RNAi-Mediated Interference with EonuGR1 Affects the Recognition of Phenylacetaldehyde by Empoasca onukii Matsuda (Hemiptera: Cicadellidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Culture and Reagent Materials
2.2. Total RNA Isolation
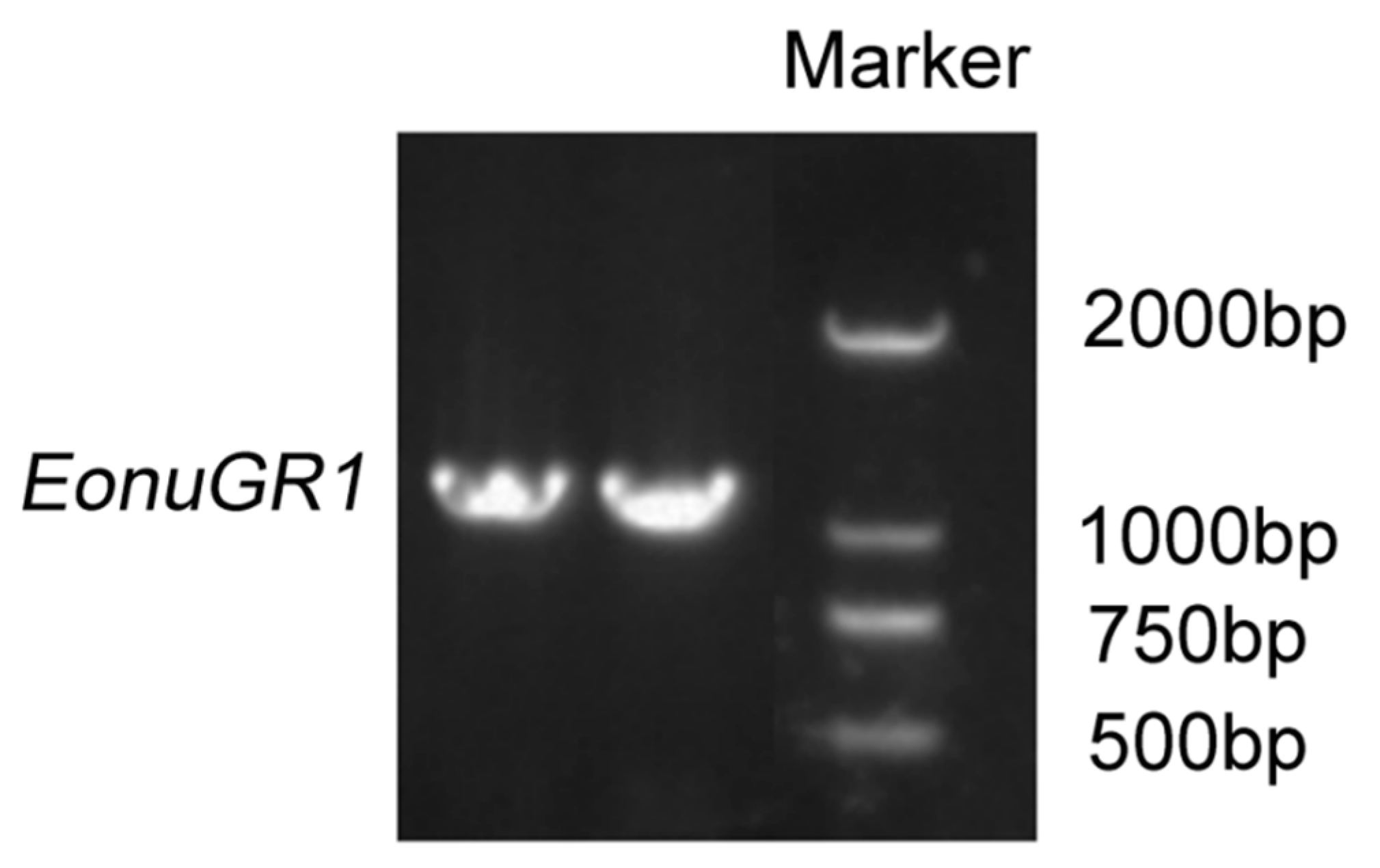
2.3. Genetic Cloning
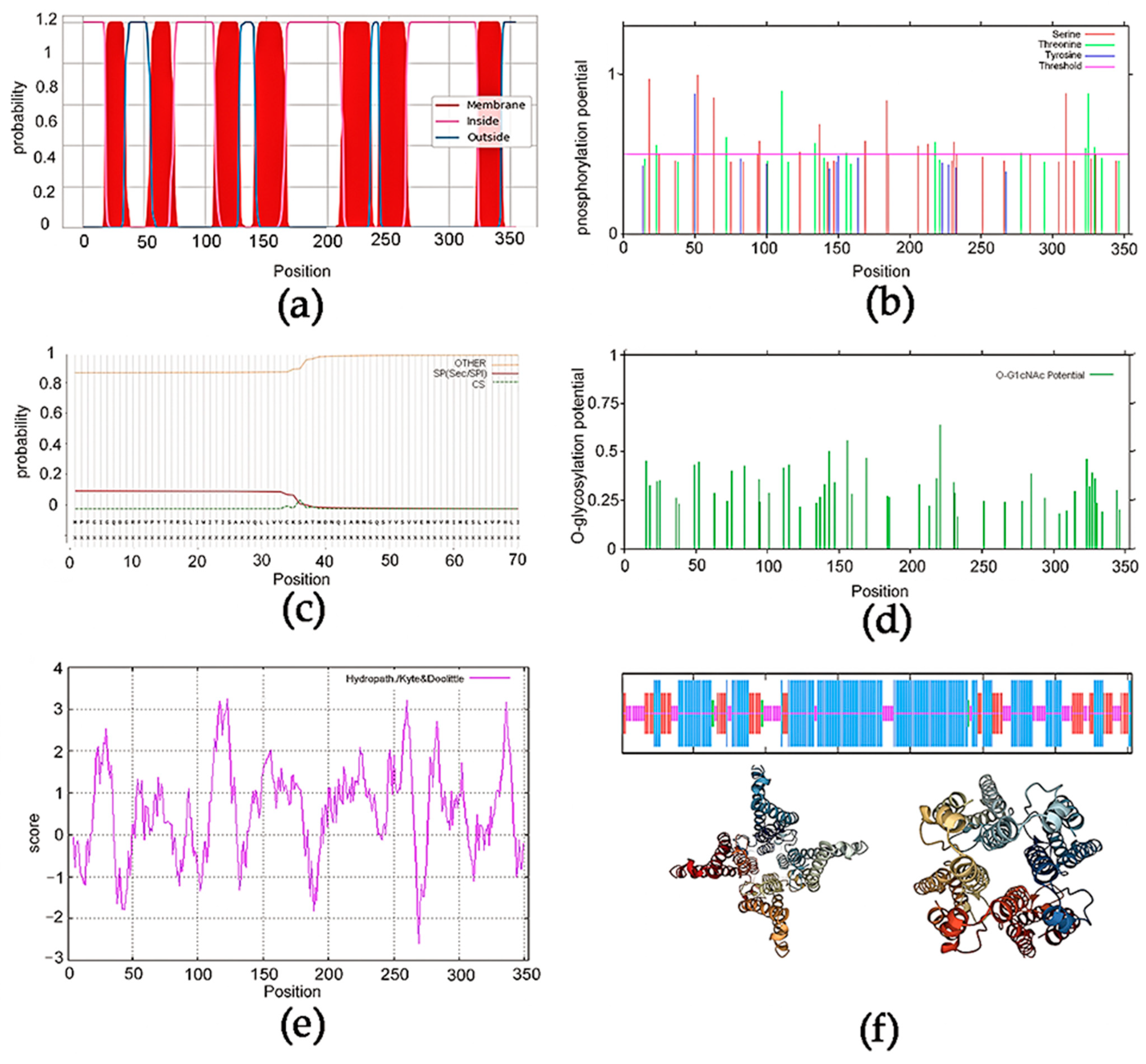
2.4. Bioinformatic Techniques
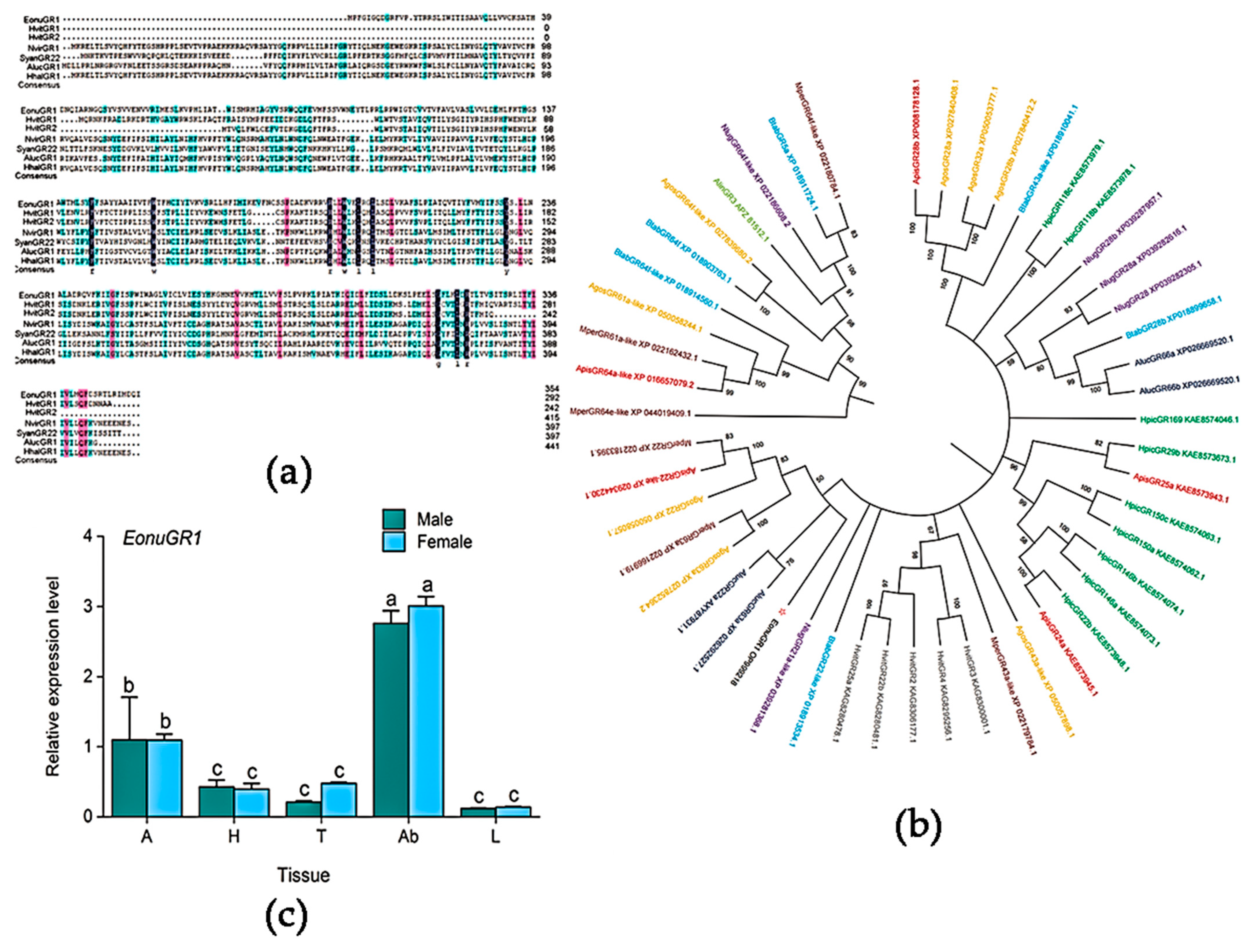
2.5. Expression Profiles of EonuGR1 in Diverse Tissues
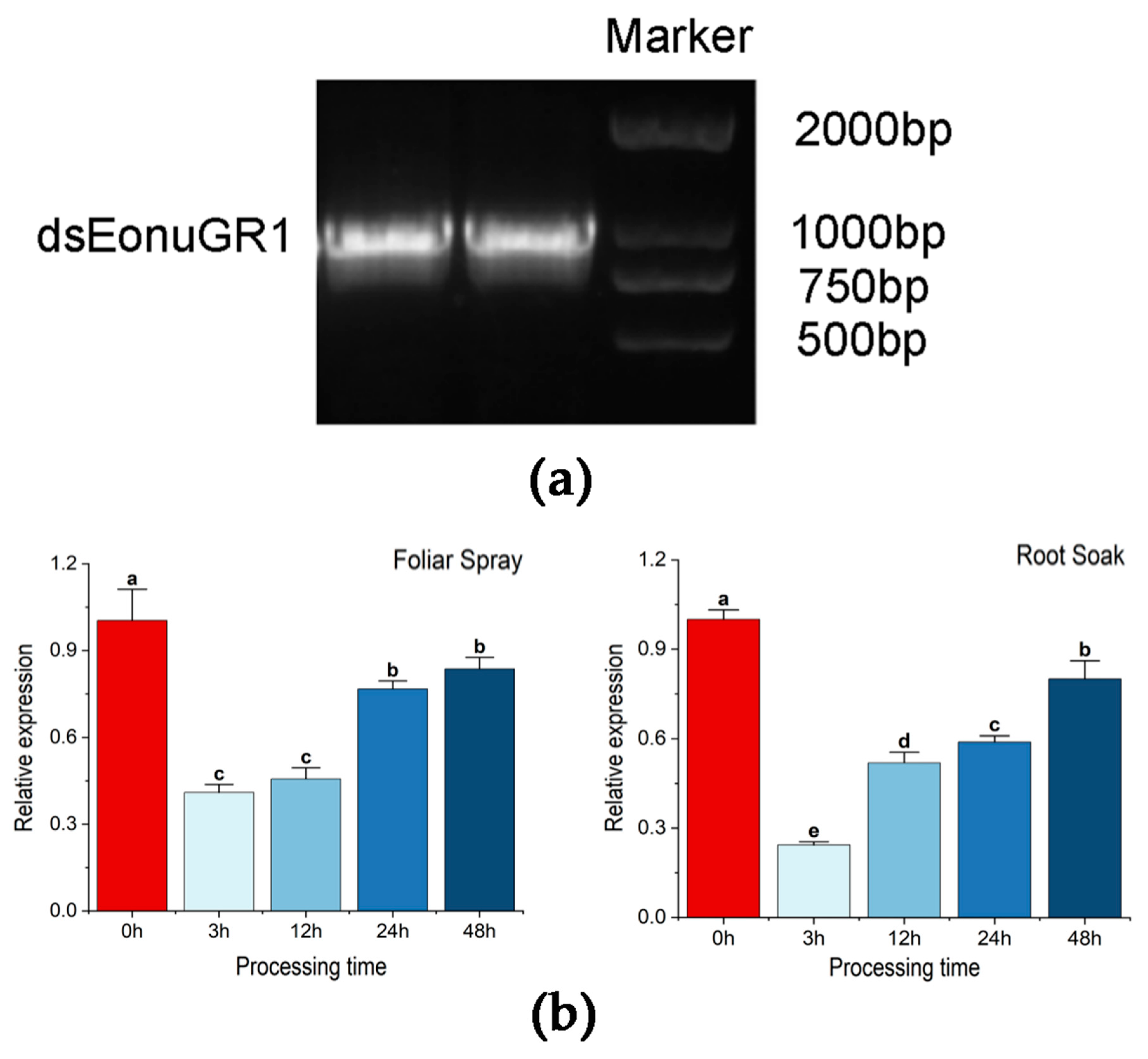
2.6. Synthesis of dsRNA
2.7. Delivery of dsRNA via Foliar Spray and Root Soak
2.8. Y-Tube Olfactometer Assays
2.9. Homology Modelling and Molecular Docking
2.10. Statistical Analysis of Data
3. Results
3.1. Sequence Prediction Analysis of EonuGR1
3.2. Sequence Alignment and Tissue Expression Profiling
3.3. In Vitro Synthesis and RNAi Interference Efficiency Analysis
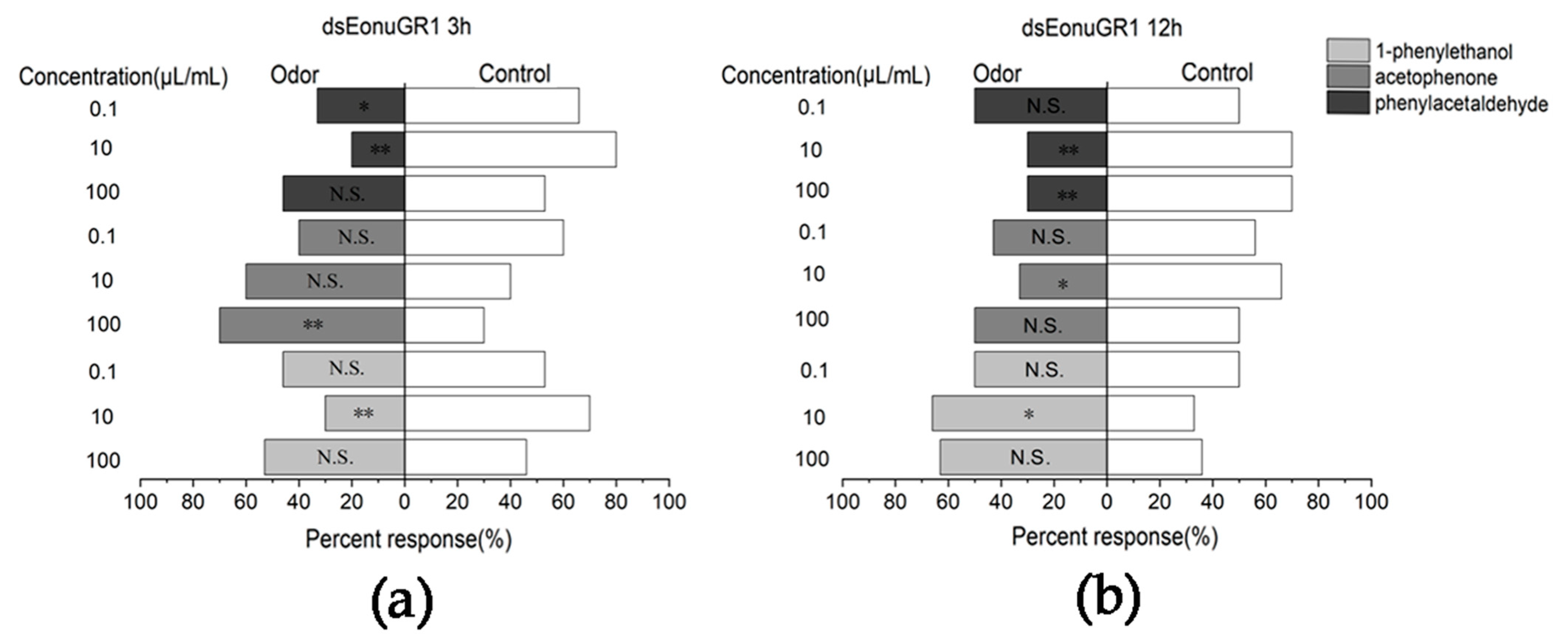
3.4. Responses to Compounds after Interferencing with EonuGR1 in E. onukii
3.5. Homology Modeling and Molecular Docking
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- da Silva Pinto, M. Tea: A New Perspective on Health Benefits. Food Res. Int. 2013, 53, 558–567. [Google Scholar] [CrossRef]
- Jin, S.; Chen, Z.M.; Backus, E.A.; Sun, X.L.; Xiao, B. Characterization of EPG Waveforms for the Tea Green Leafhopper, Empoasca vitis Göthe (Hemiptera: Cicadellidae), on Tea Plants and Their Correlation with Stylet Activities. J. Insect Physiol. 2012, 58, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Dai, J.X.; Guo, G.Y.; Wang, Z.H.; Liu, W.; Liu, X.; Chen, H.P. Residue Pattern of Chlorpyrifos and Its Metabolite in Tea from Cultivation to Consumption. J. Sci. Food Agric. 2021, 101, 4134–4141. [Google Scholar] [CrossRef]
- Lin, L.; Cheng, S.; Li, J.; Huang, M.; Zhu, D. Effects of Tea Tree Oils on Removing Pesticide Residue in Cowpea. Nongye Gongcheng Xuebao/Trans. Chin. Soc. Agric. Eng. 2013, 29, 273–278. [Google Scholar]
- Morinaga, S.; Nagata, K.; Ihara, S.; Yumita, T.; Niimura, Y.; Sato, K.; Touhara, K. Structural Model for Ligand Binding and Channel Opening of an Insect Gustatory Receptor. J. Biol. Chem. 2022, 298, 102573. [Google Scholar] [CrossRef]
- Liu, H.; Sun, X.; Shi, Z.; An, X.; Khashaveh, A.; Li, Y.; Gu, S.; Zhang, Y. Identification and Functional Analysis of Odorant-Binding Proteins Provide New Control Strategies for Apolygus lucorum. Int. J. Biol. Macromol. 2023, 224, 1129–1141. [Google Scholar] [CrossRef]
- Agnihotri, A.R.; Roy, A.A.; Joshi, R.S. Gustatory Receptors in Lepidoptera: Chemosensation and Beyond. Insect Mol. Biol. 2016, 25, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Clyne, P.J.; Warr, C.G.; Carlson, J.R. Candidate Taste Receptors in Drosophila. Science 2000, 287, 1830–1834. [Google Scholar] [CrossRef]
- Scott, K.; Brady, R.J.; Cravchik, A.; Morozov, P.; Rzhetsky, A.; Zuker, C.; Axel, R. A Chemosensory Gene Family Encoding Candidate Gustatory and Olfactory Receptors in Drosophila. Cell 2001, 104, 661–673. [Google Scholar] [CrossRef]
- Scott, K. Gustatory Processing in Drosophila melanogaster. Annu. Rev. Entomol. 2018, 63, 15–30. [Google Scholar] [CrossRef]
- Zhang, H.J.; Anderson, A.R.; Trowell, S.C.; Luo, A.R.; Xiang, Z.H.; Xia, Q.Y. Topological and Functional Characterization of an Insect Gustatory Receptor. PLoS ONE 2011, 6, e24111. [Google Scholar] [CrossRef]
- Xu, W.; Anderson, A. Carbon Dioxide Receptor Genes in Cotton Bollworm Helicoverpa armigera. Naturwissenschaften 2015, 102, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Zhu, J.L.; Zhou, W.W.; Liu, S.; Khairul, Q.M.; Ansari, N.A.; Zhu, Z.R. Identification and Expression Analysis of Putative Chemoreception Genes from Cyrtorhinus lividipennis (Hemiptera: Miridae) Antennal Transcriptome. Sci. Rep. 2018, 8, 12981. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Q.; Dong, J.F.; Chen, Q.X.; Hu, Z.J.; Sun, H.Z. Analysis of the Antennal Transcriptome and Chemoreception-Related Genes of the Bean Bug, Riptortus pedestris (Hemiptera: Alydidae). Acta Entomol. Sin. 2017, 60, 1120–1128. (In Chinese) [Google Scholar] [CrossRef]
- He, P.; Engsontia, P.; Chen, G.L.; Yin, Q.; Wang, J.; Lu, X.; Zhang, Y.N.; Li, Z.Q.; He, M. Molecular Characterization and Evolution of a Chemosensory Receptor Gene Family in Three Notorious Rice Planthoppers, Nilaparvata lugens, Sogatella furcifera and Laodelphax striatellus, Based on Genome and Transcriptome Analyses. Pest Manag. Sci. 2018, 15, 2156–2167. [Google Scholar] [CrossRef]
- Kang, K.; Yang, P.; Chen, L.E.; Pang, R.; Yu, L.J.; Zhou, W.W.; Zhu, Z.R.; Zhang, W.Q. Identification of Putative Fecundity-Related Gustatory Receptor Genes in the Brown Planthopper Nilaparvata lugens. BMC Genom. 2018, 19, 970. [Google Scholar] [CrossRef]
- Robertson, H.M.; Gadau, J.; Wanner, K.W. The Insect Chemoreceptor Superfamily of the Parasitoid Jewel Wasp Nasonia vitripennis. Insect Mol. Biol. 2010, 19, 121–136. [Google Scholar] [CrossRef]
- Ai, D.; Dong, C.; Yang, B.; Yu, C.; Wang, G. A Fructose Receptor Gene Influences Development and Feed Intake in Helicoverpa armigera. Insect Sci. 2022, 29, 993–1005. [Google Scholar] [CrossRef]
- Wu, Z.; Tong, N.; Li, Y.; Guo, J.; Lu, M.; Liu, X. Foreleg Transcriptomic Analysis of the Chemosensory Gene Families in Plagiodera versicolora (Coleoptera: Chryso, melidae). Insects 2022, 13, 763. [Google Scholar] [CrossRef]
- Dahanukar, A.; Lei, Y.T.; Kwon, J.Y.; Carlson, J.R. Two Gr Genes Underlie Sugar Reception in Drosophila. Neuron 2007, 56, 503–516. [Google Scholar] [CrossRef]
- Jiao, Y.C.; Moon, S.J.; Montell, C.A. Drosophila Gustatory Receptor Required for the Responses to Sucrose, Glucose, and Maltose Identified by Mrna Tagging. Proc. Natl. Acad. Sci. USA 2007, 104, 14110–14115. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Slone, J.; Song, X.; Amrein, H. A Fructose Receptor Functions as a Nutrient Sensor in the Drosophila Brain. Cell 2012, 151, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.D.; Cayirlioglu, P.; Kadow, I.G.; Vosshall, L.B. Two Chemosensory Receptors Together Mediate Carbon Dioxide Detection in Drosophila. Nature 2007, 445, 86–90. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, H.J.; Anderson, A. A Sugar Gustatory Receptor Identified from The Foregut of Cotton Bollworm Helicoverpa armigera. J. Chem. Ecol. 2012, 38, 1513–1520. [Google Scholar] [CrossRef]
- Montell, C. A Taste of the Drosophila Gustatory Receptors. Curr. Opin. Neurobiol. 2009, 19, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Yavuz, A.; Slone, J.; Jagge, C.; Song, X.; Amrein, H. Drosophila Sugar Receptors in Sweet Taste Perception, Olfaction, and Internal Nutrient Sensing. Curr. Biol. 2015, 25, 621–627. [Google Scholar] [CrossRef]
- Dweck, H.K.M.; Carlson, J.R. Molecular Logic and Evolution of Bitter Taste in Drosophila. Curr. Biol. 2020, 30, 17–30. [Google Scholar] [CrossRef]
- Moon, S.J.; Lee, Y.; Jiao, Y.; Montell, C. A Drosophila Gustatory Receptor Essential for Aversive Taste and Inhibiting Male-To-Male Courtship. Curr. Biol. 2009, 19, 1623–1627. [Google Scholar] [CrossRef]
- Lee, Y.; Moon, S.J.; Wang, Y.; Montell, C. A Drosophila Gustatory Receptor Required for Strychnine Sensation. Chem. Senses 2015, 40, 525–533. [Google Scholar] [CrossRef]
- Poudel, S.; Kim, Y.; Gwak, J.S.; Jeong, S.; Lee, Y. Gustatory Receptor 22e Is Essential for Sensing Chloroquine and Strychnine in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2017, 88, 30–36. [Google Scholar] [CrossRef]
- Liu, J.B.; Wu, H.; Yi, J.Q.; Zhang, G.R. Two Gustatory Receptors Are Necessary for Sensing Sucrose in an Egg Parasitoid, Trichogramma chilonis. Chemoecology 2020, 30, 103–115. [Google Scholar] [CrossRef]
- Liu, N.Y.; Xu, W.; Papanicolaou, A.; Dong, S.L.; Anderson, A. Identification and Characterization of Three Chemosensory Receptor Families in the Cotton Bollworm Helicoverpa armigera. BMC Genom. 2014, 15, 597. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, X.; Lei, C.; Zhu, F. Sensory Genes Identification with Head Transcriptome of the Migratory Armyworm, Mythimna separata. Sci. Rep. 2017, 7, 46033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, B.; Dong, S.; Cao, D.; Dong, J.; Walker, W.B.; Liu, Y.; Wang, G. Antennal Transcriptome Analysis and Comparison of Chemosensory Gene Families in Two Closely Related Noctuidae Moths, Helicoverpa armigera and H. assulta. PLoS ONE 2015, 10, e0117054. [Google Scholar] [CrossRef]
- Shim, J.; Lee, Y.; Jeong, Y.T.; Kim, Y.; Lee, M.G.; Montell, C.; Moon, S.J. The Full Repertoire of Drosophila Gustatory Receptors for Detecting an Aversive Compound. Nat. Commun. 2015, 6, 8867. [Google Scholar] [CrossRef]
- Ozaki, K.; Ryuda, M.; Yamada, A.; Utoguchi, A.; Ishimoto, H.; Calas, D.; Marion-Poll, F.; Tanimura, T.; Yoshikawa, H. A Gustatory Receptor Involved in Host Plant Recognition for Oviposition of a Swallowtail Butterfly. Nat. Commun. 2011, 2, 542. [Google Scholar] [CrossRef]
- Yang, K.; Gong, X.L.; Li, G.C.; Huang, L.Q.; Ning, C.; Wang, C.Z. A Gustatory Receptor Tuned to the Steroid Plant Hormone Brassinolide in Plutella xylostella (Lepidoptera: Plutellidae). eLife 2020, 9, e64114. [Google Scholar] [CrossRef]
- Yang, J.; Guo, H.; Jiang, N.J.; Tang, R.; Li, G.C.; Huang, L.Q.; van Loon, J.J.A.; Wang, C.Z. Identification of a Gustatory Receptor Tuned to Sinigrin in The Cabbage Butterfly Pieris rapae. PLoS Genet. 2021, 17, e1009527. [Google Scholar] [CrossRef]
- Zhang, X.Z. Identification and Expression Analysis of Olfactory ReceptorGenes in Empoasca onukii Matsuda. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2022. (In Chinese). [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2−ΔΔt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, R.R.; Lun, X.Y.; Zhang, Y.; Zhao, Y.H.; Xu, X.X.; Zhang, Z.Q. Characterization of Ionotropic Receptor Gene EonuIR25a in the Tea Green Leafhopper, Empoasca onukii Matsuda. Plants 2023, 12, 2034. [Google Scholar] [CrossRef]
- Valanciute, A.; Nygaard, L.; Zschach, H.; Maglegaard, J.M.; Lindorff-Larsen, K.; Stein, A. Accurate Protein Stability Predictions from Homology Models. Comput. Struct. Biotechnol. J. 2022, 21, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Quan, W.; Wang, Y.; Chen, Y.H.; Shao, Q.; Gong, Y.Z.; Hu, J.W.; Liu, W.H.; Wu, Z.J.; Wang, J.; Ma, S.B.; et al. Screening of Rosmarinic Acid from Salvia Miltiorrhizae Acting on the Novel Target Trpc1 Based on the ‘Homology Modelling-Virtual Screening-Molecular Docking-Affinity Assay-Activity Evaluation’ Method. Pharm. Biol. 2023, 61, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Fu, A.; Zhang, L. Progress in Molecular Docking. Quant. Biol. 2019, 7, 83–89. [Google Scholar] [CrossRef]
- Kaur, T.; Madgulkar, A.; Bhalekar, M.; Asgaonkar, K. Molecular Docking in Formulation and Development. Curr. Drug. Discov. Technol. 2019, 16, 30–39. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Autodock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and Autodocktools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Hill, C.A.; Fox, A.N.; Pitts, R.J.; Kent, L.B.; Tan, P.L.; Chrystal, M.A.; Cravchik, A.; Collins, F.H.; Robertson, H.M.; Zwiebel, L.J. G Protein-Coupled Receptors in Anopheles gambiae. Science 2002, 298, 176–178. [Google Scholar] [CrossRef]
- Robertson, H.M.; Warr, C.G.; Carlson, J.R. Molecular Evolution of the Insect Chemoreceptor Gene Superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2003, 100, 14537–14542. [Google Scholar] [CrossRef]
- Engsontia, P.; Sangket, U.; Chotigeat, W.; Satasook, C. Molecular Evolution of the Odorant and Gustatory Receptor Genes in Lepidopteran Insects Implications for Their Adaptation and Speciation. J. Mol. Evol. 2014, 79, 21–39. [Google Scholar] [CrossRef]
- Hallem, E.A.; Dahanukar, A.; Carlson, J.R. Insect odor and taste receptors. Annu. Rev. Entomol. 2006, 51, 113–135. [Google Scholar] [CrossRef] [PubMed]
- Kent, L.B.; Walden, K.K.O.; Robertson, H.I.M. The Cr Family Ofcandidate Gustatory and Olfactory Receptors in the Yellow-Fevermosquito Aedes aegypti. Chem. Senses 2008, 33, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Wanner, K.W.; Robertson, H.M. The Gustatory Receptor Family in the Silkworm Moth Bombyx mori Is Characterized by a Large Expansion of a Single Lineage of Putative Bitter Receptors. Insect Mol. Biol. 2008, 17, 621–629. [Google Scholar] [CrossRef]
- Ahmed, T.; Zhang, T.; Wang, Z.; He, K.; Bai, S. Gene Set of Chemosensory Receptors in the Polyembryonic Endoparasitoid Macrocentrus cingulum. Sci. Rep. 2016, 6, 24078. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.K.; Yang, S.; Wang, S.; Wang, J.; Zhang, X.X.; Liu, Y.; Xi, J.H. Identification of Candidate Chemosensory Receptors in The Antennal Transcriptome of the Large Black Chafer Holotrichia parallela Motschulsky (Coleoptera: Scarabaeidae). Comp. Biochem. Physiol. Part D Genom. Proteom. 2018, 28, 63–71. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Y.N.; Qian, J.L.; Kang, K.; Zhang, X.Q.; Deng, J.D.; Tang, Y.P.; Chen, C.; Hansen, L.; Xu, T.; et al. Identification and Expression Patterns of Anoplophora chinensis (Forster) Chemosensory Receptor Genes from the Antennal Transcriptome. Front. Physiol. 2018, 9, 90. [Google Scholar] [CrossRef]
- Ojha, A.; Zhang, W. Characterization of Gustatory Receptor 7 in the Brown Planthopper Reveals Functional Versatility. Insect Biochem. Mol. Biol. 2021, 132, 103567. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.; Shao, Y.; Wang, X.; Ma, Y.; Ling, E.; Xue, L. Gr33a Modulates Drosophila Male Courtship Preference. Sci. Rep. 2015, 5, 7777. [Google Scholar] [CrossRef]
- Robertson, H.M.; Wanner, K.W. The Chemoreceptor Superfamily in the Honey Bee, Apis mellifera: Expansion of the Odorant, but Not Gustatory, Receptor Family. Genome Res. 2006, 16, 1395–1403. [Google Scholar] [CrossRef]
- Krieger, J.; Raming, K.; Dewer, Y.M.; Bette, S.; Conzelmann, S.; Breer, H. A Divergent Gene Family Encoding Candidate Olfactory Receptors of the Moth Heliothis virescens. Eur. J. Neurosci. 2002, 16, 619–628. [Google Scholar] [CrossRef]
- Qu, C.; Wang, R.; Li, F.Q.; Luo, C. Cloning and Expression Profiling of Gustatory Receptor Genes BtabGR1 and BtabGR2 in Bemisia tabaci. Sci. Agric. Sin. 2022, 55, 2552–2561. (In Chinese) [Google Scholar] [CrossRef]
- Thorne, N.; Amrein, H. Atypical Expression of Drosophila Gustatory Receptor Genes in Sensory and Central Neurons. J. Comp. Neurol. 2008, 506, 548–568. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kwon, J.Y. A Systematic Analysis of Drosophila Gustatory Receptor Gene Expression in Abdominal Neurons Which Project to the Central Nervous System. Mol. Cells 2011, 32, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 9, 1912. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.; Waterhouse, P. RNAi for Insect-proof Plants. Nat. Biotechnol. 2007, 25, 1231–1232. [Google Scholar] [CrossRef]
- Chen, S.; Luo, X.; Nanda, S.; Yang, C.; Li, Z.; Zhang, Y.; Zhou, X.; Pan, H. RNAi-Based Biopesticides Against 28-Spotted Ladybeetle Henosepilachna vigintioctopunctata Does Not Harm the Insect Predator Propylea Japonica. J. Agric. Food Chem. 2023, 71, 3373–3384. [Google Scholar] [CrossRef]
- Andrade, C.E.; Hunter, W.B. RNA Interference -Natural Gene-Based Technology for Highly Specific Pest Control (HiSPeC). RNA Interf. 2016, 1, 391–409. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New Insights into an RNAi Approach for Plant Defence against Piercing-Sucking and Stem-Borer Insect Pests. Plant. Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef]
- Li, H.; Khajuria, C.; Rangasamy, M.; Gandra, P.; Fitter, M.; Geng, C.; Woosely, A.; Hasler, J.; Schulenberg, G.; Worden, S.; et al. Long dsRNA but Not siRNA Initiates RNAi in Western Corn Rootworm Larvae and Adults. J. Appl. Entomol. 2015, 139, 432–445. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full Crop Protection from an Insect Pest by Expression of Long Double-Stranded Rnas in Plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef]
- Liu, X.L.; Yan, Q.; Yang, Y.L.; Hou, W.; Miao, C.L.; Peng, Y.C.; Dong, S.L. A Gustatory Receptor Gr8 Tunes Specifically to D-Fructose in the Common Cutworm Spodoptera litura. Insects 2019, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Sun, S.J.; Hou, W.; Zhang, J.; Yan, Q.; Dong, S.L. Functional Characterization of Two Spliced Variants of Fructose Gustatory Receptor in the Diamondback Moth, Plutella xylostella. Pestic. Biochem. Physiol. 2020, 164, 7–13. [Google Scholar] [CrossRef]
- Jung, J.W.; Park, K.W.; Ahn, Y.J.; Kwon, H.W. Functional Characterization of Sugar Receptors in the Western Honeybee, Apis mellifera. J. Asia Pac. Entomol. 2015, 18, 19–26. [Google Scholar] [CrossRef]
- Mang, D.; Mayu, K.; Toyama, T.; Yamagishi, T.; Sato, R. BmGR4 Responds to Sucrose and Glucose and Expresses in Tachykinin-Related Peptide-Secreting Enteroendocrine Cells. Insect Biochem. Mol. Biol. 2022, 150, 103858. [Google Scholar] [CrossRef] [PubMed]
- Poudel, S.; Kim, Y.; Kim, Y.T.; Lee, Y. Gustatory Receptors Required for Sensing Umbelliferone in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2015, 66, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Sang, J.; Rimal, S.; Lee, Y. Gustatory Receptor 28b Is Necessary for Avoiding Saponin in Drosophila melanogaster. EMBO Rep. 2019, 20, e47328. [Google Scholar] [CrossRef]
- Liu, J.; Wu, H.; Yi, J.; Jiang, D.; Zhang, G. Identification and Functional Characterization of D-Fructose Receptor in an Egg Parasitoid, Trichogramma chilonis. PLoS ONE 2019, 14, e0217493. [Google Scholar] [CrossRef]
- Aryal, B.; Dhakal, S.; Shrestha, B.; Lee, Y. Molecular and Neuronal Mechanisms for Amino Acid Taste Perception in the Drosophila labellum. Curr. Biol. 2022, 32, 1376–1386.e4. [Google Scholar] [CrossRef]
- Shrestha, B.; Lee, Y. Mechanisms of Carboxylic Acid Attraction in Drosophila melanogaster. Mol. Cells 2021, 44, 900–910. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular Evolution of the Major Chemosensory Gene Families in Insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Forward Primer | Reverse Primer | PCR Category |
|---|---|---|---|
| EonuGR1 | GGGGTACCATGCCATTTGGTATTGGACAAG | CCGCTCGAGCTAAATTTGATCCATGATTCTTAAAGTTCTTG | RT-PCR |
| EonuGR1 | TAGCAACTCAAGTAACCATCTA | CTTAACAACCTCATTATGTCCTT | qRT-PCR |
| dsEonuGR1 | TAATACGACTCACTATAGGG ATGCCATTTGGTATTGGACAAG | TAATACGACTCACTATAGGG CTAAATTTGATCCATGATTCTTAAAGTTCTTG | RT-PCR |
| β-actin | AGCGTGGTTACTCTTTCA | GCAACTCGTAGGACTTCT | qRT-PCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Lun, X.; Zhao, Y.; Zhang, Y.; Cao, Y.; Zhang, X.; Jin, M.; Zhang, Z.; Xu, X. RNAi-Mediated Interference with EonuGR1 Affects the Recognition of Phenylacetaldehyde by Empoasca onukii Matsuda (Hemiptera: Cicadellidae). Agronomy 2023, 13, 2221. https://doi.org/10.3390/agronomy13092221
Zhang R, Lun X, Zhao Y, Zhang Y, Cao Y, Zhang X, Jin M, Zhang Z, Xu X. RNAi-Mediated Interference with EonuGR1 Affects the Recognition of Phenylacetaldehyde by Empoasca onukii Matsuda (Hemiptera: Cicadellidae). Agronomy. 2023; 13(9):2221. https://doi.org/10.3390/agronomy13092221
Chicago/Turabian StyleZhang, Ruirui, Xiaoyue Lun, Yunhe Zhao, Yu Zhang, Yan Cao, Xiangzhi Zhang, Meina Jin, Zhengqun Zhang, and Xiuxiu Xu. 2023. "RNAi-Mediated Interference with EonuGR1 Affects the Recognition of Phenylacetaldehyde by Empoasca onukii Matsuda (Hemiptera: Cicadellidae)" Agronomy 13, no. 9: 2221. https://doi.org/10.3390/agronomy13092221
APA StyleZhang, R., Lun, X., Zhao, Y., Zhang, Y., Cao, Y., Zhang, X., Jin, M., Zhang, Z., & Xu, X. (2023). RNAi-Mediated Interference with EonuGR1 Affects the Recognition of Phenylacetaldehyde by Empoasca onukii Matsuda (Hemiptera: Cicadellidae). Agronomy, 13(9), 2221. https://doi.org/10.3390/agronomy13092221







