing (e.g., recreation and leisure areas), economic reasons (e.g., income increase in mountain marginal areas), human diet, etc. [17]. In this regard, and to obtain commercial quality certification (e.g., the European Protected Designation of Origin), it is necessary to select true-to-type accessions as mother plants for further agamic nursery filiation.
Thus, chestnut varietal characterization has been carried out in the last decades using different approaches [18,19] as tools to support traditional identification based on the morphological and pomological descriptors developed by the International Union of New Varieties of Plants (UPOV) [14,20]. Many studies have focused on recognition using chemical and enzymatic assays [4,21,22,23,24,25,26]. Another interesting procedure is based on the application of molecular markers such as RAPDs, ISSRs, EST-SSRs, and AFLPs, which have been used in many pieces of research to detect the genetic diversity of different chestnut varieties and cultivars [3,27,28,29,30,31]. Among them, the most used and reliable tool is focused on simple sequence repeats (SSRs), which are broadly adopted to also assess the genetic variability of many fruit species [32,33,34,35,36,37,38]. In this context, their application in C. sativa germplasm has already been developed, leading to the genetic characterization of several varieties in the whole of Europe [3,39,40,41,42,43]. Specifically, SSR markers have been previously used to elucidate the relationships among European and Italian chestnut varieties [5,44,45,46,47] and assess the genetic diversity of cultivars and accessions across Italy, spanning from the southern to northern regions [15,19,48,49,50]. These investigations allowed for the identification of redundant accessions, homonyms, synonyms, and intra-varietal clones. Notably, the varietal panorama of Italian chestnuts is quite complex, encompassing numerous traditional dominant cultivars derived from the selection of a vast array of native ecotypes that have adapted to diverse geographic regions. Particularly in Central Italy, the application of SSR markers has enabled the accurate identification of predominant varieties from the Tuscan-Emilian Apennines area [19,48,49]. Alessandri et al. [49] examined the genetic diversity of 134 C. sativa accessions in the Emilia-Romagna region, establishing guidelines for the characterization and varietal certification of chestnut varieties in that specific area. Furthermore, Cavallini et al. [50] employed SSR markers to detect adaptive genetic variations among chestnut-grafted trees at a small geographical scale within the Tuscan region.
Tuscany is renowned for its extensive cultivation of chestnut trees, particularly in the Apennine area. It covers approximately 177,000 hectares (ha), 33,000 ha of which are used for chestnut fruit production [51]. However, only 32 ha of them are intensively cultivated, and the rest are largely abandoned. Tuscany is also the region with the highest number of cultivated varieties (26.9% of the total in Italy) [52]. It holds the highest number of associations of chestnut fruit producers (11), accounting for almost 70% of the total in Italy. Additionally, there are six consortia and co-operatives, as well as 31 municipalities, that are dedicated to the promotion of chestnut cultivation. The regional authority promotes local varieties and implements measures to prevent and safeguard the health of Tuscan chestnut orchards. Although there are five designated origins, including two for chestnut flour (PDO Garfagnana Neccio Flour and PDO Lunigiana Chestnut Flour), currently, there is no recognition for the “Sweet Chestnut Flour from the Pistoiese Mountains”, but only an “Area Mark” (“Marchio d’area”), for which the production area falls within the province of Pistoia, covering approximately 53,767 ha. The flour is produced using traditional local methods and technologies in drying facilities called “metati” and traditional mills. The flour is obtained through the processing of chestnuts from 12 local varieties, each one with a specific flavor.
This study examined a collection of 55 accessions supposedly belonging to four varieties commonly used in this area to produce chestnut flour: Carpinese, Pastinese, Rossola, and Nerattino. While it might be challenging to distinguish these varieties based on tree characteristics, their fruits exhibit various distinctive traits. There is very little information available in the literature about these cultivars, and many of the available sources on the web are unreliable and often contradict each other. Nevertheless, like other traditional cultivars, they are named according to their geographic origin, ripening period, and nut traits.
Carpinese, also known as Carrarese, is one of the most important and renowned varieties spread along the Tuscan-Romagnolo Apennines. It exhibits resistance to cold winters but is highly susceptible to wind damage [53], as well as attacks from the invasive Asian chestnut gall wasp (Dryocosmus kuriphilus) [54]. The nut has a light, glossy brown color with no striations, and its shiny appearance distinguishes it from other cultivars. It is generally an earlier variety of good quality, suitable for flour production and for fresh consumption, although the skin is difficult to peel [55]. The shelf life of the nut after treatment in a burr chestnut store or through curing is good [56]. The tree does not exhibit strong alternating fruit production, which is around 110 fruits per kilogram. Carpinese has been demonstrated to have exceptional nutritional and nutraceutical properties, including macronutrient minerals, total phenolic content, and high sucrose content [57]. It is recognized as the sweetest and most aromatic variety; however, due to its high sugar content, flour made exclusively from this variety may have abnormal flavors.
Pastinese, also known as Pastanese, Pastenese, Pastonese, or Pelosa [58], is widely cultivated in the Garfagnana area, Pistoia, Romagna, and the Monte Amiata region. It is a late-ripening variety. The leaves start to develop between late April and early May, and flowering occurs between early June and the first decade of July [53]. It produces oval-shaped fruits with a dark brown color. Notably, it has a remarkable ability to bear a substantial amount of fruit. The flour derived from Pastinese chestnuts is renowned for its excellent quality and prolonged shelf life. Nevertheless, it tends to rapidly lose its sensory attributes, which limits its utilization as a standalone ingredient.
Rossola is a cultivar with a significant number of synonyms, such as Rossole, Rossolo, Rossella, and Rossolino, depending on the cultivation area [53]. The variety is named after the reddish skin with faint striations and the round shape of its fruits. Its nuts have a good flavor, and they peel well [55]. Due to its resilient and tough leaves, this variety displays resistance against attacks from the chestnut gall wasp, making it difficult for the insect to lay its eggs [54]. It is widespread in Garfagnana, Lunigiana, and Val di Bisenzio [59]. The production is approximately 106 fruits per kilogram. It is a medium-early variety of good quality, thanks to its low internal skin presence. It is primarily used to produce dried chestnuts and flour [60]. The flour produced from Rossola chestnuts is darker in color but equally sweet and flavorful.
The Nerattino variety (or Neratina, Nerattina Sambucano) is an original variety from the Tuscan-Emilian Apennines, mainly cultivated in the province of Pistoia. It is an early variety that produces small, dark, round-shaped fruits of good quality. It has very high productivity, and the skin peels well, making it suitable for baking and it is also used for flour. In addition, the cultivar is highly resistant and adaptable to adverse conditions [61] and less susceptible to attacks from the chestnut gall wasp when compared to Carpinese and Pastinese [48].
Finally, Marrone Fiorentino, also known as Casentinese or, more generally, Tuscan chestnut, is considered the “typical” chestnut and was used in this study as a reference for the classification of other Italian varieties [62]. This group includes numerous varieties characterized by high-quality products, which have different names depending on the region but have been found to share a common origin based on genetic analysis [56]. The fruit stands out for its large size (approximately 55–60 fruits per kilogram) and is used for fresh consumption and candying. It is highly appreciated for its delicate flavor, good culinary properties, and good shelf life, and is also well-regarded in foreign markets [60].
By using SSR markers, the main objective was to investigate the genetic variability on a set of 55 accessions, named after the suggestions of local growers, and clustering this diversity according to each indicated variety. This is essential to determine the amplitude of sampling for preserving historical regions’ germplasm and also to establish traceability for chestnut specimens and products. In order to achieve this objective, a set of 21 SSR markers, previously designed for Castanea sativa, Quercus robur, and Quercus petrae [63,64,65], were employed to enhance the identification of genetic diversity among the chestnut varieties. Thus, this research can help growers, providing them with certified and selected plant material from these renowned areas for further processing, thereby ensuring the production of high-quality PGI chestnut flour that meets consumer expectations. It is worth noting that among the ancient chestnut trees selected by Beghé et al. [19] in the province of Parma, only the Carpinese and Rossellina ecotypes were included. To the best of our knowledge, no similar studies have been conducted on specimens from the Tuscan-Emilian Apennines area in the province of Pistoia.
2. Materials and Methods
2.1. Plant Material
In this study, 48 chestnut accessions were sampled in four localities in the Northern Tuscan Apennines, Pistoia, Italy (altitude ~ 500–800 m asl; latitude, DMS co-ordinates 44°3′26.64″ N), locally known as ‘Cutigliano’, ‘Torri di Popiglio’, ‘Pian del Meo’, and ‘Monte Pidocchina’. ‘Cutigliano’ and ‘Pian del Meo’ are ~5 km away from ‘Torri di Popiglio’, whereas ‘Monte Pidocchina’ is located ~16 km from the others. The work focused on characterizing four local varieties (i.e., Carpinese, Pastinese, Rossola, and Nerattino), which are the most widespread ecotypes usually used as food sources, particularly for flour production. The sampling was carried out in 2021 with the support of the Associazione dei Castanicoltori della Montagna Pistoiese. Cultivar correspondence for each accession was determined based on local farmers’ knowledge and evaluations of the morphological, phenological, and agronomic characteristics, following the UPOV descriptor list. Az. Agr. Fiori Rita (Molazzana, Lucca, Italy), provided two certified specimens, including one Rossola and one Carpinese sample. In addition, five “Marrone” accessions were also added to the analysis as outgroups. Three accessions were collected from Az. Agr. Rossi Renato (Poppi, Arezzo, Italy), while the other two specimens were taken from a local farmer (Borgo San Lorenzo, Florence, Italy). All the information about the accessions studied is reported in Table S1.
2.2. DNA Extraction and Quantification
The young leaves (collected at the end of April) from each accession were reduced to a fine powder (approximately 20–30 mg) with tungsten carbide beads in liquid nitrogen using a tissue homogenizer (Tissue Lyser, Qiagen, Hilden, Germany). DNA was extracted according to the cetyltrimethylammonium bromide (CTAB) technique [66] with minor modifications. DNA quality and quantity were determined with agarose gel electrophoresis and a Qubit 1.0 fluorometer (Invitrogen, Waltham, MA, USA), respectively.
2.3. PCR Reaction and Genotyping
The microsatellite regions of the chestnut accessions were amplified using 21 SSR primer pairs (Table 1), which were previously developed for chestnut [63], Quercus robur [64], and Quercus petraea [65].

Table 1.
List of primer sequences of Castanea sativa for microsatellite loci, with corresponding annealing temperature and length range of the amplicons.
Forward primers were labeled with a fluorescence tag (6-FAM, HEX, and TET) (Eurofins Genomics, Ebersberg, Germany), and amplifications were composed of 1X GoTaq Buffer (Promega, Madison, WI, USA), 2 mM MgCl2, 0.2 mM dNTPs, 0.2 µM of each primer, 20 ng of extracted DNA, and 1.2 U of GoTaq DNA Polymerase (Promega, Madison, WI, USA), reaching a final volume of 25 µL. Subsequently, a Primus 96 advanced (PEQLAB Biotechnologie Gmbh, Erlanger, Germany) was used to perform thermal reactions based on 5 min denaturation at 95 °C; 35 cycles of 40 s of denaturation at 95 °C, 40 s of primer annealing at the proper temperature for each pair, 40 s of extension at 72 °C and a 7 min final extension at 72 °C. The labeled PCR products were genotyped on ABI3130xl (Applied Biosystems Inc., Waltham, MA, USA) using a performance-optimized polymer (POP7). The subsequent fragment analysis was performed with GeneMapper 4.0 software (Applied Biosystems Inc., Waltham, MA, USA).
2.4. Data Analysis
2.4.1. Genetic Diversity Parameters for the SSR Loci
In the entire population, the SSRs detected for the 21 primers pairs were analyzed with GenAlEx 6.5 [67], computing the number of alleles (Na), the effective number of alleles (Ne), major allele frequency (Fa), the expected (He) and observed (Ho) heterozygosity and the Fixation index (F). Moreover, the Polymorphic Information Content (PIC) was calculated with PowerMarker 3.25 [68].
2.4.2. Population Structure Analysis
The population structure was revealed using a Bayesian approach implemented in STRUCTURE 2.3.4 [69]. The effective number of populations (K), presumably ranging from 1 to 10, was determined using an admixture model. In this model, 300,000 burn-in periods followed by 600,000 Markov chain Monte Carlo (MCMC) iterations were computed 10 times for each K. No prior information was added regarding cultivar or origin (popinfo = 0, popflag = 0). The most suitable K was identified with the ΔK of the Evanno method [70] using the software STRUCTURE HARVESTER 0.6.93 [71]. This result was converted into a bar plot using the R-package pophelper 2.3.1 [72]. Accessions were grouped in a specific cluster with a coefficient of membership greater than 0.65.
Moreover, the STRUCTURE results were validated using clustering analysis on the basis of the genetic distances among the specimens. Particularly, an unweighted-pair group method with arithmetic mean (UPGMA) phylogenetic tree was assembled, in accordance with Bruvo’s genetic distance [73], with 1000 bootstraps permutations using the R-packages adegenet 2.1.3 [74] and poppr 2.9.3 [75]. The resulting dendrogram was graphically edited with the software Interactive Tree OF Life (iTOL) 5 [76]. Furthermore, a matrix based on Jaccard genetic distances was estimated with the R-package vegan 2.5.7 [77], and a principal co-ordinates analysis (PCoA) was constructed using adegenet 2.1.3 [74], ade4 [78], and adegraphics 1.0.16 [79] in R-project.
2.4.3. Genetic Distances and Analysis of Molecular Variance
The coefficient of membership obtained using STRUCTURE was used to subdivide the specimens into populations and to calculate the genetic diversity among the groups. Particularly, GenAlEx 6.5 software [67] was also used to compute the pairwise Nei’s unbiased genetic distances as well as to determine the variation among and within populations through the analysis of molecular variance (AMOVA), FST, and gene flow (Nm). AMOVA was computed with 999 permutations. Nm was calculated from FST using the formula Nm = [(1/FST) − 1]/4. In addition, two pairwise matrices for both FST and Nm were generated using GenAlEx 6.5 software [67].
3. Results
3.1. SSRs Descriptive Genetic Parameters
All the 21 SSR primer pairs provided polymorphic loci for a total of 98 detected alleles, with an average of 4.667 alleles per locus (Na), varying from 2.000 (CsCAT15) to 9.000 (CsCAT3), as shown in Table 2. Moreover, the effective number of alleles (Ne) showed a mean of 2.965, ranging from 1.855 (QrZAG121) to 4.608 (CsCAT14). The major allele frequency (Fa) ranged from 0.291 (CsCAT14) to 0.717 (QrZAG121), with an overall average of 0.492. The mean observed heterozygosity (Ho) was 0.475, varying from 0.000 (QrZAG75) to 0.800 (CsCAT7, CsCAT8), and was lower than the expected in heterozygosity (He), showing values in the range between 0.461 (QrZAG121) and 0.783 (CsCAT14) with an average of 0.634. Furthermore, the mean fixation index (F) was 0.247, with values extending from −0.102 (QpZAG36) to 1.000 (QrZAG75). Finally, with regard to the polymorphism information content (PIC), for which the average amount was 0.582, CsCAT15 showed the lowest value (0.374), whereas CsCAT14 had the highest value (0.748).

Table 2.
Descriptive genetic parameters for each SSR locus in the 56 C. sativa accessions. The number of alleles (Na), the effective number of alleles (Ne), the major allele frequency (Fa), the observed (Ho) and the expected heterozygosity (He), the fixation index (F), and the polymorphism information content (PIC) are detailed.
3.2. Population Structure Analysis
The population structure of 55 C. sativa accessions was determined using the STRUCTURE 2.3.4 software [69]. The optimal number of populations detected was five, according to the ΔK of the Evanno method [70], which showed a sharp peak at K = 5 (Figure S1). The result almost confirmed the varietal correspondence, as indicated by the local farmers. In this context, the STRUCTURE analysis was able to identify the five considered varieties: Nerattino, Pastinese, Rossola, Carpinese, and Marrone (Figure 1). Indeed, the first cluster was composed of all Nerattino accessions, which showed a coefficient of membership greater than 0.90. In addition, all Pastinese samples were gathered in the second arrangement, with a high coefficient of membership (~greater than 0.88), except for Past_7, which revealed an association equal to 0.70. Surprisingly, Carp_5, which was previously established as Carpinese, exhibited a genetic fingerprinting similar to Pastinese, being clustered with the second group. Moreover, the third group comprised all the Rossola specimens (membership coefficient >0.95) collected in the Pistoia area (Ross_1, Ross_2, Ross_3, and Ross_4), as well as the sample originated from a local nursery (Ross_N). The fourth cluster was composed of all Marrone outgroups, while the fifth group consisted of all Carpinese accessions with a membership greater than 0.65.
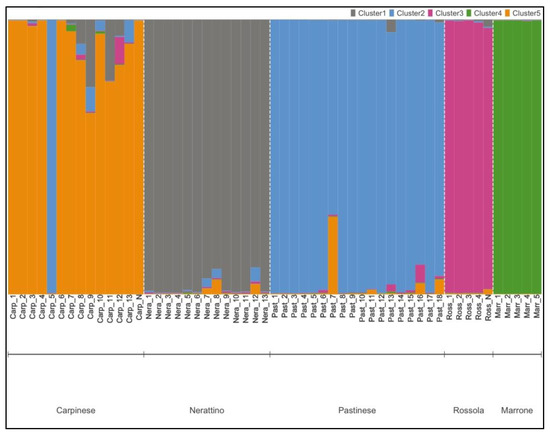
Figure 1.
Bar plot of the estimated five populations predicted by STRUCTURE software. Each color shows the coefficient of membership to the five clusters for the 55 C. sativa accessions. The population name at the bottom of the plot is the previous assignment carried out at the sampling time.
The result obtained by a Bayesian approach was confirmed using a clustering analysis based on the genetic distances among the 55 C. sativa specimens, as revealed by the UPGMA dendrogram generated in accordance with Bruvo’s genetic distances (Figure 2). The overall average genetic distance was equal to 0.166, ranging from 0.000 (observed in the pairs Carp_1 and Carp_2, Carp_4 and Carp_N, Ross_3, and Ross_1, as well as the group Past_2, Past_3, Past_4, Past_5) to 0.823, as was noticed between Marr_4 and Past_10. The UPGMA dendrogram showed the same clustering for STRUCTURE. The first cluster (I) was composed of all Marrone accessions, which resulted in being the most distant group in the considered population. Otherwise, the four other groups comprised the chestnut ecotypes. Particularly, the second arrangement (II) included all Carpinese specimens except for Carp_5, which was also clustered in the dendrogram with all Pastinese samples (V), even though it was recognized. The other two groups (III and IV) consisted of all Rossola and Nerattino accessions, respectively. The cophenetic correlation coefficient was equal to 0.95, confirming the robust consistency and reliability of the dendrogram.
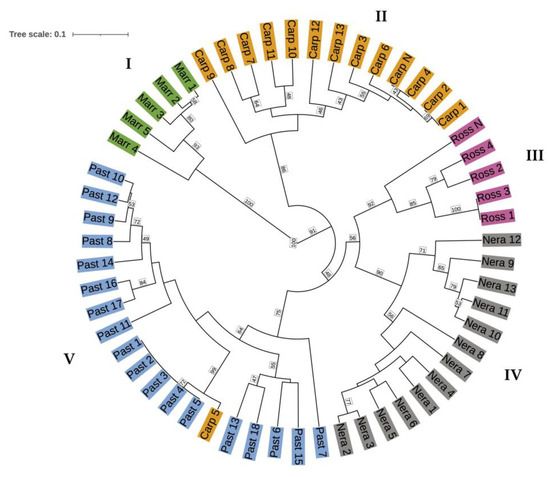
Figure 2.
UPGMA dendrogram of the 55 C. sativa accessions according to Bruvo’s genetic distances. Accessions are colored according to the varieties association previously carried out by local farmers. Branches are colored differently to highlight the five different clusters obtained. Main clusters are indicated with I, II, III, IV, and V.
Another clustering analysis was achieved using principal co-ordinates analysis (PCoA) according to Jaccard distances, as shown in Figure 3. Figure 3a reveals the plot between the first two co-ordinates, while Figure 3b displays the representation between Co-ordinate 1 and Co-ordinate 3. The first three co-ordinates explained 21.18%, 18.30%, and 13.04% of the total variation, respectively, describing 52.52% of the whole variance together. When considering both plots, PCoA evidently separated the 55 C. sativa into five distinct groups, confirming the results previously observed, as well as the assignment performed by the local farmers. The first two co-ordinates distinguished the Carpinese, Pastinese, and Nerattino varieties, located at the top left, top right, and middle bottom left of the plot, respectively (Figure 3a). In addition, the Carp_5 accession was grouped with the Pastinese cultivars, as was already noticed in the STRUCTURE and UPGMA dendrogram. Otherwise, Marrone and Rossola were not clearly separated by the first two co-ordinates, overlapping in the plot’s center. The segregation of Rossola and Marrone can be observed in Figure 3b, displaying the first and third co-ordinates. Indeed, Marrone was located at the bottom of the plot, evidencing the greatest distance among the groupings, while the other four groups were found around the center of the figure.
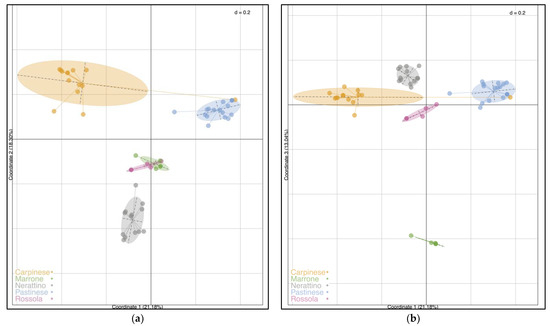
Figure 3.
Principal co-ordinates analysis (PCoA) of the 55 C. sativa accessions according to Jaccard distances. (a) Displays the plot between the first and second co-ordinates, whereas (b) displays the first and third co-ordinates. Accessions are colored according to the varieties association previously carried out by local farmers.
3.3. Nei’s Unbiased Genetic Diversity among the Five Varieties
The five populations obtained from the STRUCTURE and the clustering analyses were considered to assess Nei’s genetic diversity among groups. Particularly, Table 3 describes the pairwise matrices of Nei’s unbiased genetic distances (above diagonal) and Nei’s unbiased genetic identity (below diagonal). The maximum distance was detected between Marrone and Carpinese (1.239). Conversely, the lowest distance was observed between Nerattino and Pastinese (0.280). Marrone showed a high genetic distance from all the other varieties, particularly Nerattino (1.078), Rossola (1.062), and Pastinese (0.977). The greatest genetic identity (below diagonal) was found among the Nerattino and Pastinese cultivars (0.756), with the lowest one between Marrone and Carpinese (0.290).

Table 3.
Pairwise Nei’s unbiased genetic diversity (above diagonal) and genetic identity (below diagonal) among the five analyzed C. sativa varieties.
3.4. Differentiation Among and Within Populations
The interpopulation divergence was evaluated through the AMOVA, FST, and gene flow (Nm). The AMOVA uncovered that 33% of the total genetic variation was detected among the five varieties, whereas 67% was observed within populations (Table 4). Moreover, a low overall Nm value (0.498) was noticed, with FST showing a considerably high value of 0.334 (p-value < 0.001).

Table 4.
Analysis of the molecular variance (AMOVA), FST, and Nm of the five populations of C. sativa.
Regarding the pairwise analysis, the pairwise FST (above diagonal) and Nm (below diagonal) indices among the five populations are shown in Table 5. The pairwise FST (above diagonal) revealed the lowest value between Rossola and Pastinese (0.184), whereas the highest FST index was noticed between Marrone and Carpinese varieties (0.534). On the other hand, the pairwise gene flow (Nm) reported below in the diagonal was the greatest among Rossola and Pastinese (1.111), with Marrone and Carpinese having the lowest pairwise value, accounting for 0.218.

Table 5.
Pairwise FST (above diagonal) and pairwise Nm (below diagonal) among the five chestnut populations (p-value < 0.001).
4. Discussion
This research was based on molecular analysis using a set of 21 SSRs. This marker set differed, in part, from those considered in previous studies [15,19,44,49] due to the absence of markers belonging to the EMCs series, which have been reported to result in lower polymorphism. Particularly, this set has a reduced mutation rate because of their intrinsic characteristic of trinucleotide SSRs in comparison with dinucleotide microsatellites [19]. Thus, considering the relatively small number of accessions analyzed, dinucleotide markers from the QpZAGs and QrZAGs series, previously used for SSR genotyping in Castanea spp. and other related species [80,81,82], were added to the 13 CsCAT primers, increasing the total number of markers to 21. In this context, the use of SSRs designed in a different genus, such as Quercus, may increase the uncovering of diversity due to a major phylogenetic distance and a diverse homozygote pattern [39]. Indeed, while this number exceeds the ideal range of 15–16 markers suggested by Urrestarazu et al. [83] for this type of analysis, it allowed for the amplification and visualization of a high number of alleles (4.7 alleles per locus on average), characterizing all the considered accessions. This average number of alleles observed was in line with the results reported by Beghè et al. [19], who used 8 SSRs to analyze 54 ancient local ecotypes from the Emilian Apennines in Northern Italy (4.7 alleles per locus). However, it was lower compared to the results obtained by Martin et al. [15], with 7 SSRs for 94 individuals supposedly belonging to 26 Italian traditional varieties (7.4 alleles per locus) by Marinoni et al. [84], who examined the genetic variability of 68 Piedmont chestnut individuals using 10 SSRs (8.0 alleles per locus), and by Alessandri et al. [49], who noted 16 SSRs on 134 chestnut accessions from the Tuscan-Emilian Apennines supposedly corresponding to 21 varieties (8.2 alleles per locus). The discovered difference in allele richness was influenced by the number of accessions analyzed. Indeed, when a large number of samples and varieties were considered, there was a notable increase in the total number of alleles. However, the number of effective alleles (Ne), which represents the expected number of alleles in a population with equal allele frequency distribution and the same heterozygosity [85,86], aligned with values reported in other studies. For instance, although Mattioni et al. [87] observed a higher total number of alleles (7.6 alleles per locus) in their study, which included 223 accessions from 10 different sites across Italy, the Ne (averaging 4.2 alleles per locus) aligned with the mean value we observed (2.97 alleles per locus). Moreover, our result was in line with the one obtained by Beghè et al. [19], who detected 3.04 effective alleles per locus.
The good quality of this microsatellite set was confirmed by polymorphism information content (PIC), which indicates a marker’s informativeness. Indeed, a PIC value greater than 0.5 is considered highly descriptive, while values lower than 0.25 do not properly explain the considered population [88]. Specifically, the average PIC value (0.582) observed in this study was comparable to the one detected by Alessandri et al. [49], who noticed a mean of 0.683. Additionally, our result was in line with the ones obtained by Alessandri et al. [5], who revealed an average PIC value of 0.735 in a study focused on the genetic characterization of 630 accessions of wild chestnut trees widespread throughout Italy and Spain. In our study, CsCAT3 emerged as one of the most polymorphic loci (PIC = 0.746), confirming the data previously observed by many authors [5,15,49,89,90]. By concentrating on the specific CsCAT3 locus, it was possible to differentiate the studied ecotypes by identifying six unique alleles. Hence, this locus proved crucial in distinguishing these ecotypes as it exhibited alleles that were absent in other accessions. The presence of private alleles was also found in other 13 SSR loci (i.e., CsCAT1, CsCAT2, CsCAT5, CsCAT8, CsCaT14, CsCAT16, CsCAT41, QrZAG20, QrZAG75, QrZAG121, and QpZAG119) for a total of 23 private alleles, including the ones for CsCAT3. In our study area, where we aimed to identify genotypes within a geographically limited germplasm, the presence of loci with rare or unique alleles holds significant importance. This underscores the effectiveness of discrimination and the informative nature of these 21 SSRs. The efficiency of the microsatellite set was further supported by the average heterozygosity values (Ho = 0.475 and He = 0.634). These values align with those obtained by Beghè et al. [19], Mattioni et al. [87], and Cavallini et al. [50] from several chestnut populations in Central Italy by means of six to eight SSRs.
The cluster analyses (Figure 1, Figure 2 and Figure 3) confirmed a significant level of genetic diversity among the five ecotypes under study, with the Marrone type exhibiting particularly high diversity. Each cluster consisted of accessions from a specific chestnut ecotype, except for Carp_5, which was grouped with the Pastinese types, leading to the hypothesis of incorrect labeling by the growers or a potential mistake during plant propagation through grafting. Therefore, the reliability of SSR markers in accurately identifying C. sativa varieties functions as valuable complements to traditional morphological and pomological analyses, which are not always accurate. The AMOVA, FST, and Nm results observed in the accessions from the Pistoia Apennines evidenced that the considered varieties were clearly genetically distinct, with low gene flow between chestnut populations. Indeed, the detected FST value of 0.334 is considerably high compared to the 0.25 threshold proposed by Wright [91] for differentiation among populations.
In our study, we observed that the within-population genetic distance was minimal (0.12) for the Marrone group, while it remained reduced but ranged between 0.18 and 0.20 for the Rossola, Nerattino, Pastinese, and Carpinese collections. Interestingly, prior studies have demonstrated a notable level of genetic uniformity within the Marrone group, which comprises different varieties and ecotypes known for producing high-quality, mono-embryonic fruits [3,15,92]. This Marrone group is believed to have emerged through intentional selection by growers aiming to preserve desirable traits and facilitate clonal propagation. During this process, growers chose mother plants according to unique traits found exclusively in specific varieties. This selection might have significantly contributed to restricting hybridization between accessions, thus reducing gene flow. The subsequent cultivation of these selected clones in diverse geographical areas has led to variations in the morphological characteristics of the nuts, resulting in distinct denominations, but preserving restricted genetic variability, generating mostly synonyms [5]. However, the mountain region of Central Italy is rich in autochthonous chestnut populations, and the domestication of chestnuts for flour production was achieved through the selection of wild individuals, originating from seeds that were subsequently grafted by farmers to produce fruits with desired characteristics. These practices have played a significant role in preserving a considerable level of genetic diversity, as observed in our investigation, and explain why trees of the same variety do not possess identical genotypes as they were not grafted from the same mother plant [50]. Pereira-Lorenzo et al. [90] and Alessandri et al. [5] also suggested that hybridization may have been a significant factor in the diversification process, which also explains the remarkable diversity observed in small geographical regions.
Our study provides additional evidence supporting the prevalence of polyclonal cultivars in chestnut species, as we observed several cases where specific accessions were identified as heterogeneous clones that were all clustered within the same variety. Despite the limited genetic dissimilarity observed among these accessions, the exact causes of these variations remain uncertain. It is challenging to determine whether these variations arise from somatic mutations, which are commonly observed in species propagated vegetatively over extended periods [93], or if they are consequences of using scions taken for grafting many decades ago, which might have originated from different mother plants. These mother plants may have exhibited slight genetic variations among themselves, but the resulting offspring plants share the same desirable characteristics that distinguish them as a specific variety.
5. Conclusions
This study focused on genetic characterization by means of SSR markers of four old chestnut varieties, which have local importance for flour production, in the Pistoia Apennine mountains: Pastinese, Carpinese, Nerattino, and Rossola. Our findings highlight the invaluable importance of the Tuscan Apennine Mountain as a vital reservoir of genetic diversity within the chestnut populations. Preserving this diversity is crucial for the long-term maintenance of biodiversity, especially in the face of expected adverse conditions arising from climate change and its associated biotic and abiotic impacts. Hence, the implementation of in situ conservation practices becomes crucial in preserving genetic variability within chestnut forests and their natural habitats. By adopting sustainable management and protection strategies, such as establishing protected areas, devising sustainable forest plans, and promoting natural regeneration, we can effectively safeguard the diverse genetic pool of chestnut varieties within their native ecosystems.
In this context, employing efficient and reliable molecular markers plays a pivotal role in identifying genetic variations and improving the sustainable management of chestnut forests. In addition, these markers enable the traceability of chestnut products, particularly flour, leading to an increase in their economic value by incorporating quality and origin markers (i.e., high-quality PGI chestnut flour), which are currently underexplored in various Italian chestnut-growing territories.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13071947/s1, Figure S1: The ΔK of the Evanno method [70] calculated for the 55 C. sativa accessions; Table S1: List of 55 Castanea sativa accessions sampled in this study, their identification (ID) code, putative cultivar name, location, GPS co-ordinates, and the province of geographic origin.
Author Contributions
Conceptualization, E.M., E.G. and S.B.; methodology, L.B., M.G., E.M., E.G. and S.B.; software, L.B.; validation, L.B., M.G., S.N., R.N. and S.B.; formal analysis, L.B., M.G., S.N. and R.N.; investigation, L.B., M.G. and R.N.; resources, E.M. and E.G.; data curation, L.B., M.G., S.N. and R.N.; writing—original draft preparation, L.B. and S.N.; writing—review and editing, L.B., M.G., S.N., R.N., E.M., E.G. and S.B.; supervision, E.G. and S.B.; funding acquisition, E.M., E.G. and S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are contained within the article or Supplementary Material.
Acknowledgments
The authors thank the Fondazione Cassa di Risparmio di Pistoia e Pescia for supporting the research and the Associazione dei Castanicoltori della Montagna Pistoiese for providing samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fernández-López, J.; Alía, R. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Chestnut (Castanea sativa); International Plant Genetic Resources Institute: Rome, Italy, 2003; p. 6. [Google Scholar]
- Conedera, M.; Tinner, W.; Krebs, P.; de Rigo, D.; Caudullo, G. Castanea sativa in Europe: Distribution, Habitat, Usage and Threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Durrant, T.H., Mauri, A., Eds.; Publication Office of the European Union: Luxembourg, 2016; pp. 78–79. [Google Scholar]
- Gobbin, D.; Hohl, L.; Conza, L.; Jermini, M.; Gessler, C.; Conedera, M. Microsatellite-Based Characterization of the Castanea sativa Cultivar Heritage of Southern Switzerland. Genome 2007, 50, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Neri, L.; Dimitri, G.; Sacchetti, G. Chemical Composition and Antioxidant Activity of Cured Chestnuts from Three Sweet Chestnut (Castanea sativa Mill.) Ecotypes from Italy. J. Food Compos. Anal. 2010, 23, 23–29. [Google Scholar] [CrossRef]
- Alessandri, S.; Cabrer, A.M.R.; Martìn, M.A.; Mattioni, C.; Pereira-Lorenzo, S.; Dondini, L. Genetic Characterization of Italian and Spanish Wild and Domesticated Chestnut Trees. Sci. Hortic. 2022, 295, 110882. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Oxford University Press: Oxford, UK, 2012; ISBN 019162425X. [Google Scholar]
- Conedera, M.; Krebs, P.; Tinner, W.; Pradella, M.; Torriani, D. The Cultivation of Castanea sativa (Mill.) in Europe, from Its Origin to Its Diffusion on a Continental Scale. Veg. Hist. Archaeobot. 2004, 13, 161–179. [Google Scholar] [CrossRef]
- Roces-Díaz, J.V.; Jiménez-Alfaro, B.; Chytrý, M.; Díaz-Varela, E.R.; Álvarez-Álvarez, P. Glacial Refugia and Mid-Holocene Expansion Delineate the Current Distribution of Castanea sativa in Europe. Palaeogeogr Palaeoclim. Palaeoecol 2018, 491, 152–160. [Google Scholar] [CrossRef]
- Krebs, P.; Pezzatti, G.B.; Beffa, G.; Tinner, W.; Conedera, M. Revising the Sweet Chestnut (Castanea sativa Mill.) Refugia History of the Last Glacial Period with Extended Pollen and Macrofossil Evidence. Quat. Sci. Rev. 2019, 206, 111–128. [Google Scholar] [CrossRef]
- Catalano, M. To Safeguard and to Make the Most of the Rural Environment by Means of a “Sustainable Agro-Environmental Systems” Study. Ital. J. Agron. 2010, 5, 295–299. [Google Scholar] [CrossRef]
- Casasoli, M.; Mattioni, C.; Cherubini, M.; Villani, F. A Genetic Linkage Map of European Chestnut (Castanea sativa Mill.) Based on RAPD, ISSR and Isozyme Markers. Theor. Appl. Genet. 2001, 102, 1190–1199. [Google Scholar] [CrossRef]
- Piccioli, L. Monografia Del Castagno: Suoi Caratteri, Morfologici, Varietà, Coltivazione, Prodotti e Nemici… Studio Fatto per Incarico Dei Fabbricanti Italiani Di Estratto Di Castagno; Stab. tipo-litografico G. Spinelli & C.: Florence, Italy, 1922; p. 397. [Google Scholar]
- Bagnaresi, U.; Bassi, D.; Casini, E.; Conticini, L.; Magnani, G.P. Contributo Alla Individuazione Delle Cultivar Di Castagno Tosco-Emiliane. In Atti Del Convegno “Giorn. Del Castagno”; Caprese Michelangelo (Arezzo): Florence, Italy, 1977; pp. 165–234. [Google Scholar]
- Bounous, G.; Beccaro, G.L.; Barrel, A.; Lovisolo, C. Inventory of Chestnut Research, Germplasm and References; FAO Ciheam Reu Technical Series; FAO: Rome, Italy, 2001; Volume 65, pp. 1–174. [Google Scholar]
- Martín, M.A.; Mattioni, C.; Cherubini, M.; Taurchini, D.; Villani, F. Genetic Characterisation of Traditional Chestnut Varieties in Italy Using Microsatellites (Simple Sequence Repeats) Markers. Ann. Appl. Biol. 2010, 157, 37–44. [Google Scholar] [CrossRef]
- Breviglieri, N. Indagini e Osservazioni Sulle Cultivar Di Castagno. Studio Monografico sul Castagno Nella Provincia di Lucca; Centro di Studio sul Castagno: Marradi, Italy, 1958; pp. 65–137. [Google Scholar]
- Mattioli, W.; Mancini, L.D.; Portoghesi, L.; Corona, P. Biodiversity Conservation and Forest Management: The Case of the Sweet Chestnut Coppice Stands in Central Italy. Plant Biosyst.—Int. J. Deal. All Asp. Plant Biol. 2016, 150, 592–600. [Google Scholar] [CrossRef]
- Martin, M.A.; Alvarez, J.B.; Mattioni, C.; Cherubini, M.; Villani, F.; Martin, L.M. Identification and Characterisation of Traditional Chestnut Varieties of Southern Spain Using Morphological and Simple Sequence Repeat (SSRs) Markers. Ann. Appl. Biol. 2009, 154, 389–398. [Google Scholar] [CrossRef]
- Beghè, D.; Ganino, T.; Dall’Asta, C.; Silvanini, A.; Cirlini, M.; Fabbri, A. Identification and Characterization of Ancient Italian Chestnut Using Nuclear Microsatellite Markers. Sci. Hortic. 2013, 164, 50–57. [Google Scholar] [CrossRef]
- Bellini, E.; Giordani, E.; Giannelli, G.; Picardi, E. Chestnut. In The Fruit Woody Species; Descriptor List: Tuscan, Italy, 2007; pp. 417–437. [Google Scholar]
- Ramos-Cabrer, A.M.; Pereira-Lorenzo, S. Genetic Relationship between Castanea sativa Mill. Trees from North-Western to South Spain Based on Morphological Traits and Isoenzymes. Genet. Resour. Crop. Evol. 2005, 52, 879–890. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Díaz-Hernández, M.B.; Ciordia-Ara, M.; Ríos-Mesa, D. Chemical Composition of Chestnut Cultivars from Spain. Sci. Hortic. 2006, 107, 306–314. [Google Scholar] [CrossRef]
- Cirlini, M.; Dall’Asta, C.; Silvanini, A.; Begh, D.; Fabbri, A.; Galaverna, G.; Ganino, T. Volatile Fingerprinting of Chestnut Flours from Traditional Emilia Romagna (Italy) Cultivars. Food Chem. 2012, 134, 662–668. [Google Scholar] [CrossRef]
- Cosmulescu, S.; Trandafir, I.; Nour, V.; Botu, M. Physical and Compositional Characteristics of Chestnut Fruits. Rom. J. Hortic. 2020, 1, 51–58. [Google Scholar] [CrossRef]
- Poljak, I.; Vahčić, N.; Liber, Z.; Šatović, Z.; Idžojtić, M. Morphological and Chemical Variation of Wild Sweet Chestnut (Castanea sativa Mill.) Populations. Forests 2022, 13, 55. [Google Scholar] [CrossRef]
- Santos, M.J.; Pinto, T.; Vilela, A. Sweet Chestnut (Castanea sativa Mill.) Nutritional and Phenolic Composition Interactions with Chestnut Flavor Physiology. Foods 2022, 11, 4052. [Google Scholar] [CrossRef]
- Galderisi, U.; Cipollaro, M.; Di Bernardo, G.; De Masi, L.; Galano, G.; Cascino, A. Molecular Typing of Italian Sweet Chestnut Cultivars by Random Amplified Polymorphic DNA Analysis. J. Hortic. Sci. Biotechnol. 1998, 73, 259–263. [Google Scholar] [CrossRef]
- Goulao, L.; Valdiviesso, T.; Santana, C.; Oliveira, C.M. Comparison between Phenetic Characterisation Using RAPD and ISSR Markers and Phenotypic Data of Cultivated Chestnut (Castanea sativa Mill.). Genet. Resour. Crop Evol. 2001, 48, 329–338. [Google Scholar] [CrossRef]
- Abdelhamid, S.; Küpfer, P.; Conedera, M. Identification of Chestnut (C. sativa Mill.) Cultivars Using RAPD and AFLP Markers in Switzerland. Rev. Suisse De Vitic. Arboric. Et Hortic. 2004, 36, 349–354. [Google Scholar]
- Mattioni, C.; Cherubini, M.; Micheli, E.; Villani, F.; Bucci, G. Role of Domestication in Shaping Castanea sativa Genetic Variation in Europe. Tree Genet. Genomes 2008, 4, 563–574. [Google Scholar] [CrossRef]
- Martin, M.A.; Mattioni, C.; Cherubini, M.; Taurchini, D.; Villani, F. Genetic Diversity in European Chestnut Populations by Means of Genomic and Genic Microsatellite Markers. Tree Genet. Genomes 2010, 6, 735–744. [Google Scholar] [CrossRef]
- del Mar Naval, M.; Zuriaga, E.; Pecchioli, S.; Llácer, G.; Giordani, E.; Badenes, M.L. Analysis of Genetic Diversity among Persimmon Cultivars Using Microsatellite Markers. Tree Genet. Genomes 2010, 6, 677–687. [Google Scholar] [CrossRef]
- Erfani, J.; Ebadi, A.; Abdollahi, H.; Fatahi, R. Genetic Diversity of Some Pear Cultivars and Genotypes Using Simple Sequence Repeat (SSR) Markers. Plant Mol. Biol. Rep. 2012, 30, 1065–1072. [Google Scholar] [CrossRef]
- Gürcan, K.; Önal, N.; Yilmaz, K.U.; Ullah, S.; Erdoğan, A.; Zengin, Y. Evaluation of Turkish Apricot Germplasm Using SSR Markers: Genetic Diversity Assessment and Search for Plum Pox Virus Resistance Alleles. Sci. Hortic. 2015, 193, 155–164. [Google Scholar] [CrossRef]
- Çalişkan, O.; Bayazit, S.; Öktem, M.; Ergül, A. Evaluation of the Genetic Diversity of Pomegranate Accessions from Turkey Using New Microsatellite Markers. Turk. J. Agric. For. 2017, 41, 142–153. [Google Scholar] [CrossRef]
- Pérez, V.; Larrañaga, N.; Abdallah, D.; Wünsch, A.; Hormaza, J.I. Genetic Diversity of Local Peach (Prunus persica) Accessions from La Palma Island (Canary Islands, Spain). Agronomy 2020, 10, 457. [Google Scholar] [CrossRef]
- López, M.; Gori, M.; Bini, L.; Ordoñez, E.; Durán, E.; Gutierrez, O.; Masoni, A.; Giordani, E.; Biricolti, S.; Palchetti, E. Genetic Purity of Cacao Criollo from Honduras Is Revealed by SSR Molecular Markers. Agronomy 2021, 11, 225. [Google Scholar] [CrossRef]
- Zuriaga, E.; Pintová, J.; Bartual, J.; Badenes, M.L. Characterization of the Spanish Pomegranate Germplasm Collection Maintained at the Agricultural Experiment Station of Elche to Identify Promising Breeding Materials. Plants 2022, 11, 1257. [Google Scholar] [CrossRef] [PubMed]
- Beccaro, G.L.; Torello-Marinoni, D.; Binelli, G.; Donno, D.; Boccacci, P.; Botta, R.; Cerutti, A.K.; Conedera, M. Insights in the Chestnut Genetic Diversity in Canton Ticino (Southern Switzerland). Silvae Genet. 2012, 61, 292–300. [Google Scholar] [CrossRef]
- Quintana, J.; Contreras, A.; Merino, I.; Vinuesa, A.; Orozco, G.; Ovalle, F.; Gomez, L. Genetic Characterization of Chestnut (Castanea sativa Mill.) Orchards and Traditional Nut Varieties in El Bierzo, a Glacial Refuge and Major Cultivation Site in Northwestern Spain. Tree Genet. Genomes 2015, 11, 1–12. [Google Scholar] [CrossRef]
- Janfaza, S.; Yousefzadeh, H.; Hosseini Nasr, S.M.; Botta, R.; Asadi Abkenar, A.; Torello Marinoni, D. Genetic Diversity of Castanea sativa an Endangered Species in the Hyrcanian Forest. Silva Fenn. 2017, 51, 15. [Google Scholar] [CrossRef]
- El Chami, M.A.; Tourvas, N.; Kazakis, G.; Kalaitzis, P.; Aravanopoulos, F.A. Genetic Characterisation of Chestnut Cultivars in Crete. Forests 2021, 12, 1659. [Google Scholar] [CrossRef]
- Tumpa, K.; Šatović, Z.; Liber, Z.; Vidaković, A.; Idžojtić, M.; Ježić, M.; Ćurković-Perica, M.; Poljak, I. Gene Flow between Wild Trees and Cultivated Varieties Shapes the Genetic Structure of Sweet Chestnut (Castanea sativa Mill.) Populations. Sci. Rep. 2022, 12, 15007. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Barreneche, T.; Mattioni, C.; Villani, F.; Díaz-Hernández, M.B.; Martín, L.M.; Martín, Á. Database of European Chestnut Cultivars and Definition of a Core Collection Using Simple Sequence Repeats. Tree Genet. Genomes 2017, 13, 114. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; Barreneche, T.; Mattioni, C.; Villani, F.; Díaz-Hernández, B.; Martín, L.M.; Robles-Loma, A.; Cáceres, Y.; Martín, A. Instant Domestication Process of European Chestnut Cultivars. Ann. Appl. Biol. 2019, 174, 74–85. [Google Scholar] [CrossRef]
- Bouffartigue, C.; Debille, S.; Fabreguettes, O.; Cabrer, A.R.; Pereira-Lorenzo, S.; Flutre, T.; Harvengt, L. Two Main Genetic Clusters with High Admixture between Forest and Cultivated Chestnut (Castanea sativa Mill.) in France. Ann. Sci. 2020, 77, 74. [Google Scholar] [CrossRef]
- Castellana, S.; Martin, M.Á.; Solla, A.; Alcaide, F.; Villani, F.; Cherubini, M.; Neale, D.; Mattioni, C. Signatures of Local Adaptation to Climate in Natural Populations of Sweet Chestnut (Castanea sativa Mill.) from Southern Europe. Ann. Sci. 2021, 78, 27. [Google Scholar] [CrossRef]
- Bracalini, M.; Croci, F.; Turchi, A.; Giordani, E.; Tiberi, R.; Panzavolta, T. The Asian Chestnut Gall Wasp in Italy: Surveys on Its Native and Exotic Parasitoids as Well as on Chestnut Cultivar Susceptibility. Asian J. Adv. Agric. Res. 2019, 11, 1–8. [Google Scholar] [CrossRef]
- Alessandri, S.; Krznar, M.; Ajolfi, D.; Cabrer, A.M.R.; Pereira-Lorenzo, S.; Dondini, L. Genetic Diversity of Castanea sativa Mill. Accessions from the Tuscan-Emilian Apennines and Emilia Romagna Region (Italy). Agronomy 2020, 10, 1319. [Google Scholar] [CrossRef]
- Cavallini, M.; Lombardo, G.; Binelli, G.; Cantini, C. Assessing the Genetic Identity of Tuscan Sweet Chestnut (Castanea sativa Mill.). Forests 2022, 13, 0967. [Google Scholar] [CrossRef]
- Bini, C. Castagna, Eccellenza Toscana: Protocollo Regione-Anci per Valorizzare Settore. Toscana Notizie, 2021. Available online: https://www.toscana-notizie.it/-/castagna-eccellenza-toscana-protocollo-regione-anci-per-valorizzare-settore (accessed on 16 May 2023).
- Fideghelli, C. Aspetti Pomologici e Qualitativi Dei Materiali Di Propagazione; I Georgofili; Polistampa: Florence, Italy, 2016; Volume 2, pp. 55–61. [Google Scholar]
- Di Gioia, F. Recupero Delle Varietà Di Castagno Autoctone Della Garfagnana, Collana Secondo Natura. In Edizioni Andromeda; Beniamini s.r.l.: Rome, Italy, 2018; pp. 1–172. [Google Scholar]
- Panzavolta, T.; Croci, F.; Bracalini, M.; Melika, G.; Benedettelli, S.; Tellini Florenzano, G.; Tiberi, R. Population Dynamics of Native Parasitoids Associated with the Asian Chestnut Gall Wasp (Dryocosmus kuriphilus) in Italy. Psyche 2018, 2018, 8078049. [Google Scholar] [CrossRef]
- Bellini, E.; Giordani, E.; Morelli, D.; Ferri, A.; Paradisi, G.; Fattorini, M.; Autino, A.; Cresti, M. Le Varietà Locali Di Castagno Della Garfagnana Nel Repertorio Regionale Toscano (L.R. 64/04), I Castagni Della Garfagnana, Studi per La Tracciabilità Di Filiera e La Caratterizzazione Qualitativa Della Farina Di Neccio Della Garfagnana DOP; ARSIA (Agenzia Regionale per Lo Sviluppo e l’Innovazione Nel Settore Agricolo-Forestale): Florence, Italy, 2009; pp. 1–160. [Google Scholar]
- Bianchi, L.; Maltoni, A.; Mariotti, B.; Paci, M. La Selvicoltura Dei Castagneti Da Frutto Abbandonati Della Toscana DISTAF—Dipartimento Di Scienze e Tecnologie Ambientali Forestali Università Degli Studi Di Firenze; ARSIA (Agenzia Regionale per Lo Sviluppo e l’Innovazione Nel Settore Agricolo-Forestale): Florence, Italy, 2009; pp. 1–140. [Google Scholar]
- Lo Piccolo, E.; Landi, M.; Ceccanti, C.; Mininni, A.N.; Marchetti, L.; Massai, R.; Guidi, L.; Remorini, D. Nutritional and Nutraceutical Properties of Raw and Traditionally Obtained Flour from Chestnut Fruit Grown in Tuscany. Eur. Food Res. Technol. 2020, 246, 1867–1876. [Google Scholar] [CrossRef]
- Antonaroli, R.; Bellini, E. Pastinese, Regione Emilia-Romagna – Agricoltura, 2014. Available online: https://agricoltura.regione.emilia-romagna.it/produzioni-agroalimentari/temi/agrobiodiversita/schede-specie-vegetali/castagno/pastinese (accessed on 16 May 2023).
- Antonaroli, R.; Bellini, E. Scheda Tecnica per l’iscrizione al Repertorio, Rossola RER V077, Regione Emilia-Romagna L.R. N. 1/2008, 2022. Available online: https://agricoltura.regione.emilia-romagna.it/produzioni-agroalimentari/temi/agrobiodiversita/schede-specie-vegetali/castagno/rossola (accessed on 16 May 2023).
- Bellini, E.; Giordani, E.; Marinelli, C.; Migliorini, M.; Funghini, L. Marrone Del Mugello PGI: Nutritional and Organoleptic Quality of European Chestnut (Castanea sativa Mill.). In Proceedings of the IV International Chestnut Symposium, Beijing, China, 25–28 September 2008; Volume 844, pp. 61–68. [Google Scholar]
- Giannini, M.; Del Biondo, A. La Castanicoltura Toscana: Aspetti Tecnici e Produttivi; L’Informatore Agrario: Verona, Italy, 2010. [Google Scholar]
- Stival, O. Il Castagno Da Frutto. In Rubrica Verde; Sebino Bresciano: Brescia, Italy, 2007. [Google Scholar]
- Marinoni, D.; Akkak, A.; Bounous, G.; Edwards, K.J.; Botta, R. Development and Characterization of Microsatellite Markers in Castanea sativa (Mill.). Mol. Breed. 2003, 11, 127–136. [Google Scholar] [CrossRef]
- Kampfer, S.; Lexer, C.; Glössl, J.; Steinkellner, H. Characterization of (GA)(n) Microsatellite Loci from Quercus robur. Hereditas 1998, 129, 183–186. [Google Scholar] [CrossRef]
- Steinkellner, H.; Fluch, S.; Turetschek, E.; Lexer, C.; Streiff, R.; Kremer, A.; Burg, K.; Glössl, J. Identification and Characterization of (GA/CT) n-Microsatellite Loci from Quercus petraea. Plant Mol. Biol. 1997, 33, 1093–1096. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J. Isolation of Plant DNA from Fresh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar]
- Liu, K.; Muse, S. V PowerMarker: An Integrated Analysis Environment for Genetic Marker Analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Francis, R.M. Pophelper: An R Package and Web App to Analyse and Visualize Population Structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Bruvo, R.; Michiels, N.K.; D’souza, T.G.; Schulenburg, H. A Simple Method for the Calculation of Microsatellite Genotype Distances Irrespective of Ploidy Level. Mol. Ecol. 2004, 13, 2101–2106. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Oksanen, J. Vegan: Ecological Diversity. R Proj. 2013, 368, 1–11. [Google Scholar]
- Chessel, D.; Dufour, A.B.; Thioulouse, J. The Ade4 Package-I-One-Table Methods. R News 2004, 4, 5–10. [Google Scholar]
- Siberchicot, A.; Julien-Laferrière, A.; Dufour, A.-B.; Thioulouse, J.; Dray, S. Adegraphics: An S4 Lattice-Based Package for the Representation of Multivariate Data. R J. 2017, 9, 198–212. [Google Scholar] [CrossRef]
- Sisco, P.H.; Kubisiak, T.L.; Casasoli, M.; Barreneche, T.; Kremer, A.; Clark, C.; Sederoff, R.R.; Hebard, F.V.; Villani, F. An Improved Genetic Map for Castanea mollissima/Castanea dentata and Its Relationship to the Genetic Map of Castanea sativa. In Proceedings of the III International Chestnut Congress 693; Abreu, C.G., Rosa, E., Monteiro, A.A., Eds.; Acta Horticulturae: Leuven, Belgium, 2005; pp. 491–495. [Google Scholar]
- Akkak, A.; Boccacci, P.; Torello Marinoni, D. Cross-Species Amplification of Microsatellite Markers in Castanea Spp. and Other Related Species. In Proceedings of the I European Congress on Chestnut-Castanea 866; Acta Horticulturae: Leuven, Belgium, 2009; pp. 195–201. [Google Scholar]
- Medina-Mora, C.; Fulbright, D.W.; Jarosz, A.M. SSR Genotyping of Progeny from a Chestnut Orchard in Michigan. In Proceedings of the V International Chestnut Symposium 1019, Shepherdstown, WV, USA, 4–8 September 2012; pp. 173–178. [Google Scholar]
- Urrestarazu, J.; Royo, J.B.; Santesteban, L.G.; Miranda, C. Evaluating the Influence of the Microsatellite Marker Set on the Genetic Structure Inferred in Pyrus communis L. PLoS ONE 2015, 10, e0138417. [Google Scholar] [CrossRef] [PubMed]
- Torello Marinoni, D.; Akkak, A.; Beltramo, C.; Guaraldo, P.; Boccacci, P.; Bounous, G.; Ferrara, A.M.; Ebone, A.; Viotto, E.; Botta, R. Genetic and Morphological Characterization of Chestnut (Castanea sativa Mill.) Germplasm in Piedmont (North-Western Italy). Tree Genet. Genomes 2013, 9, 1017–1030. [Google Scholar] [CrossRef]
- Petit, R.J.; El Mousadik, A.; Pons, O. Identifying Populations for Conservation on the Basis of Genetic Markers. Conserv. Biol. 1998, 12, 844–855. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Luikart, G.H.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; Blackwell Publishing: Malden, MA, USA, 2012; p. 624. [Google Scholar]
- Mattioni, C.; Martin, M.A.; Chiocchini, F.; Cherubini, M.; Gaudet, M.; Pollegioni, P.; Velichkov, I.; Jarman, R.; Chambers, F.M.; Paule, L.; et al. Landscape Genetics Structure of European Sweet Chestnut (Castanea sativa Mill): Indications for Conservation Priorities. Tree Genet. Genomes 2017, 13, 39. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar]
- Pereira-Lorenzo, S.; Costa, R.M.L.; Ramos-Cabrer, A.M.; Ribeiro, C.A.M.; da Silva, M.F.S.; Manzano, G.; Barreneche, T. Variation in Grafted European Chestnut and Hybrids by Microsatellites Reveals Two Main Origins in the Iberian Peninsula. Tree Genet. Genomes 2010, 6, 701–715. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Costa, R.M.L.; Ramos-Cabrer, A.M.; Ciordia-Ara, M.; Ribeiro, C.A.M.; Borges, O.; Barreneche, T.; Donini, P. Chestnut Cultivar Diversification Process in the Iberian Peninsula, Canary Islands, and Azores. Genome 2011, 54, 301–315. [Google Scholar] [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 395–420. [Google Scholar] [CrossRef]
- Mellano, M.G.; Beccaro, G.L.; Donno, D.; Marinoni, D.T.; Boccacci, P.; Canterino, S.; Cerutti, A.K.; Bounous, G. Castanea spp. Biodiversity Conservation: Collection and Characterization of the Genetic Diversity of an Endangered Species. Genet. Resour. Crop. Evol. 2012, 59, 1727–1741. [Google Scholar] [CrossRef]
- Hocquigny, S.; Pelsy, F.; Dumas, V.; Kindt, S.; Heloir, M.C.; Merdinoglu, D. Diversification within Grapevine Cultivars Goes through Chimeric States. Genome 2004, 47, 579–589. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).