Identification of Soybean Germplasm and Associated Molecular Markers with Resistance to Fusarium graminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Inoculum
2.2. Plant Material
2.3. Phenotyping
2.4. Genome-Wide Association Study
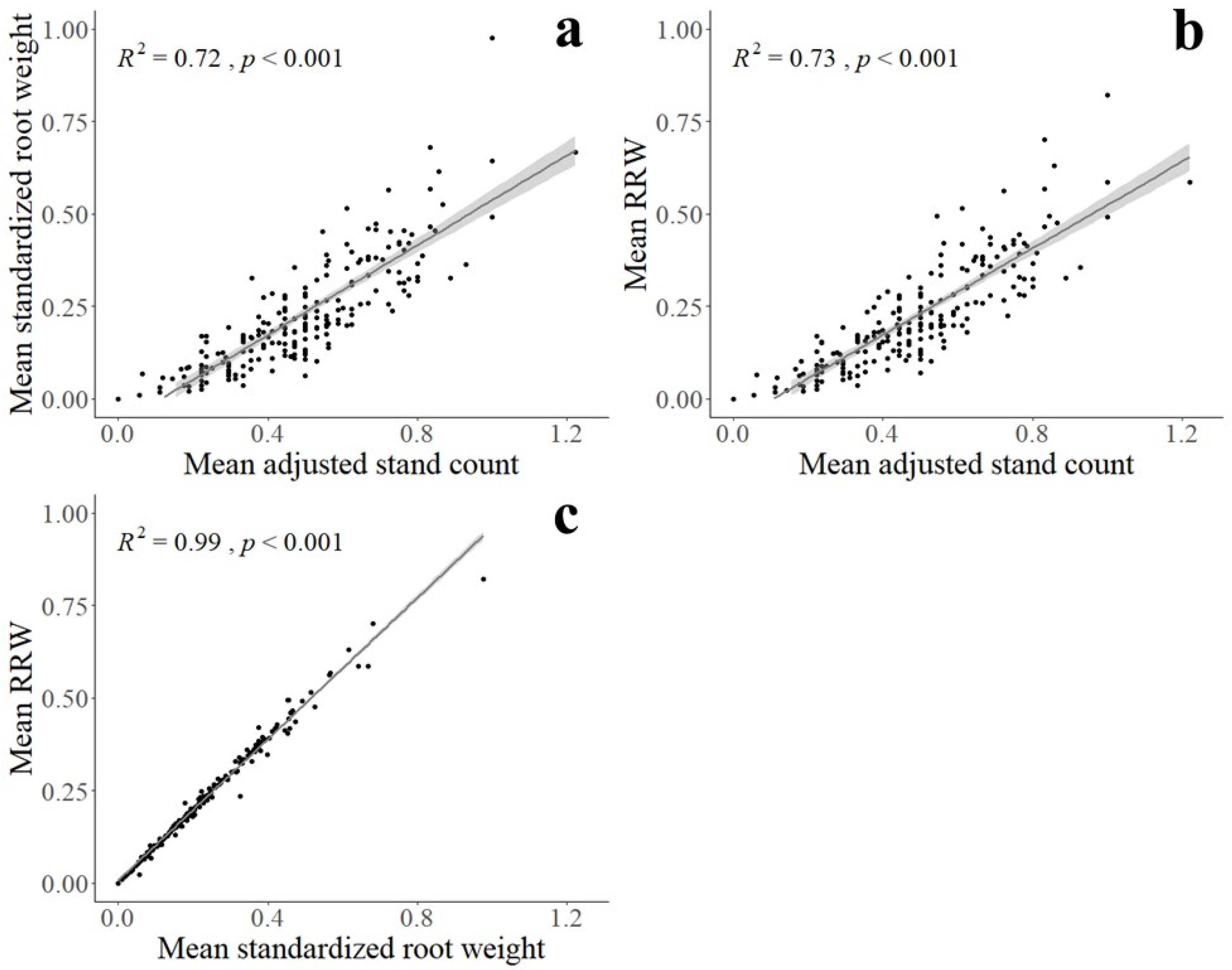
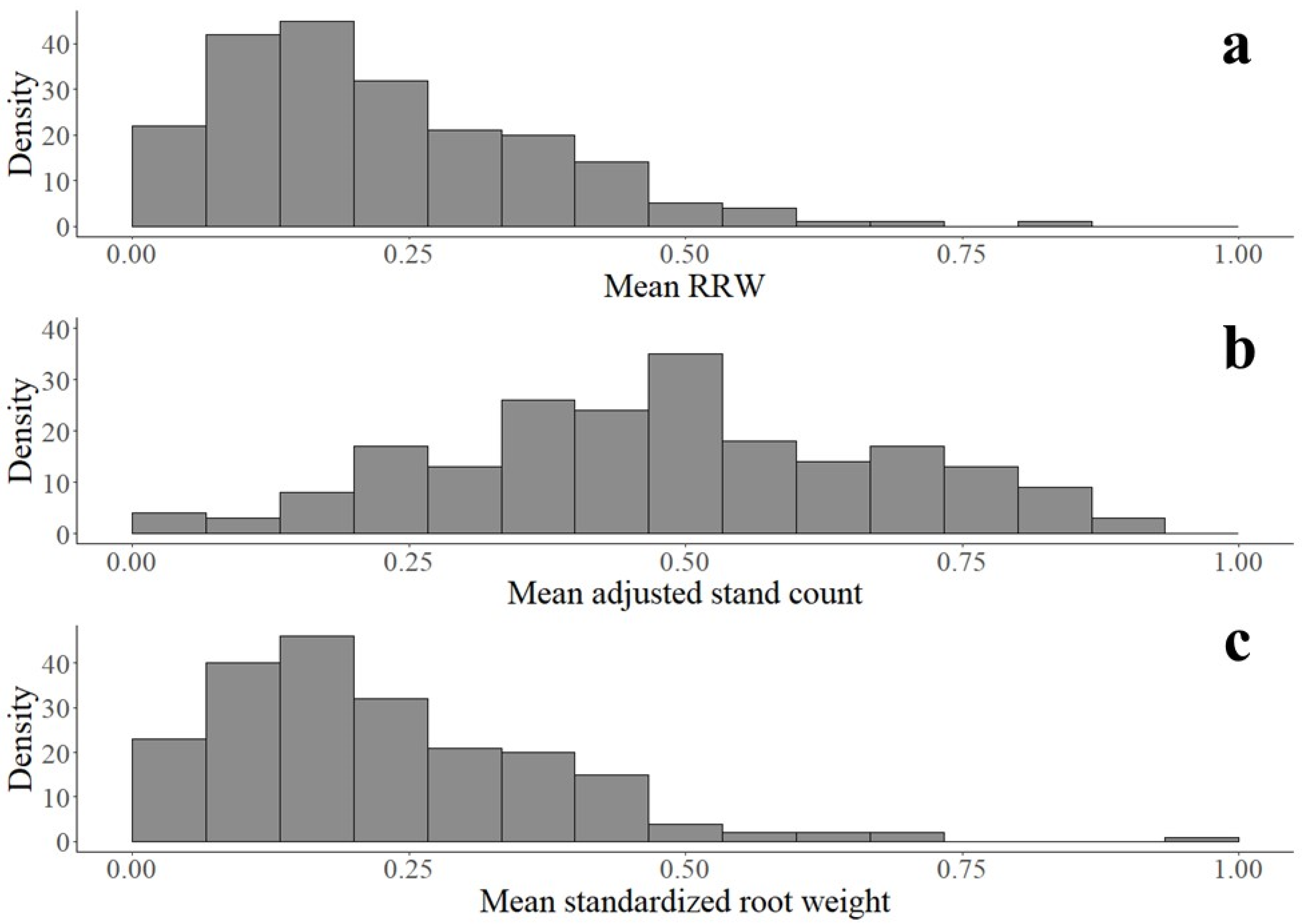
2.5. Statistical Analysis
3. Results
3.1. Soybean Accessions Resistant to Fusarium graminearum
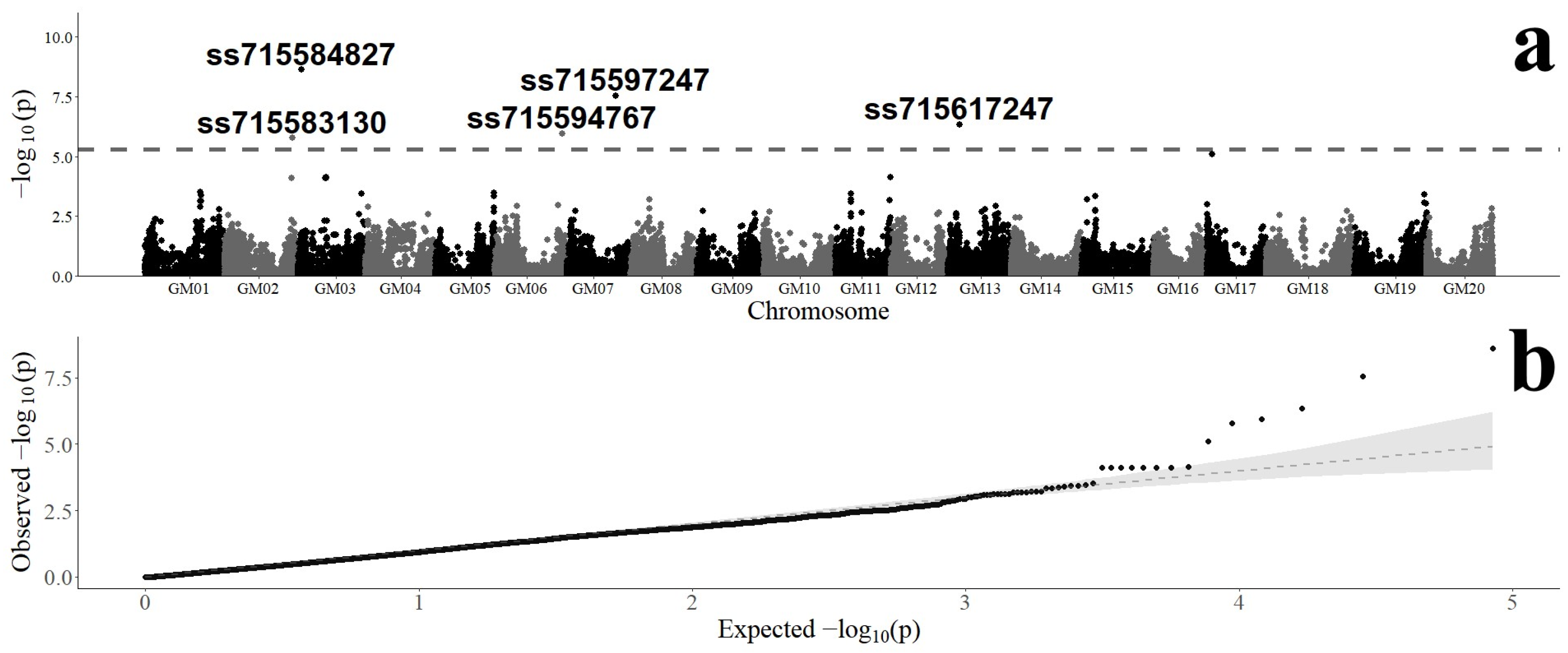
3.2. Genome-Wide Association of SNP Markers with Fusarium graminearum Resistance
4. Conclusions and Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agrios, G.N. Plant Pathology, 5th ed.; Elsevier Academic Press: Burlington, MA, USA, 2005. [Google Scholar]
- Lamichhane, J.R.; Dürr, C.; Schwanck, A.A.; Robin, M.-H.; Sarthou, J.-P.; Cellier, V.; Messéan, A.; Aubertot, J.-N. Integrated management of damping-off diseases. A review. Agron. Sustain. Dev. 2017, 37, 10. [Google Scholar] [CrossRef]
- Pimentel, M.F.; Srour, A.Y.; Warner, A.J.; Bond, J.P.; Bradley, C.A.; Rupe, J.; Chilvers, M.I.; Rojas, J.A.; Jacobs, J.L.; Little, C.R.; et al. Ecology and diversity of culturable fungal species associated with soybean seedling diseases in the Midwestern United States. J. Appl. Microbiol. 2022, 132, 3797–3811. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.A.; Jacobs, J.L.; Napieralski, S.; Karaj, B.; Bradley, C.A.; Chase, T.; Esker, P.D.; Giesler, L.J.; Jardine, D.J.; Malvick, D.K.; et al. Oomycete species associated with soybean seedlings in North America—Part I: Identification and pathogenicity characterization. Phytopathology 2017, 107, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.A.; Jacobs, J.L.; Napieralski, S.; Karaj, B.; Bradley, C.A.; Chase, T.; Esker, P.D.; Giesler, L.J.; Jardine, D.J.; Malvick, D.K.; et al. Oomycete species associated with soybean seedlings in North America-part II: Diversity and ecology in relation to environmental and edaphic factors. Phytopathology 2017, 107, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.W.; Bradley, C.A.; Sisson, A.J.; Byamukama, E.; Chilvers, M.I.; Coker, C.M.; Collins, A.A.; Damicone, J.P.; Dorrance, A.E.; Dufault, N.S.; et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 2017, 18, 19–27. [Google Scholar] [CrossRef]
- Bradley, C.A.; Allen, T.W.; Sisson, A.J.; Bergstrom, G.C.; Bissonnette, K.M.; Bond, J.; Byamukama, E.; Chilvers, M.I.; Collins, A.A.; Damicone, J.P.; et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2015 to 2019. Plant Health Prog. 2021, 22, 483–495. [Google Scholar] [CrossRef]
- Koenning, S.R.; Wrather, J.A. Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009. Plant Health Prog. 2010, 11, 5. [Google Scholar] [CrossRef]
- Wrather, J.A.; Koenning, S.R. Estimates of disease effects on soybean yields in the United States 2003 to 2005. J. Nematol. 2006, 38, 173–180. [Google Scholar]
- Winsor, S. Keep your eyes open for these wet-season soybean diseases. Crops Soils Mag. 2020, 53, 16–23. [Google Scholar] [CrossRef]
- Bandara, A.Y.; Weerasooriya, D.K.; Conley, S.P.; Allen, T.W.; Esker, P.D. Modeling the relationship between estimated fungicide use and disease-associated yield losses of soybean in the United States II: Seed-applied fungicides vs. seedling diseases. PLoS ONE 2020, 15, e0244424. [Google Scholar] [CrossRef]
- Becher, R.; Hettwer, U.; Karlovsky, P.; Deising, H.B.; Wirsel, S.G.R. Adaptation of Fusarium graminearum to tebuconazole yielded descendants diverging for levels of fitness, fungicide resistance, virulence, and mycotoxin production. Phytopathology 2010, 100, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Broders, K.D.; Lipps, P.E.; Paul, P.A.; Dorrance, A.E. Evaluation of Fusarium graminearum associated with corn and soybean seed and seedling disease in Ohio. Plant Dis. 2007, 91, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.L.; Broders, K.D.; Paul, P.A.; Dorrance, A.E. Infection of soybean seed by Fusarium graminearum and effect of seed treatments on disease under controlled conditions. Plant Dis. 2011, 95, 401–407. [Google Scholar] [CrossRef]
- Talas, F.; McDonald, B.A. Significant variation in sensitivity to a DMI fungicide in field populations of Fusarium graminearum. Plant Pathol. 2015, 64, 664–670. [Google Scholar] [CrossRef]
- Pioli, R.N.; Mozzoni, L.; Morandi, E.N. First report of pathogenic association between Fusarium graminearum and soybean. Plant Dis. 2004, 88, 220. [Google Scholar] [CrossRef] [PubMed]
- Díaz Arias, M.M.; Munkvold, G.P.; Ellis, M.L.; Leandro, L.F.S. Distribution and frequency of Fusarium species associated with soybean roots in Iowa. Plant Dis. 2013, 97, 1557–1562. [Google Scholar] [CrossRef]
- Díaz Arias, M.M.; Leandro, L.F.; Munkvold, G.P. Aggressiveness of Fusarium species and impact of root infection on growth and yield of soybeans. Phytopathology 2013, 103, 822–832. [Google Scholar] [CrossRef]
- Okello, P.N.; Petrovic, K.; Singh, A.K.; Kontz, B.; Mathew, F.M. Characterization of species of Fusarium causing root rot of Soybean (Glycine max L.) in South Dakota, USA. Can. J. Plant Pathol. 2020, 42, 560–571. [Google Scholar] [CrossRef]
- Zhang, J.X.; Xue, A.G.; Zhang, H.J.; Nagasawa, A.E.; Tambong, J.T. Response of soybean cultivars to root rot caused by Fusarium species. Can. J. Plant Sci. 2010, 90, 767–776. [Google Scholar] [CrossRef]
- Ellis, M.L.; Wang, H.; Paul, P.A.; St. Martin, S.K.; McHale, L.K. Identification of soybean genotypes resistant to Fusarium graminearum and genetic mapping of resistance quantitative trait loci in the cultivar conrad. Crop Sci. 2012, 52, 2224–2233. [Google Scholar] [CrossRef]
- Stasko, A.K.; Wickramasinghe, D.; Nauth, B.J.; Acharya, B.; Ellis, M.L.; Taylor, C.G.; McHale, L.K.; Dorrance, A.E. High-Density mapping of resistance QTL toward Phytophthora sojae, Pythium irregulare, and Fusarium graminearum in the same soybean population. Crop Sci. 2016, 56, 2476–2492. [Google Scholar] [CrossRef]
- Acharya, B.; Lee, S.; Rouf Mian, M.A.; Jun, T.-H.; McHale, L.K.; Michel, A.P.; Dorrance, A.E. Identification and mapping of quantitative trait loci (QTL) conferring resistance to Fusarium graminearum from soybean PI 567301B. Theor. Appl. Genet. 2015, 128, 827–838. [Google Scholar] [CrossRef]
- Million, C.R.; Wijeratne, S.; Cassone, B.J.; Lee, S.; Rouf Mian, M.A.; McHale, L.K.; Dorrance, A.E. Hybrid genome assembly of a major quantitative disease resistance locus in soybean toward Fusarium graminearum. Plant Genome 2019, 12, 180102. [Google Scholar] [CrossRef]
- Cheng, P.; Gedling, C.R.; Patil, G.; Vuong, T.D.; Shannon, J.G.; Dorrance, A.E.; Nguyen, H.T. Genetic mapping and haplotype analysis of a locus for quantitative resistance to Fusarium graminearum in soybean accession PI 567516C. Theor. Appl. Genet. 2017, 130, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhao, X.; Qu, Y.; Teng, W.; Qiu, L.; Zheng, H.; Wang, Z.; Han, Y.; Li, W. Loci and candidate genes in soybean that confer resistance to Fusarium graminearum. Theor. Appl. Genet. 2019, 132, 431–441. [Google Scholar] [CrossRef]
- Zhang, C.; Han, Y.; Qu, Y.; Teng, W.; Zhao, X.; Morris, B. Identification of quantitative trait loci underlying resistance of soybean to Fusarium graminearum. Plant Breed. 2020, 139, 141–147. [Google Scholar] [CrossRef]
- Detranaltes, C.; Cai, G. First report of Mycoleptodiscus terrestris causing root rot of soybean in Indiana. Plant Dis. 2021, 105, 1194. [Google Scholar] [CrossRef]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Fingerprinting Soybean Germplasm and Its Utility in Genomic Research. G3 Genes Genomes Genet. 2015, 5, 1999–2006. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Wani, S.H.; Collins, P.J.; Wen, Z.; Gu, C.; Chilvers, M.I.; Wang, D. Mapping quantitative trait loci for tolerance to Pythium irregulare in soybean (Glycine max L.). G3 Genes Genomes Genet 2018, 8, 3155–3161. [Google Scholar] [CrossRef]
- Ellis, M.L.; McHale, L.K.; Paul, P.A.; St. Martin, S.K.; Dorrance, A.E. Soybean germplasm resistant to Pythium irregulare and molecular mapping of resistance quantitative trait loci derived from the soybean accession PI 424354. Crop Sci. 2013, 53, 1008–1021. [Google Scholar] [CrossRef]
- Paul, C.; Walker, D.R. Aggressiveness of isolates of five Pythium species on seeds and seedlings of six North American soybean cultivars. Can. J. Plant Pathol. 2022, 44, 596–614. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Version 4.2.1; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Z. GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Okello, P.N.; Solanki, S.; Rafi, N.; Mathew, F. Sources of resistance, effect of maturity groups and marker-trait associations associated with Fusarium graminearum causing root rot of soybean (Glycine max). Plant Health Prog. 2023. [Google Scholar] [CrossRef]
- Bolanos-Carriel, C.; Balk, C.; Wickramasinghe, D.; Acharya, B.; Dorrance, A.E. Screening the soybean nested association mapping (SoyNAM) parents for resistance towards isolates of Phytophthora sojae, Fusarium graminearum, and species of Globisporangium. Plant Health Prog. 2023, in press. [CrossRef]
- McConaughy, S.; Amundsen, K.; Song, Q.; Pantalone, V.; Hyten, D. Recombination hotspots in soybean [Glycine max (L.) Merr.]. G3 Genes Genomes Genet. 2023, 13, jkad075. [Google Scholar] [CrossRef]
- Lin, F.; Chhapekar, S.S.; Vieira, C.C.; Da Silva, M.P.; Rojas, A.; Lee, D.; Liu, N.; Pardo, E.M.; Lee, Y.-C.; Dong, Z.; et al. Breeding for disease resistance in soybean: A global perspective. Theor. Appl. Genet. 2022, 135, 3773–3872. [Google Scholar] [CrossRef]
- Scott, K.; Balk, C.; Veney, D.; McHale, L.K.; Dorrance, A.E. Quantitative Disease Resistance Loci towards Phytophthora sojae and Three Species of Pythium in Six Soybean Nested Association Mapping Populations. Crop Sci. 2019, 59, 605–623. [Google Scholar] [CrossRef]
- Almeida-Silva, F.; Venancio, T.M. Integration of genome-wide association studies and gene coexpression networks unveils promising soybean resistance genes against five common fungal pathogens. Sci. Rep. 2021, 11, 24453. [Google Scholar] [CrossRef] [PubMed]
| Variety a | RRW b | Stand Count c | Seed Coat Color d | Origin d |
|---|---|---|---|---|
| PI 548311 | 0.823 ± 0.46 | 1 ± 0.16 | Yellow | Ontario, Canada |
| PI 438500 | 0.7 ± 0.13 | 0.833 ± 0.08 | Greenish brown | United States |
| PI 561318 A | 0.631 ± 0.27 | 0.857 ± 0.06 | Black | Beijing city, China |
| PI 547690 | 0.587 ± 0.05 | 1.222 ± 0.14 | Yellow | Illinois, United States |
| PI 391577 | 0.586 ± 0.11 | 1 ± 0.13 | Brown | Jilin Province, China |
| PI 157484 | 0.568 ± 0.17 | 0.833 ± 0.08 | Light green | South Korea |
| PI 632418 | 0.564 ± 0 | 0.722 ± 0.12 | Yellow | Maryland, United States |
| PI 70466 -3 | 0.515 ± 0.1 | 0.611 ± 0.12 | Yellow | Jilin Province, China |
| PI 54615 -1 | 0.495 ± 0.22 | 0.545 ± 0.31 | Yellow | Heilongjiang Province, China |
| PI 549040 | 0.494 ± 0.19 | 0.846 ± 0.11 | Green | Liaoning Province, China |
| PI 81785 | 0.492 ± 0.13 | 1 ± 0 | Brown | Hokkaidô, Japan |
| PI 603420 | 0.476 ± 0.01 | 0.867 ± 0.27 | Black | China |
| PI 578375 B | 0.465 ± 0.08 | 0.833 ± 0 | Black | China |
| PI 378680 E | 0.46 ± 0.19 | 0.667 ± 0.27 | Yellow | Russian Federation |
| PI 594451 | 0.46 ± 0.12 | 0.667 ± 0.14 | Yellow | Sichuan Province, China |
| PI 639528 B | 0.444 ± 0.11 | 0.765 ± 0.04 | Brown | Primorsky krai, Russian Federation |
| PI 89005 -5 | 0.437 ± 0.12 | 0.688 ± 0.12 | Yellow | China |
| PI 547779 | 0.429 ± 0.14 | 0.75 ± 0.15 | Yellow | Illinois, United States |
| PI 548540 | 0.422 ± 0.13 | 0.778 ± 0.12 | Yellow | Iowa, United States |
| PI 592937 | 0.422 ± 0.19 | 0.563 ± 0.21 | Yellow | China |
| PI 567262 A | 0.419 ± 0.17 | 0.611 ± 0.18 | Yellow | Fujian Province, China |
| PI 458505 | 0.419 ± 0.02 | 0.75 ± 0.19 | Yellow | Liaoning Province, China |
| PI 507088 | 0.418 ± 0.15 | 0.688 ± 0.23 | Yellow | Japan |
| PI 567,685 | 0.412 ± 0.08 | 0.786 ± 0.08 | Yellow | Henan Province, China |
| PI 518750 | 0.41 ± 0.15 | 0.722 ± 0.05 | Yellow | Former Serbia and Montenegro |
| PI 68604 -1 | 0.405 ± 0.1 | 0.727 ± 0.24 | Yellow | China |
| PI 567361 | 0.395 ± 0.11 | 0.813 ± 0.01 | Yellow | Ningxia Hui Autonomous Region, China |
| PI 548634 | 0.391 ± 0.11 | 0.765 ± 0.12 | Yellow | Ohio, United States |
| PI 518751 | 0.391 ± 0.25 | 0.556 ± 0.09 | Yellow | Former Serbia and Montenegro |
| PI 84987 A | 0.385 ± 0.1 | 0.647 ± 0.17 | Yellow | Saitama, Japan |
| PI 594301 | 0.385 ± 0.12 | 0.667 ± 0.14 | Yellow | Japan |
| PI 594777 | 0.384 ± 0.07 | 0.706 ± 0.14 | Yellow | Yunnan Province, China |
| PI 84987 | 0.376 ± 0.13 | 0.667 ± 0.21 | Yellow | Saitama, Japan |
| PI 154189 | 0.373 ± 0.05 | 0.643 ± 0.07 | Yellow | Netherlands |
| PI 603424 A | 0.367 ± 0.1 | 0.8 ± 0 | Yellow | China |
| PI 567353 | 0.362 ± 0.04 | 0.667 ± 0.12 | Brown | Gansu Province, China |
| PI 514671 | 0.362 ± 0.19 | 0.556 ± 0.12 | Yellow | Heilongjiang Province, China |
| PI 567346 | 0.361 ± 0.14 | 0.75 ± 0.12 | Yellow | Gansu Province, China |
| PI 567558 | 0.357 ± 0.15 | 0.688 ± 0.3 | Yellow | Shandong Province, China |
| PI 639543 | 0.356 ± 0.04 | 0.929 ± 0.05 | Greenish brown | Primorsky krai, Russian Federation |
| PI 603399 | 0.353 ± 0.16 | 0.611 ± 0.25 | Yellow | China |
| PI 437991 B | 0.347 ± 0.14 | 0.625 ± 0.21 | Yellow | China |
| PI 475820 | 0.346 ± 0.12 | 0.722 ± 0.16 | Yellow | Xinjiang Uygur Autonomous Region, China |
| PI 567604 A | 0.339 ± 0.16 | 0.529 ± 0.13 | Yellow | Shandong Province, China |
| PI 437110 A | 0.336 ± 0.13 | 0.667 ± 0.21 | Brown | Russian Federation |
| PI 479735 | 0.333 ± 0.16 | 0.556 ± 0.24 | Yellow | Jilin Province, China |
| PI 437505 | 0.33 ± 0.13 | 0.765 ± 0.22 | Black | Primorsky krai, Russian Federation |
| PI 603494 | 0.33 ± 0.15 | 0.471 ± 0.13 | Greenish brown | China |
| PI 605765 B | 0.326 ± 0.04 | 0.889 ± 0.05 | Greenish brown | Tuyên Quang, Vietnam |
| PI 437169 B | 0.325 ± 0.1 | 0.778 ± 0.12 | Yellow | Krasnodar, Russian Federation |
| PI 603426 G | 0.323 ± 0.06 | 0.8 ± 0.17 | Yellow | China |
| PI 567418 A | 0.304 ± 0.02 | 0.625 ± 0.02 | Yellow | Shanxi Province, China |
| PI 171451 | 0.303 ± 0.02 | 0.8 ± 0.28 | Yellow | Kanagawa, Japan |
| PI 603442 | 0.303 ± 0.1 | 0.75 ± 0.1 | Black | China |
| PI 567416 | 0.302 ± 0.2 | 0.5 ± 0.21 | Yellow | Shanxi Province, China |
| PI 591431 | 0.301 ± 0.12 | 0.625 ± 0.15 | Yellow | Ontario, Canada |
| PI 157421 | 0.291 ± 0.17 | 0.412 ± 0.2 | Black | South Korea |
| PI 547460 | 0.285 ± 0.11 | 0.5 ± 0.14 | Yellow | Illinois, United States |
| PI 438239 B | 0.283 ± 0.06 | 0.765 ± 0.09 | Brown | China |
| PI 437776 | 0.281 ± 0.12 | 0.588 ± 0.24 | Yellow | China |
| PI 548512 | 0.28 ± 0.09 | 0.444 ± 0.12 | Yellow | Indiana, United States |
| PI 424391 | 0.28 ± 0.12 | 0.688 ± 0.16 | Light green | Jeollabuk-do, South Korea |
| PI 437788 A | 0.279 ± 0.03 | 0.778 ± 0.05 | Black | China |
| PI 209334 | 0.277 ± 0.12 | 0.5 ± 0.21 | Brown | Hokkaidô, Japan |
| PI 507681 B | 0.275 ± 0.12 | 0.389 ± 0.16 | Yellow | Uzbekistan |
| PI 548561 | 0.273 ± 0.09 | 0.444 ± 0.12 | Yellow | Minnesota, United States |
| PI 437160 | 0.268 ± 0.11 | 0.556 ± 0.2 | Yellow | Krasnodar, Russian Federation |
| PI 483464 A | 0.265 ± 0.01 | 0.722 ± 0.05 | Black | Ningxia Hui Autonomous Region, China |
| PI 437485 | 0.262 ± 0.06 | 0.647 ± 0.1 | Green | Primorsky krai, Russian Federation |
| PI 398296 | 0.26 ± 0.07 | 0.529 ± 0.06 | Yellow | Kyonggi, South Korea |
| PI 603345 | 0.258 ± 0.03 | 0.667 ± 0.08 | Yellow | China |
| PI 378663 | 0.256 ± 0.17 | 0.625 ± 0.32 | Greenish brown | Russian Federation |
| PI 438323 | 0.249 ± 0.14 | 0.375 ± 0.16 | Yellow | France |
| PI 87620 | 0.248 ± 0.08 | 0.5 ± 0.14 | Yellow | Hamkyeongpukto, North Korea |
| PI 438112 B | 0.247 ± 0.08 | 0.6 ± 0.16 | Yellow | China |
| PI 91160 | 0.246 ± 0.11 | 0.444 ± 0.2 | Yellow | Liaoning Province, China |
| PI 603290 | 0.238 ± 0.09 | 0.444 ± 0.12 | Yellow | China |
| PI 591432 | 0.236 ± 0.09 | 0.5 ± 0.12 | Yellow | Ontario, Canada |
| PI 437653 | 0.236 ± 0.08 | 0.529 ± 0.16 | Yellow | China |
| PI 561701 | 0.235 ± 0.12 | 0.357 ± 0.12 | Yellow | Georgia, United States |
| PI 468408 B | 0.235 ± 0.1 | 0.588 ± 0.25 | Yellow | China |
| PI 561389 B | 0.234 ± 0.04 | 0.429 ± 0.08 | Yellow | Japan |
| PI 437695 A | 0.233 ± 0.1 | 0.529 ± 0.21 | Yellow | China |
| PI 567525 | 0.231 ± 0.08 | 0.5 ± 0.08 | Yellow | Shandong Province, China |
| PI 548182 | 0.228 ± 0.09 | 0.588 ± 0.23 | Imperfect black | Illinois, United States |
| PI 548198 | 0.225 ± 0.09 | 0.556 ± 0.16 | Gray | Illinois, United States |
| PI 548427 | 0.225 ± 0.06 | 0.733 ± 0.27 | Black | Liaoning Province, China |
| PI 578493 | 0.22 ± 0.08 | 0.5 ± 0.16 | Yellow | China |
| PI 88479 | 0.217 ± 0.11 | 0.444 ± 0.18 | Yellow | Jilin Province, China |
| PI 548356 | 0.217 ± 0.06 | 0.625 ± 0.21 | Yellow | Pyongyang, North Korea |
| PI 490766 | 0.216 ± 0.12 | 0.444 ± 0.2 | Black | Hebei Province, China |
| PI 591541 | 0.215 ± 0.03 | 0.556 ± 0.16 | Yellow | Illinois, United States |
| PI 639559 B | 0.206 ± 0.04 | 0.389 ± 0.05 | Black | Ukraine |
| PI 567298 | 0.206 ± 0.09 | 0.471 ± 0.24 | Yellow | Gansu Province, China |
| PI 416751 | 0.204 ± 0.08 | 0.5 ± 0.21 | Yellow | Japan |
| PI 567293 | 0.202 ± 0.04 | 0.438 ± 0.03 | Yellow | Gansu Province, China |
| PI 89138 | 0.201 ± 0.08 | 0.5 ± 0.21 | Yellow | Hamkyeongpukto, North Korea |
| PI 567780 B | 0.201 ± 0.08 | 0.529 ± 0.13 | Yellow | Jiangsu Province, China |
| PI 467343 | 0.2 ± 0.1 | 0.556 ± 0.23 | Yellow | Jilin Province, China |
| PI 88468 | 0.199 ± 0.04 | 0.563 ± 0.11 | Black | Liaoning Province, China |
| PI 508083 | 0.199 ± 0.03 | 0.588 ± 0.04 | Yellow | Minnesota, United States |
| PI 567258 | 0.198 ± 0.07 | 0.5 ± 0.16 | Brown | Jiangxi Province, China |
| PI 597464 | 0.198 ± 0.07 | 0.611 ± 0.18 | Yellow | Zhejiang Province, China |
| PI 542403 | 0.196 ± 0.08 | 0.556 ± 0.12 | Yellow | Minnesota, United States |
| PI 548402 S | 0.194 ± 0.08 | 0.294 ± 0.09 | Black | Beijing city, China |
| PI 54614 | 0.193 ± 0.08 | 0.444 ± 0.2 | Yellow | Jilin Province, China |
| PI 567439 | 0.193 ± 0.07 | 0.529 ± 0.06 | Yellow | Shanxi Province, China |
| PI 603549 | 0.191 ± 0.05 | 0.444 ± 0.16 | Black | China |
| PI 81041 -1 | 0.187 ± 0.11 | 0.471 ± 0.06 | Reddish brown | Hokkaidô, Japan |
| PI 603389 | 0.187 ± 0.08 | 0.5 ± 0.21 | Yellow | China |
| PI 548402 | 0.187 ± 0.05 | 0.4 ± 0.14 | Black | Beijing city, China |
| PI 291310 C | 0.185 ± 0.03 | 0.5 ± 0.06 | Yellow | Heilongjiang Province, China |
| PI 506933 | 0.181 ± 0.07 | 0.471 ± 0.13 | Yellow | Japan |
| PI 507467 | 0.18 ± 0.1 | 0.375 ± 0.15 | Yellow | Japan |
| PI 438309 | 0.18 ± 0.05 | 0.625 ± 0.14 | Yellow | China |
| PI 548298 | 0.179 ± 0.09 | 0.389 ± 0.2 | Yellow | China |
| PI 103088 | 0.177 ± 0.08 | 0.444 ± 0.18 | Yellow | Henan Province, China |
| PI 476352 B | 0.176 ± 0.12 | 0.444 ± 0.24 | Yellow | Kyrgyzstan |
| PI 438230 A | 0.175 ± 0.07 | 0.556 ± 0.24 | Yellow | China |
| PI 438019 B | 0.173 ± 0.07 | 0.333 ± 0.16 | Yellow | China |
| PI 404188 A | 0.17 ± 0.07 | 0.222 ± 0.09 | Yellow | China |
| PI 84631 | 0.17 ± 0.08 | 0.375 ± 0.15 | Green | Kyonggi, South Korea |
| PI 548521 | 0.17 ± 0.08 | 0.412 ± 0.11 | Yellow | Iowa, United States |
| PI 612730 | 0.169 ± 0.06 | 0.333 ± 0.08 | Yellow | China |
| PI 603675 | 0.167 ± 0.09 | 0.529 ± 0.24 | Yellow | China |
| PI 81041 | 0.167 ± 0.12 | 0.357 ± 0.17 | Yellow | Hokkaidô, Japan |
| PI 549021 A | 0.162 ± 0.05 | 0.333 ± 0.04 | Black | Liaoning Province, China |
| PI 86904 -1 | 0.161 ± 0.07 | 0.5 ± 0.21 | Yellow | Chungcheongbuk-do, South Korea |
| PI 417398 | 0.16 ± 0.12 | 0.333 ± 0.21 | Yellow | China |
| PI 68521 -1 | 0.157 ± 0.11 | 0.412 ± 0.2 | Yellow | China |
| PI 547686 | 0.157 ± 0.07 | 0.353 ± 0.15 | Yellow | Illinois, United States |
| PI 84946 -2 | 0.157 ± 0.09 | 0.471 ± 0.2 | Yellow | Busan-gwangyeoksi, South Korea |
| PI 437793 | 0.155 ± 0.06 | 0.563 ± 0.15 | Yellow | China |
| PI 88313 | 0.155 ± 0.09 | 0.235 ± 0.15 | Yellow | China |
| PI 567395 | 0.155 ± 0.08 | 0.4 ± 0.2 | Grayish green | Shaanxi Province, China |
| PI 497953 | 0.155 ± 0.03 | 0.333 ± 0.08 | Yellow | Bihar, India |
| PI 548391 | 0.154 ± 0.07 | 0.471 ± 0.2 | Yellow | Liaoning Province, China |
| PI 404182 | 0.153 ± 0.07 | 0.333 ± 0.16 | Yellow | China |
| PI 592960 | 0.151 ± 0.02 | 0.235 ± 0.06 | Yellow | Heilongjiang Province, China |
| PI 62203 | 0.148 ± 0.06 | 0.444 ± 0.12 | Yellow | Hebei Province, China |
| PI 567782 | 0.146 ± 0.04 | 0.471 ± 0.11 | Yellow | Ontario, Canada |
| PI 548411 | 0.146 ± 0.07 | 0.389 ± 0.16 | Yellow | China |
| PI 438496 C | 0.139 ± 0.05 | 0.389 ± 0.12 | Black | United States |
| PI 567225 | 0.138 ± 0.06 | 0.563 ± 0.24 | Yellow | Moldova |
| FC 33243 -1 | 0.132 ± 0.05 | 0.5 ± 0.21 | Yellow | Unknown |
| PI 547716 | 0.13 ± 0.05 | 0.444 ± 0.2 | Yellow | Illinois, United States |
| PI 54591 | 0.13 ± 0.07 | 0.429 ± 0.2 | Yellow | Liaoning Province, China |
| PI 548360 | 0.129 ± 0.05 | 0.333 ± 0.08 | Yellow | North Korea |
| PI 547459 | 0.129 ± 0.05 | 0.333 ± 0.08 | Yellow | Illinois, United States |
| PI 548571 | 0.128 ± 0.05 | 0.353 ± 0.15 | Yellow | Ontario, Canada |
| PI 592940 | 0.127 ± 0.06 | 0.222 ± 0.12 | Yellow | China |
| PI 548193 | 0.127 ± 0.02 | 0.5 ± 0.14 | Yellow | Iowa, United States |
| PI 437685 D | 0.126 ± 0.06 | 0.278 ± 0.12 | Yellow | China |
| PI 437814 A | 0.123 ± 0.06 | 0.267 ± 0.11 | Yellow | China |
| PI 253661 B | 0.12 ± 0.04 | 0.5 ± 0.16 | Yellow | China |
| PI 417381 | 0.119 ± 0.08 | 0.286 ± 0.14 | Yellow | Hokkaidô, Japan |
| PI 58955 | 0.118 ± 0.05 | 0.444 ± 0.18 | Yellow | Shandong Province, China |
| PI 525453 | 0.118 ± 0.01 | 0.471 ± 0.15 | Yellow | Iowa, United States |
| PI 86024 | 0.116 ± 0.05 | 0.471 ± 0.2 | Grayish green | Hokkaidô, Japan |
| PI 464896 | 0.109 ± 0.05 | 0.412 ± 0.17 | Yellow | Jilin Province, China |
| PI 567698 A | 0.108 ± 0.02 | 0.5 ± 0.16 | Yellow | Anhui Province, China |
| PI 438498 | 0.105 ± 0.03 | 0.462 ± 0.2 | Black | United States |
| PI 533655 | 0.103 ± 0.07 | 0.235 ± 0.1 | Yellow | Illinois, United States |
| PI 291294 | 0.102 ± 0.02 | 0.375 ± 0.1 | Yellow | Heilongjiang Province, China |
| PI 424354 | 0.102 ± 0.04 | 0.182 ± 0.06 | Black | Chungcheongnam-do, South Korea |
| PI 603463 | 0.101 ± 0.03 | 0.529 ± 0.06 | Yellow | China |
| PI 464912 | 0.101 ± 0.04 | 0.294 ± 0.16 | Green | Liaoning Province, China |
| PI 507293 B | 0.099 ± 0.04 | 0.278 ± 0.09 | Yellow | Japan |
| PI 68732 -1 | 0.098 ± 0.04 | 0.353 ± 0.15 | Yellow | Heilongjiang Province, China |
| PI 587588 A | 0.095 ± 0.06 | 0.294 ± 0.12 | Yellow | Jiangsu Province, China |
| PI 437127 A | 0.091 ± 0.05 | 0.222 ± 0.12 | Yellow | Georgia |
| PI 407701 | 0.09 ± 0.05 | 0.25 ± 0.12 | Yellow | China |
| PI 361066 B | 0.089 ± 0.03 | 0.333 ± 0.14 | Yellow | Romania |
| PI 83881 A | 0.088 ± 0.03 | 0.235 ± 0.04 | Yellow | Kangweonto, North Korea |
| PI 54608 -1 | 0.082 ± 0.06 | 0.294 ± 0.16 | Yellow | Liaoning Province, China |
| PI 90479 P | 0.082 ± 0.06 | 0.167 ± 0.08 | Yellow | China |
| PI 548362 | 0.081 ± 0.06 | 0.222 ± 0.12 | Yellow | Illinois, United States |
| PI 594456 A | 0.078 ± 0.02 | 0.412 ± 0.18 | Yellow | Sichuan Province, China |
| PI 437265 D | 0.074 ± 0.05 | 0.294 ± 0.16 | Yellow | Moldova |
| PI 417242 | 0.074 ± 0.04 | 0.353 ± 0.16 | Green | China |
| PI 578503 | 0.074 ± 0.03 | 0.235 ± 0.12 | Yellow | China |
| PI 417479 | 0.073 ± 0.06 | 0.222 ± 0.18 | Yellow | Japan |
| PI 437838 | 0.071 ± 0.05 | 0.313 ± 0.16 | Yellow | Russian Federation |
| PI 587804 | 0.07 ± 0.04 | 0.5 ± 0.18 | Yellow | Hubei Province, China |
| PI 407716 | 0.067 ± 0.01 | 0.222 ± 0.05 | Yellow | Jilin Province, China |
| PI 232992 | 0.067 ± 0.05 | 0.188 ± 0.12 | Black | Saga, Japan |
| PI 548631 | 0.066 ± 0.04 | 0.294 ± 0.12 | Yellow | Illinois, United States |
| PI 391583 | 0.064 ± 0.05 | 0.063 ± 0.05 | Yellow | Jilin Province, China |
| FC 29333 | 0.064 ± 0.03 | 0.222 ± 0.09 | Yellow | Unknown |
| PI 438083 | 0.061 ± 0.03 | 0.333 ± 0.16 | Yellow | China |
| PI 464923 | 0.061 ± 0.03 | 0.176 ± 0.09 | Yellow | Liaoning Province, China |
| PI 79862 -1 | 0.058 ± 0.03 | 0.118 ± 0.05 | Yellow | China |
| PI 548190 | 0.056 ± 0.01 | 0.313 ± 0.08 | Yellow | Illinois, United States |
| PI 592954 | 0.051 ± 0.02 | 0.294 ± 0.12 | Yellow | China |
| PI 578412 | 0.049 ± 0.03 | 0.222 ± 0.12 | Yellow | China |
| PI 548520 | 0.048 ± 0.03 | 0.235 ± 0.12 | Yellow | Iowa, United States |
| PI 398881 | 0.039 ± 0.03 | 0.176 ± 0.14 | Yellow | Kyonggi, South Korea |
| PI 548565 | 0.037 ± 0.01 | 0.222 ± 0.05 | Yellow | Ohio, United States |
| PI 567532 | 0.036 ± 0.02 | 0.333 ± 0.16 | Yellow | Shandong Province, China |
| PI 594880 | 0.035 ± 0.02 | 0.188 ± 0.09 | Reddish brown | Yunnan Province, China |
| PI 506942 | 0.032 ± 0.03 | 0.111 ± 0.09 | Yellow | Japan |
| PI 458510 | 0.025 ± 0.01 | 0.222 ± 0.09 | Yellow | Liaoning Province, China |
| PI 567307 | 0.024 ± 0.02 | 0.143 ± 0.27 | Black | Gansu Province, China |
| PI 83881 | 0.02 ± 0.01 | 0.188 ± 0.08 | Black | Kangweonto, North Korea |
| PI 591511 | 0.019 ± 0.01 | 0.111 ± 0.05 | Yellow | Illinois, United States |
| PI 567722 | 0.01 ± 0.01 | 0.056 ± 0.05 | Yellow | Anhui Province, China |
| PI 548348 | 0.01 ± 0.01 | 0.056 ± 0.05 | Yellow | China |
| PI 548162 | 0 ± 0 | 0 ± 0 | Yellow | Illinois, United States |
| Predictor | SS | MS | Numerator DF | Denominator DF | F-Value | p-Value a |
|---|---|---|---|---|---|---|
| Accession | 12.89 | 0.062271 | 207 | 414 | 2.1874 | 9.06 × 10−12 * |
| Estimator | Estimated Effect | T-Value | p-Value a |
|---|---|---|---|
| Intercept | 0.225 | 1.9115 | 7.07 × 10−2 |
| PI 548311 | 0.598 | 4.3395 | 1.80 × 10−5 * |
| PI 438500 | 0.469 | 3.4058 | 7.24 × 10−4 * |
| PI 561318 A | 0.406 | 2.9492 | 3.37 × 10−3 * |
| PI 547690 | 0.362 | 2.6272 | 8.93 × 10−3 * |
| PI 391577 | 0.362 | 2.6247 | 8.99 × 10−3 * |
| PI 157484 | 0.343 | 2.4898 | 1.32 × 10−2 * |
| PI 632418 | 0.339 | 2.4633 | 1.42 × 10−2 * |
| PI 70466 -3 | 0.291 | 2.1096 | 3.55 × 10−2 * |
| SNP Marker | Chromosome | Position (bp) a | Variants | Minor Allele | MAF b | B&H p-Value c | Effect d |
|---|---|---|---|---|---|---|---|
| ss715583130 | 2 | 46,379,738 | A/G | G | 0.48 | 1.41 × 10−2 | 0.0301 |
| ss715584827 | 3 | 2,309,023 | A/G | G | 0.39 | 1.05 × 10−4 | −0.0484 |
| ss715594767 | 6 | 47,356,804 | C/A | A | 0.24 | 1.18 × 10−2 | 0.0418 |
| ss715597247 | 7 | 34,593,871 | C/T | T | 0.37 | 6.15 × 10−4 | −0.0431 |
| ss715617247 | 13 | 12,663,715 | C/A | A | 0.34 | 6.60 × 10−3 | 0.0381 |
| SNP Marker | Favorable Allele | Effect | PI 548311 | PI 438500 | PI 561318A | PI 547690 | PI 391577 | PI 157484 | PI 632418 | PI 70466-3 |
|---|---|---|---|---|---|---|---|---|---|---|
| ss715583130 | G | 3.00% | G | G | G | A | G | A | A | G |
| ss715584827 | A | 4.80% | A | A | A | A | A | A | A | A |
| ss715594767 | A | 4.20% | A | C | A | A | C | C | A | A |
| ss715597247 | C | 4.30% | C | T | C | C | C | C | C | C |
| ss715617247 | A | 3.80% | A | A | A | A | A | A | C | C |
| Sum of allelic effects: | 20.20% | 20.2% | 11.7% | 20.2% | 17.1% | 16.0% | 13.0% | 13.3% | 16.3% | |
| Mean RRW score: | - | 0.823 | 0.7 | 0.631 | 0.587 | 0.586 | 0.568 | 0.564 | 0.515 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Detranaltes, C.; Ma, J.; Cai, G. Identification of Soybean Germplasm and Associated Molecular Markers with Resistance to Fusarium graminearum. Agronomy 2023, 13, 2376. https://doi.org/10.3390/agronomy13092376
Detranaltes C, Ma J, Cai G. Identification of Soybean Germplasm and Associated Molecular Markers with Resistance to Fusarium graminearum. Agronomy. 2023; 13(9):2376. https://doi.org/10.3390/agronomy13092376
Chicago/Turabian StyleDetranaltes, Christopher, Jianxin Ma, and Guohong Cai. 2023. "Identification of Soybean Germplasm and Associated Molecular Markers with Resistance to Fusarium graminearum" Agronomy 13, no. 9: 2376. https://doi.org/10.3390/agronomy13092376
APA StyleDetranaltes, C., Ma, J., & Cai, G. (2023). Identification of Soybean Germplasm and Associated Molecular Markers with Resistance to Fusarium graminearum. Agronomy, 13(9), 2376. https://doi.org/10.3390/agronomy13092376






