Molecular Characterization of Mitogenome of Cacopsylla picta and Cacopsylla melanoneura, Two Vector Species of ‘Candidatus Phytoplasma mali’
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. DNA Isolation
2.3. PCR Amplification and Sanger Sequencing
2.4. High-Throughput Sequencing
2.5. Assembly of HTS Reads and the Genome Annotation
2.6. Phylogenetic Analysis
3. Results
3.1. HTS Analysis
3.2. Mitogenome Structure of C. picta and C. melanoneura
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreno, A.; Miranda, M.P.; Fereres, A. Psyllids as major vectors of plant pathogens. Entomol. Gen. 2021, 41, 419–438. [Google Scholar] [CrossRef]
- Ouvrard, D.; Chalise, P.; Percy, D.M. Host-plant leaps versus host-plant shuffle: A global survey reveals contrasting patterns in an oligophagous insect group (Hemiptera, Psylloidea). Syst. Biodivers. 2015, 13, 434–454. [Google Scholar] [CrossRef]
- Burckhardt, D.; Ouvrard, D.; Percy, D.M. An updated classification of the jumping plant-lice (Hemiptera: Psylloidea) integrating molecular and morphological evidence. Eur. J. Taxon. 2021, 736, 137–182. [Google Scholar] [CrossRef]
- Bertaccini, A. Plants and Phytoplasmas: When bacteria modify plants. Plants 2022, 11, 1425. [Google Scholar] [CrossRef] [PubMed]
- Namba, S. Molecular and biological properties of phytoplasmas. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019, 95, 401–418. [Google Scholar] [CrossRef] [PubMed]
- Jarausch, B.; Tedeschi, R.; Sauvion, N.; Gross, J.; Jarausch, W. Psyllid Vectors. In Phytoplasmas: Plant Pathogenic Bacteria—II: Transmission and Management of Phytoplasma—Associated Diseases; Bertaccini, A., Weintraub, P.G., Rao, G.P., Mori, N., Eds.; Springer: Singapore, 2019; pp. 53–78. [Google Scholar]
- Bertaccini, A.; Duduk, B.; Paltrinieri, S.; Contaldo, N. Phytoplasmas and phytoplasma diseases: A severe threat to agriculture. Am. J. Plant Sci. 2014, 5, 1763–1788. [Google Scholar] [CrossRef]
- Tedeschi, R.; Alma, A. Transmission of apple proliferation phytoplasma by Cacopsylla melanoneura (Homoptera: Psyllidae). J. Econ. Entomol. 2004, 97, 8–13. [Google Scholar] [CrossRef]
- Janik, K.; Barthel, D.; Oppedisano, T.; Anfora, G.; Schuler, H. Apple Proliferation—A Joint Review; Laimburg Research Centre: Stadio-Laimburg, Italy, 2020. [Google Scholar]
- Fischnaller, S.; Parth, M.; Messner, M.; Stocker, R.; Kerschbamer, C.; Janik, K. Surveying potential vectors of apple proliferation phytoplasma: Faunistic analysis and infection status of selected Auchenorrhyncha species. Insects 2021, 12, 12. [Google Scholar] [CrossRef]
- Tedeschi, R.; Bosco, D.; Alma, A. Population dynamics of Cacopsylla melanoneura (Homoptera: Psyllidae), a vector of apple proliferation phytoplasma in northwestern Italy. J. Econ. Entomol. 2002, 95, 544–551. [Google Scholar] [CrossRef]
- Safarova, D.; Stary, M.; Valova, P.; Opatikova, M.; Bilkova, L.; Navratil, M. Impact of insecticides treatment on phytoplasma infection risk in apple orchards. Hortic. Sci. 2016, 43, 112–116. [Google Scholar] [CrossRef]
- Cermak, V.; Lauterer, P. Overwintering of psyllids in South Moravia (Czech Republic) with respect to the vectors of the apple proliferation cluster phytoplasmas. Bull. Insectol. 2008, 61, 147–148. [Google Scholar]
- Malenovsky, I.; Lauterer, P. Jumping plant-lice (Hemiptera: Psylloidea) of the Bílé Karpaty Protected Landscape Area and Biosphere Reserve (Czech Republic). Acta Musei Moraviae Sci. Biol. 2012, 96, 105–154. [Google Scholar]
- Barthel, D.; Schuler, H.; Galli, J.; Borruso, L.; Geier, J.; Heer, K.; Burckhardt, D.; Janik, K. Identification of plant DNA in adults of the phytoplasma vector Cacopsylla picta helps understanding its feeding behavior. Insects 2020, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Jarausch, B.; Fuchs, A.; Schwind, N.; Krczal, G.; Jarausch, W. Cacopsylla picta as most important vector for ‘Candidatus Phytoplasma mali’ in Germany and neighbouring regions. Bull. Insectol. 2007, 60, 189–190. [Google Scholar]
- Mattedi, L.; Forno, F.; Cainelli, C.; Grando, M.S.; Jarausch, W. Research on Candidatus phytoplasma mali transmission by insect vectors in Trentino. Acta Hortic. 2008, 781, 369–374. [Google Scholar] [CrossRef]
- Jarausch, B.; Schwind, N.; Jarausch, W.; Krczal, G.; Dickler, E.; Seemuller, E. First report of Cacopsylla picta as a vector of apple proliferation phytoplasma in Germany. Plant Dis. 2003, 87, 101. [Google Scholar] [CrossRef]
- Mittelberger, C.; Obkircher, L.; Oettl, S.; Oppedisano, T.; Pedrazzoli, F.; Panassiti, B.; Kerschbamer, C.; Anfora, G.; Janik, K. The insect vector Cacopsylla picta vertically transmits the bacterium ‘Candidatus Phytoplasma mali’ to its progeny. Plant Pathol. 2017, 66, 1015–1021. [Google Scholar] [CrossRef]
- Mayer, C.J.; Vilcinskas, A.; Gross, J. Chemically mediated multitrophic interactions in a plant-insect vector-phytoplasma system compared with a partially nonvector species. Agric. Forest Entomol. 2011, 13, 25–35. [Google Scholar] [CrossRef]
- Fialova, R.; Navratil, M.; Valova, P.; Lauterer, P. Molecular tests to determine apple proliferation phytoplasma presence in psyllid vectors from apple tree orchards in the Czech Republic. Acta Hortic. 2008, 781, 471–476. [Google Scholar] [CrossRef]
- Tedeschi, R.; Nardi, F. DNA-based discrimination and frequency of phytoplasma infection in the two hawthorn-feeding species, Cacopsylla melanoneura and Cacopsylla affinis, in northwestern Italy. Bull. Entomol. Res. 2010, 100, 741–747. [Google Scholar] [CrossRef][Green Version]
- Nokkala, C.; Kuznetsova, V.G.; Rinne, V.; Nokkala, S. Description of two new species of the genus Cacopsylla Ossiannilsson, 1970 (Hemiptera, Psylloidea) from northern Fennoscandia recognized by morphology, cytogenetic characters and COI barcode sequence. Comp. Cytogenet. 2019, 13, 367–382. [Google Scholar] [CrossRef]
- Kang, A.R.; Baek, J.Y.; Lee, S.H.; Cho, Y.S.; Kim, W.S.; Han, Y.S.; Kim, I. Geographic homogeneity and high gene flow of the pear psylla, Cacopsylla pyricola (Hemiptera: Psyllidae), detected by mitochondrial COI gene and nuclear ribosomal internal transcribed spacer 2. Anim. Cells Syst. 2012, 16, 145–153. [Google Scholar] [CrossRef]
- Cho, G.; Malenovsky, I.; Burckhardt, D.; Inoue, H.; Lee, S. DNA barcoding of pear psyllids (Hemiptera: Psylloidea: Psyllidae), a tale of continued misidentifications. Bull. Entomol. Res. 2020, 110, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, Q.Z.; Qiao, X.F.; Wang, J.W.; Zhang, T. Identification and phylogenetic analysis of pear psyllids (Hemiptera: Psyllidae) in Chinese pear orchards. J. Econ. Entomol. 2018, 111, 2908–2913. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Inoue, H.; Kuchiki, F.; Ide, Y.; Uechi, N.; Iwanami, T. Identification of a distinct lineage of Cacopsylla chinensis (Hemiptera: Psyllidae) in Japan on the basis of two mitochondrial DNA sequences. J. Econ. Entomol. 2013, 106, 536–542. [Google Scholar] [CrossRef]
- Oettl, S.; Schlink, K. Molecular identification of two vector species, Cacopsylla melanoneura and Cacopsylla picta (Hemiptera: Psyllidae), of apple proliferation disease and further common psyllids of Northern Italy. J. Econ. Entomol. 2015, 108, 2174–2183. [Google Scholar] [CrossRef]
- Percy, D.M.; Crampton-Platt, A.; Sveinsson, S.; Lemmon, A.R.; Lemmon, E.M.; Ouvrard, D.; Burckhardt, D. Resolving the psyllid tree of life: Phylogenomic analyses of the superfamily Psylloidea (Hemiptera). Syst. Entomol. 2018, 43, 762–776. [Google Scholar] [CrossRef]
- Wang, Y.; Cen, Y.; He, Y.; Wu, Y.; Huang, S.; Lu, J. The first complete mitochondrial genome sequence of Cacopsylla citrisuga (Yang & Li), a new insect vector of Huanglongbing in Yunnan Province, China. Mitochondrial DNA B Resour. 2021, 6, 575–577. [Google Scholar] [CrossRef]
- Zhao, Y.S.; Luo, X.Y.; Liu, Y.Q.; Shi, A.X.; Cai, W.Z.; Song, F. Cacopsylla fuscicella Sp. Nov. (Hemiptera, Psyllidae), a New Loquat Pest in China dagger. Insects 2023, 14, 414. [Google Scholar] [CrossRef]
- Que, S.Q.; Yu, L.P.; Xin, T.R.; Zou, Z.W.; Hu, L.X.; Xia, B. Complete mitochondrial genome of Cacopsylla coccinae (Hemiptera: Psyllidae). Mitochondrial DNA Part A 2016, 27, 3169–3170. [Google Scholar] [CrossRef]
- Kang, A.R.; Kim, M.J.; Park, J.S.; Seo, H.J.; Song, J.H.; Won, K.H.; Choi, E.D.; Kim, I. Comparative analysis of two pear pests, Cacopsylla jukyungi and Cacopsylla burckhardti (Hemiptera: Psyllidae), based on complete mitochondrial genomes and comparison to confamilial species. Agronomy 2022, 12, 37. [Google Scholar] [CrossRef]
- Jo, E.; Cho, G. The complete mitochondrial genome of Cacopsylla burckhardti (Hemiptera, Psylloidea, Psyllidae). Biodivers. Data J. 2022, 10, e85094. [Google Scholar] [CrossRef]
- Zahradnicek, P.; Brazdil, R.; Stepanek, P.; Trnka, M. Reflections of global warming in trends of temperature characteristics in the Czech Republic, 1961–2019. Int. J. Climatol. 2021, 41, 1211–1229. [Google Scholar] [CrossRef]
- Rousi, E.; Fink, A.H.; Andersen, L.S.; Becker, F.N.; Beobide-Arsuaga, G.; Breil, M.; Cozzi, G.; Heinke, J.; Jach, L.; Niermann, D.; et al. The extremely hot and dry 2018 summer in central and northern Europe from a multi-faceted weather and climate perspective. Nat. Hazard. Earth Syst. 2023, 23, 1699–1718. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- The World Psylloidea Database (from Psyl’list). Available online: https://data.nhm.ac.uk/dataset/psyl-list/resource/8746ceec-4846-4899-b607-9ba603002033 (accessed on 12 July 2023).
- Tedeschi, R.; Lauterer, P.; Brusetti, L.; Tota, F.; Alma, A. Composition, abundance and phytoplasma infection in the hawthorn psyllid fauna of northwestern Italy. Eur. J. Plant Pathol. 2009, 123, 301–310. [Google Scholar] [CrossRef]
- Tsai, C.L.; Lee, H.C.; Cho, G.; Liao, Y.C.; Yang, M.M.; Yeh, W.B. Invasive and quarantine risks of Cacopsylla chinensis (Hemiptera: Psyllidae) in East Asia: Hybridization or gene flow between differentiated lineages. J. Econ. Entomol. 2020, 113, 2890–2899. [Google Scholar] [CrossRef]
- Sauvion, N.; Peccoud, J.; Meynard, C.N.; Ouvrard, D. Occurrence data for the two cryptic species of Cacopsylla pruni (Hemiptera: Psylloidea). Biodivers. Data J. 2021, 9, e68860. [Google Scholar] [CrossRef]
- Peccoud, J.; Labonne, G.; Sauvion, N. Molecular test to assign individuals within the Cacopsylla pruni complex. PLoS ONE 2013, 8, e72454. [Google Scholar] [CrossRef]
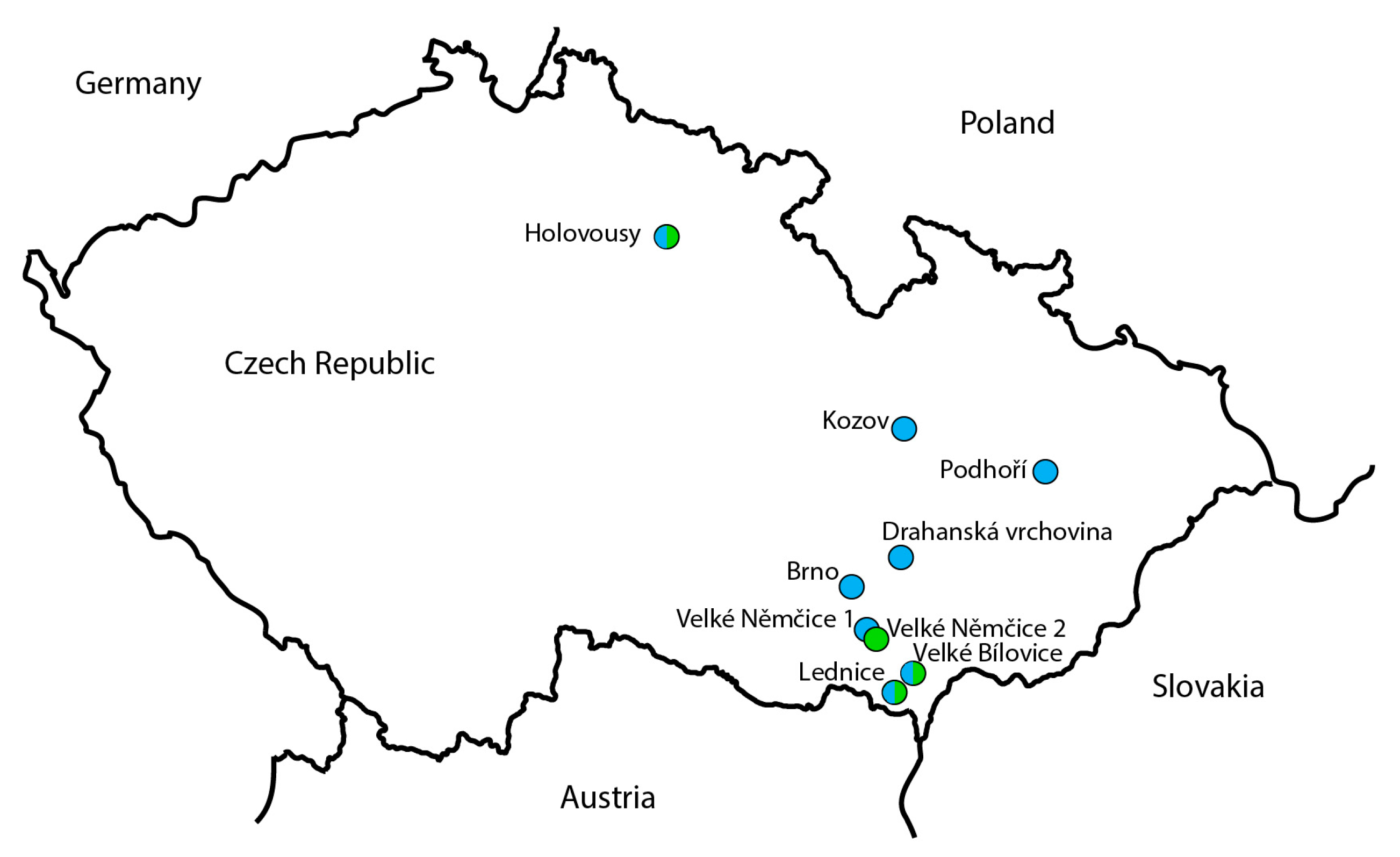



| ID | Locality (GPS Location) | Date of Collection | Determined by |
|---|---|---|---|
| C. picta | |||
| 499-2 | Velké Němčice 1 (49°0′10.538″ N, 16°39′10.617″ E) | 16 May 2006 | P. Lauterer |
| 457-1 | Podhoří (49°33′43.939″ N, 17°37′1.056″ E) | 26 April 2013 | P. Lauterer |
| S7 | Lednice (48°47′24.188″ N, 16°48′5.808″ E) | 28 April 2022 | M. Starý |
| AO353 | Holovousy (50°22′48.503″ N, 15°34′28.442″ E) | 29 April 2022 | J. Ouředníčková |
| 51-4,6,10,11,20 | Brno (49°9′27.367″ N, 16°34′20.229″ E) | 5 April 2005 | P. Lauterer |
| 61-1,4,8,12 | Velké Bílovice (48°51′19.028″ N, 16°54′46.743″ E) | 27 April 2005 | P. Lauterer |
| 357-1,2,3,4,5,6 | Drahanská vrchovina (49°15′40.686″ N, 16°50′40.728″ E) | 10 February 2012 | P. Lauterer |
| 359-1,2,3,4 | Drahanská vrchovina (49°15′40.686″ N, 16°50′40.728″ E) | 1 November 2012 | P. Lauterer |
| 491-13,14,15,16 | Kozov (49°42′9.601″ N, 16°51′8.130″ E) | 6 May 2015 | P. Lauterer |
| 498-7,8 | Lednice (48°47′24.188″ N, 16°48′5.808″ E) | 28 April 2022 | M. Starý |
| AO283, AO286 | Holovousy (50°22′48.503″ N, 15°34′28.442″ E) | 22 March 2022 | J. Ouředníčková |
| AO350-352,354-356 | Holovousy (50°22′48.503″ N, 15°34′28.442″ E) | 29 April 2022 | J. Ouředníčková |
| C. melanoneura | |||
| S3 (male) | Lednice (48°47′24.188″ N, 16°48′5.808″ E) | 13 June 2006 | P. Lauterer |
| 498-2 | Lednice (48°47′24.188″ N, 16°48′5.808″ E) | 28 April 2022 | M. Starý |
| AO282 | Holovousy (50°22′41.76″ N, 15°34′33.405″ E) | 22 March 2022 | J. Ouředníčková |
| 495-1,3 | Velké Němčice 2 (48°58′47.055″ N, 16°42′2.957″ E) | 29 May 2006 | P. Lauterer |
| 495-2 | Velké Bílovice (48°51′19.028″ N, 16°54′46.743″ E) | 18 April 2006 | P. Lauterer |
| 495-4; 496-7,10 | Lednice (48°47′24.188″ N, 16°48′5.808″ E) | 13 June 2006 | P. Lauterer |
| 496-1,5,6 | Velké Němčice 2 (48°58′47.055″ N, 16°42′2.957″ E) | 29 May 2006 | P. Lauterer |
| 497-3,4,6,7 | Lednice (48°47′24.188″ N, 16°48′5.808″ E) | 28 April 2022 | M. Starý |
| 498-3,4,7,8 | Lednice (48°47′24.188″ N, 16°48′5.808″ E) | 28 April 2022 | M. Starý |
| Sample (Acc. No.) | mtDNA Length | Reads Coverage (Average ± stdev) | HTS/mtDNA-Specific Reads |
|---|---|---|---|
| C. picta | |||
| 499-2 (OR346839) | 14,801 | 1202 ± 272 | 14,439,480/83,388 |
| AO353 (OR346838) | 14,801 | 644.6 ± 209.9 | 8,074,058/43,722 |
| 457-1 (OR346836) | 14,802 | 476.3 ± 122.7 | 7,902,966/31,389 |
| S7 (OR346837) | 14,802 | 754.9 ± 256.6 | 15,544,208/43,722 |
| C. melanoneura | |||
| S3 (OR346835) | 14,881 | 1243.3 ± 196.1 | 19,038,072/118,977 |
| 498-2 (OR346833) | 14,879 | 513.1 ± 76.4 | 5,920,406/32,409 |
| AO282 (OR346834) | 14,880 | 56.8 ± 17.2 | 3,732,814/3662 |
| Name | C. picta (499-2, AO353) | C. melanoneura (S3) | |||||
|---|---|---|---|---|---|---|---|
| Start | End | Length | Start | End | Length | ||
| trnI(gat) | Fw | 1 | 66 | 66 | 1 | 66 | 66 |
| trnQ(ttg) | Rev | 71 | 136 | 66 | 71 | 136 | 66 |
| trnM(cat) | Fw | 142 | 205 | 64 | 142 | 207 | 66 |
| ND2 | Fw | 206 | 1177 | 972 | 208 | 1179 | 972 |
| trnW(tca) | Fw | 1176 | 1237 | 62 | 1178 | 1240 | 63 |
| trnC(gca) | Rev | 1240 | 1302 | 63 | 1243 | 1305 | 63 |
| trnY(gta) | Rev | 1303 | 1364 | 62 | 1306 | 1367 | 62 |
| COI | Fw | 1366 | 2895 | 1530 | 1369 | 2901 | 1533 |
| trnL2(taa) | Fw | 2895 | 2958 | 64 | 2901 | 2965 | 65 |
| COII | Fw | 2959 | 3622 | 664 | 2966 | 3629 | 664 |
| trnK(ctt) | Fw | 3623 | 3692 | 70 | 3630 | 3699 | 70 |
| trnD(gtc) | Fw | 3691 | 3752 | 62 | 3701 | 3761 | 61 |
| ATP8 | Fw | 3753 | 3902 | 150 | 3762 | 3914 | 153 |
| ATP6 | Fw | 3902 | 4573 | 672 | 3911 | 4582 | 672 |
| COIII | Fw | 4573 | 5350 | 778 | 4582 | 5359 | 778 |
| trnG(tcc) | Fw | 5351 | 5408 | 58 | 5360 | 5419 | 60 |
| ND3 | Fw | 5409 | 5759 | 351 | 5420 | 5770 | 351 |
| trnA(tgc) | Fw | 5761 | 5821 | 61 | 5770 | 5829 | 60 |
| trnR(tcg) | Fw | 5826 | 5886 | 61 | 5840 | 5900 | 61 |
| trnN(gtt) | Fw | 5886 | 5951 | 66 | 5900 | 5964 | 65 |
| trnS1(gct) | Fw | 5952 | 6005 | 54 | 5965 | 6018 | 54 |
| trnE(ttc) | Fw | 6006 | 6066 | 61 | 6019 | 6078 | 60 |
| trnF(gaa) | Rev | 6055 | 6117 | 63 | 6068 | 6130 | 63 |
| ND5 | Rev | 6118 | 7735 | 1618 | 6131 | 7748 | 1618 |
| trnH(gtg) | Rev | 7736 | 7796 | 61 | 7749 | 7809 | 61 |
| ND4 | Rev | 7797 | 9045 | 1249 | 7810 | 9055 | 1246 |
| ND4l | Rev | 9039 | 9326 | 288 | 9049 | 9336 | 288 |
| trnT(tgt) | Fw | 9328 | 9388 | 61 | 9338 | 9397 | 60 |
| trnP(tgg) | Rev | 9389 | 9451 | 63 | 9398 | 9463 | 66 |
| ND6 | Fw | 9454 | 9939 | 486 | 9466 | 9951 | 486 |
| CYTB | Fw | 9939 | 11,075 | 1137 | 9951 | 11,087 | 1137 |
| trnS2(tga) | Fw | 11,077 | 11,14 | 64 | 11,091 | 11,153 | 63 |
| GCCTA motif | Rev | 11,152 | 11,156 | 5 | 11,165 | 11,169 | 5 |
| ND1 | Rev | 11,168 | 12,082 | 915 | 11,181 | 12,095 | 915 |
| trnL1(tag) | Rev | 12,083 | 12,146 | 64 | 12,096 | 12,158 | 63 |
| 16S rRNA | Rev | 12,147 | 13,303 | 1157 | 12,159 | 13,31 | 1152 |
| trnV(tac) | Rev | 13,304 | 13,365 | 62 | 13,311 | 13,373 | 63 |
| 12S rRNA | Rev | 13,366 | 14,112 | 747 | 13,375 | 14,122 | 748 |
| A+T rich | -- | 14,113 | 14,801 | 689 | 14,123 | 14,881 | 759 |
| polyT motif | -- | 14,531 | 14,550 | 20 | 14,608 | 14,630 | 23 |
| Gene | C. picta | C. melanoneura | Both Species | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Start/Stop Codons | Protein Mw [kDa] | Distance [%] | dN-dS | Start/Stop Codons | Protein Mw [kDa] | Distance [%] | dN-dS | Distance [%] | dN-dS | |
| ND2 | ATA/TAA | 37.68 | 0–0.1 | 0 | ATA/TAA | 37.5 | 0–0.2 | 0 | 17.1–17.3 | −0.760 |
| COI | ATG/TAA | 57.02 | 0–0.3 | −0.005 | ATG/TAA | 57.14 | 0–0.1 | −0.002 | 12.3–12.4 | −0.760 |
| COII | ATA/T | 25.53 | 0–0.3 | −0.010 | ATA/T | 25.61 | 0 | 0 | 12.8–13.1 | −0.942 |
| ATP6 | ATG/TAA | 25.37 | 0–0,3 | 0 | ATG/TAA | 25.37 | 0–0.3 | 0 | 15.6–15.7 | −0.933 |
| ATP8 | ATC/TAA | 6.03 | 0 | −0.010 | ATC/TAA | 5.89 | 0 | 0 | 27.2 | −0.810 |
| COIII | ATG/T | 30.65 | 0–0.1 | −0.010 | ATG/T | 30.56 | 0.1–0.3 | 0 | 13.8–14.0 | −0.701 |
| ND3 | ATA/TAA | 13.65 | 0–0.3 | −0.009 | ATT/TAA | 13.58 | 0 | 0 | 17.4–17.7 | −1.197 |
| ND5 | TTG/T | 60.29 | 0–0.2 | −0.005 | TTG/T | 60.27 | 0–0.1 | −0.002 | 15.2 | −0.271 |
| ND4 | ATG/T | 47.83 | 0–0.1 | 0.000 | ATG/T | 47.44 | 0–0.2 | −0.004 | 16.1–16.3 | −0.513 |
| ND4L | TTG/TAG | 11.12 | 0 | 0 | TTG/TAG | 11.01 | 0–0.3 | −0.010 | 21.7–24.1 | −0.225 |
| ND6 | ATA/TAA | 18.66 | 0–0.6 | −0.006 | ATA/TAA | 18.64 | 0–0.2 | 0.002 | 20.4–20.6 | −0.705 |
| CYTB | ATG/TAA | 43.42 | 0–0.2 | −0.010 | ATG/TAA | 43.3 | 0–0.2 | 0 | 14.6–14.8 | −0.959 |
| ND1 | ATA/TAA | 34.96 | 0 | 0 | ATA/TAG | 34.92 | 0–0.4 | −0.008 | 13.2–13.4 | −0.500 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šafářová, D.; Zrníková, E.; Holušová, K.; Ouředníčková, J.; Starý, M.; Navrátil, M. Molecular Characterization of Mitogenome of Cacopsylla picta and Cacopsylla melanoneura, Two Vector Species of ‘Candidatus Phytoplasma mali’. Agronomy 2023, 13, 2210. https://doi.org/10.3390/agronomy13092210
Šafářová D, Zrníková E, Holušová K, Ouředníčková J, Starý M, Navrátil M. Molecular Characterization of Mitogenome of Cacopsylla picta and Cacopsylla melanoneura, Two Vector Species of ‘Candidatus Phytoplasma mali’. Agronomy. 2023; 13(9):2210. https://doi.org/10.3390/agronomy13092210
Chicago/Turabian StyleŠafářová, Dana, Erika Zrníková, Kateřina Holušová, Jana Ouředníčková, Martin Starý, and Milan Navrátil. 2023. "Molecular Characterization of Mitogenome of Cacopsylla picta and Cacopsylla melanoneura, Two Vector Species of ‘Candidatus Phytoplasma mali’" Agronomy 13, no. 9: 2210. https://doi.org/10.3390/agronomy13092210
APA StyleŠafářová, D., Zrníková, E., Holušová, K., Ouředníčková, J., Starý, M., & Navrátil, M. (2023). Molecular Characterization of Mitogenome of Cacopsylla picta and Cacopsylla melanoneura, Two Vector Species of ‘Candidatus Phytoplasma mali’. Agronomy, 13(9), 2210. https://doi.org/10.3390/agronomy13092210






