Identification and Genetic Diversity of Spodoptera frugiperda J. E. Smith (Lepidoptera: Noctuidae) in Egypt
Abstract
:1. Introduction
2. Materials and Methods
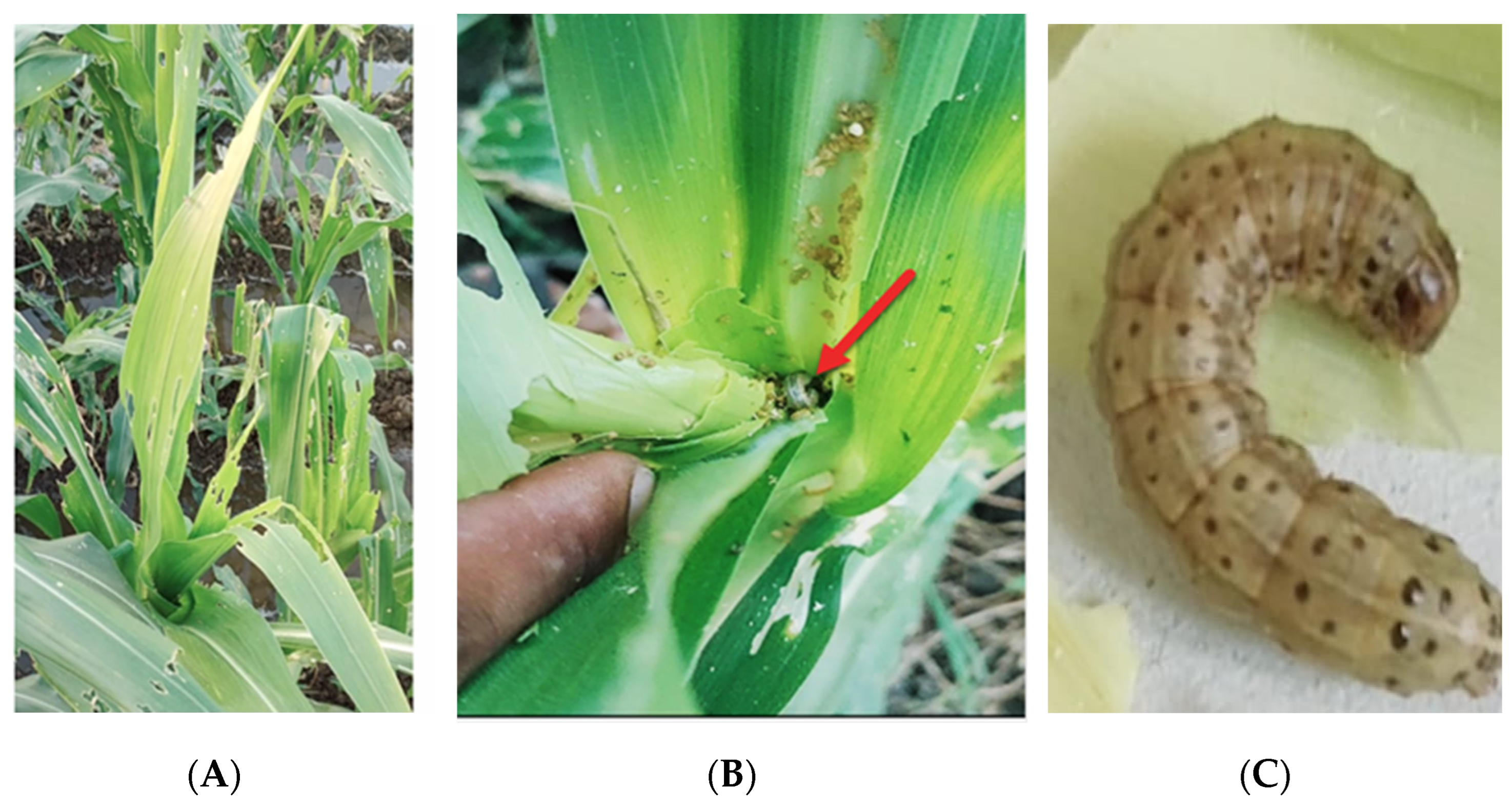
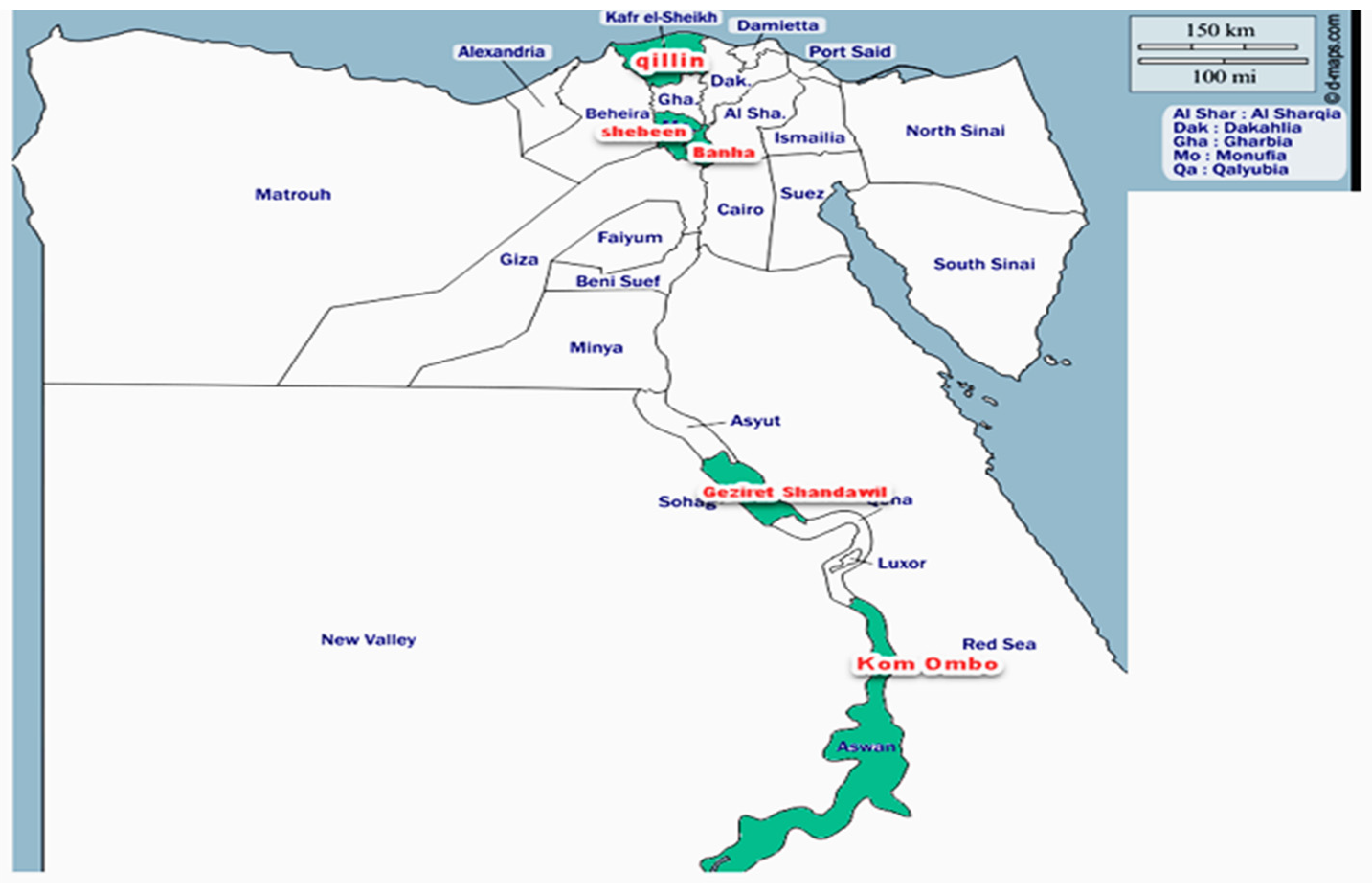
2.1. Insect Collection
2.2. Molecular Identification
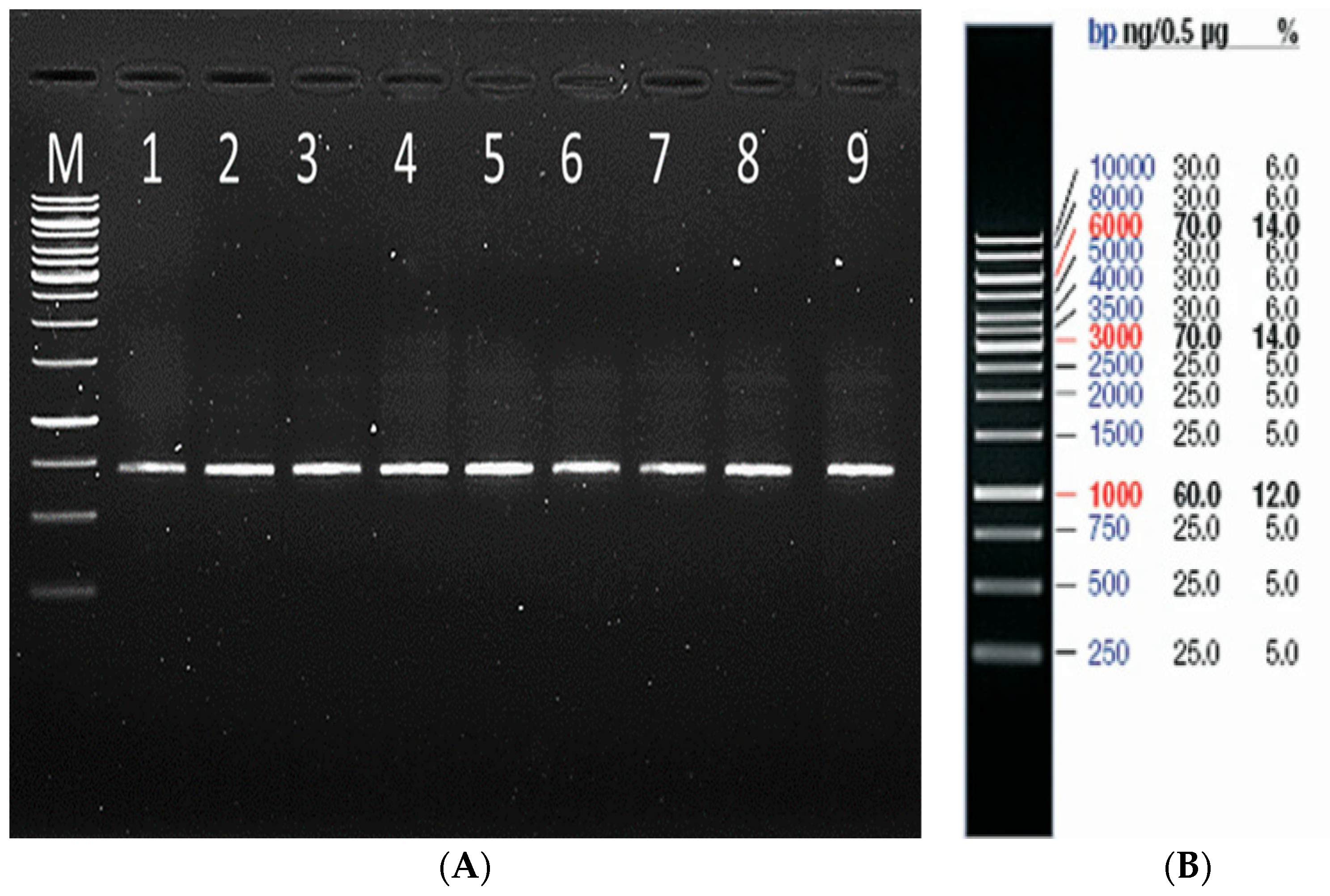
2.2.1. Genomic DNA Extraction, PCR Amplification, and Sequencing of mtCOXI Gene
2.2.2. Sequence Analysis and Phylogeny Construction
2.2.3. Genetic Population Structure Analyses
2.2.4. Genotyping of Isolated S. frugipedra by RFLP-PCR
3. Results
3.1. Molecular Identification of Spodoptera Frugiperda and Construction of Phylogenetic Tree
3.2. Polymorphism Results
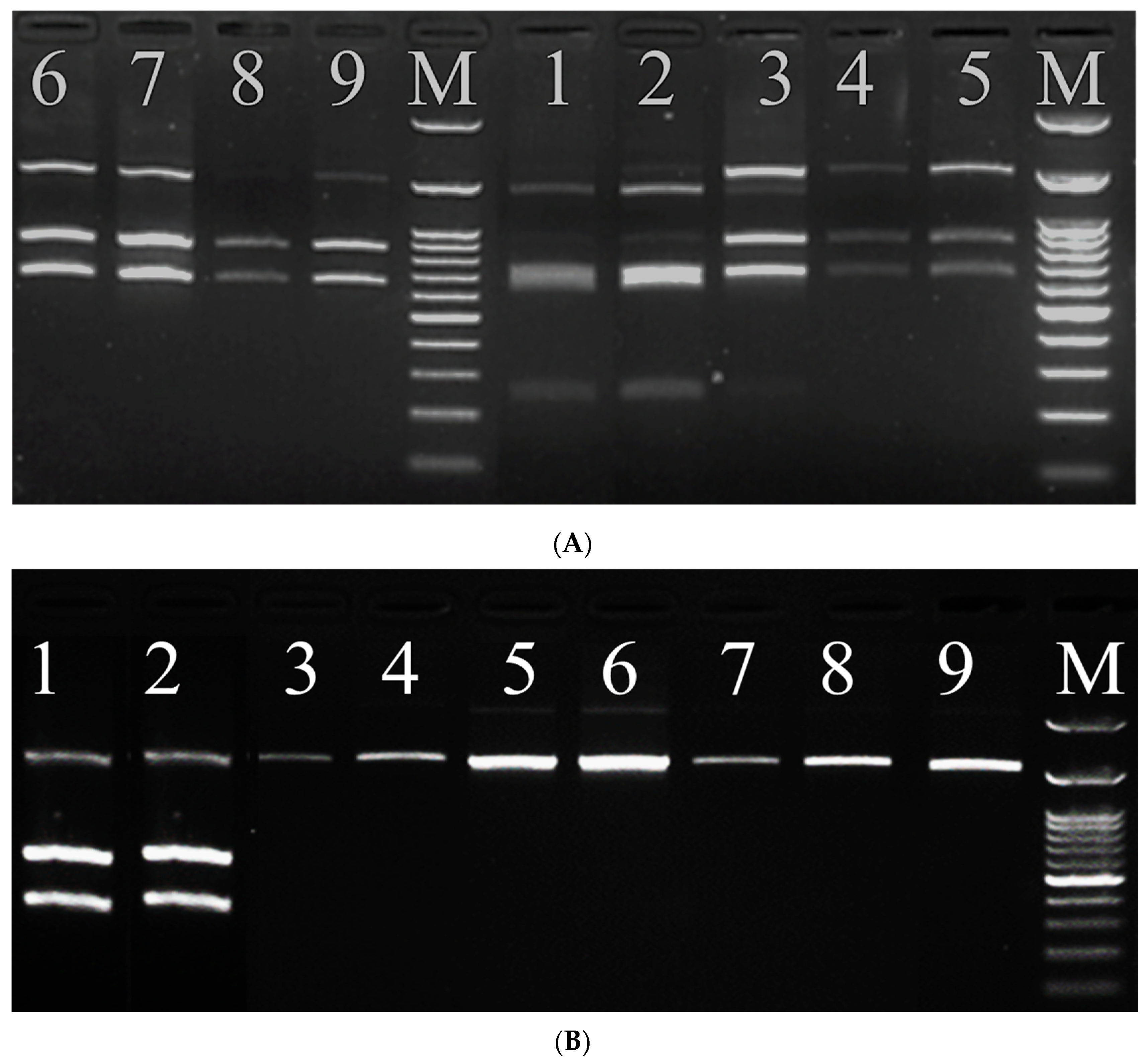
3.3. Analysis of PCR-RFLP Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stevenson, P.; Isman, M.; Belmain, S. Pesticidal plants in Africa: A global vision of new biological control products from local uses. Ind. Crop. Prod. 2017, 110, 2–9. [Google Scholar] [CrossRef]
- Kumela, T.; Simiyu, J.; Sisay, B.; Likhayo, P.; Mendesil, E.; Gohole, L.; Tefera, T. Farmers’ knowledge, perceptions, and management practices of the new invasive pest, fall armyworm (Spodoptera frugiperda) in Ethiopia and Kenya. Int. J. Pest Manag. 2018, 65, 1–9. [Google Scholar] [CrossRef]
- Hardke, J.; Lorenz, G.; Leonard, B. Fall Armyworm (Lepidoptera: Noctuidae) Ecology in Southeastern Cotton. J. Integr. Pest Manag. 2015, 6, 10. [Google Scholar] [CrossRef]
- De Groote, H.; Kimenju, S.C.; Munyua, B.; Palmas, S.; Kassie, M.; Bruce, A. Spread and impact of fall armyworm (Spodoptera frugiperda J.E. Smith) in maize production areas of Kenya. Agric. Ecosyst. Environ. 2020, 292, 106804. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, H.; El-Heneidy, A.; Dahi, H.; Awad, A. First Record of the Fall Armyworm, Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae) on Sorghum Plants, A new invasive pest in Upper Egypt. Egypt. Acad. J. Biol. Sci. Entomol. 2022, 15, 15–23. [Google Scholar] [CrossRef]
- Cope, R.C.; Ross, J.V.; Wittmann, T.A.; Watts, M.J.; Cassey, P. Predicting the Risk of Biological Invasions Using Environmental Similarity and Transport Network Connectedness. Risk Anal. Off. Publ. Soc. Risk Anal. 2019, 39, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Cock, M.J.W.; Beseh, P.K.; Buddie, A.G.; Cafá, G.; Crozier, J. Molecular methods to detect Spodoptera frugiperda in Ghana, and implications for monitoring the spread of invasive species in developing countries. Sci. Rep. 2017, 7, 4103. [Google Scholar] [CrossRef] [PubMed]
- Omuut, G.; Mollel, H.G.; Kanyesigye, D.; Akohoue, F.; Adumo Aropet, S.; Wagaba, H.; Otim, M.H. Genetic analyses and detection of point mutations in the acetylcholinesterase-1 gene associated with organophosphate insecticide resistance in fall armyworm (Spodoptera frugiperda) populations from Uganda. BMC Genom. 2023, 24, 22. [Google Scholar] [CrossRef]
- Gopalakrishnan, R.; Kalia, V.K. Biology and biometric characteristics of Spodoptera frugiperda (Lepidoptera: Noctuidae) reared on different host plants with regard to diet. Pest Manag. Sci. 2022, 78, 2043–2051. [Google Scholar] [CrossRef]
- Lewter, J.; Szalanski, A.; Nagoshi, R.; Meagher, R.; Cottone, C.; Luttrell, R. Genetic variation within and between strains of the fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). Fla. Entomol. 2006, 89, 63–68. [Google Scholar] [CrossRef]
- Blas, G.S.; Baudino, E.M.; Dias, F.M.S.; Dolibaina, D.R.; Specht, A.; Casagrande, M.M.; Cornejo, P.; Giraudo, W.G.; Mielke, O.H.H. Molecular characterization and phylogenetic assessment of agricultural-related noctuids (Lepidoptera: Noctuidae) of South America. Rev. Bras. Entomol. 2021, 65, e20210104. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H. Confidence Limits of the Maximum-Likelihood Estimate of the Hominoid three from Mitochondrial-DNA Sequences. Evolution 1989, 43, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Evolutionary relationship of DNA sequences in finite populations. Genetics 1983, 105, 437–460. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Malekera, M.J.; Mamba, D.M.; Bushabu, G.B.; Murhula, J.C.; Hwang, H.-S.; Lee, K.-Y. Genetic Diversity of the Fall Armyworm Spodoptera frugiperda (J.E. Smith) in the Democratic Republic of the Congo. Agronomy 2023, 13, 2175. [Google Scholar] [CrossRef]
- Yainna, S.; Tay, W.T.; Durand, K.; Fiteni, E.; Hilliou, F.; Legeai, F.; Clamens, A.L.; Gimenez, S.; Asokan, R.; Kalleshwaraswamy, C.M.; et al. The evolutionary process of invasion in the fall armyworm (Spodoptera frugiperda). Sci. Rep. 2022, 12, 21063. [Google Scholar] [CrossRef]
- Tay, W.T.; Gordon, K.H.J. Going global—Genomic insights into insect invasions. Curr. Opin. Insect Sci. 2019, 31, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Prentis, P.J.; Wilson, J.R.U.; Dormontt, E.E.; Richardson, D.M.; Lowe, A.J. Adaptive evolution in invasive species. Trends Plant Sci. 2008, 13, 288–294. [Google Scholar] [CrossRef]
- Bertelsmeier, C. Globalization and the anthropogenic spread of invasive social insects. Curr. Opin. Insect Sci. 2021, 46, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, H.; Crawley, M.J.; Lawton, J.H.; Usher, M.B.; Southwood, R.; O’Connor, R.J.; Gibbs, A. The Population Biology of Invaders: Discussion. Philos. Trans. R. Soc. B Biol. Sci. 1986, 314, 730–731. [Google Scholar] [CrossRef]
- Petren, K.; Case, T.J. An Experimental Demonstration of Exploitation Competition in an Ongoing Invasion. Ecology 1996, 77, 118–132. [Google Scholar] [CrossRef]
- Kowarik, I. Time lags in biological invasions with regard to the success and failure of alien species. In Time Lags in Biological Invasions with Regard to the Success and Failure of Alien Species; SPB Publishing: Amsterdam, The Netherlands, 1995; pp. 15–38. [Google Scholar]
- Acharya, R.; Akintola, A.A.; Malekera, M.J.; Kamulegeya, P.; Nyakunga, K.B.; Mutimbu, M.K.; Shrestha, Y.K.; Hemayet, J.S.; Hoat, T.X.; Dao, H.T.; et al. Genetic Relationship of Fall Armyworm (Spodoptera frugiperda) Populations That Invaded Africa and Asia. Insects 2021, 12, 439. [Google Scholar] [CrossRef] [PubMed]
- Dahi, H.F.; Salem, S.A.R.; Gamil, W.E.; Mohamed, H.O. Heat Requirements for the Fall Armyworm Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae) as a New Invasive Pest in Egypt. Egypt. Acad. J. Biol. Sci. Entomol. 2020, 13, 73–85. [Google Scholar] [CrossRef]
- Gamil, W. Fall Armyworm Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae) Biological Aspects as a New Alien Invasive Pest in Egypt. Egypt. Acad. J. Biol. Sci. Entomol. 2020, 13, 189–196. [Google Scholar] [CrossRef]
- Hailu, G.; Niassy, S.; Bässler, T.; Ochatum, N.; Studer, C.; Salifu, D.; Agbodzavu, M.K.; Khan, Z.R.; Midega, C.; Subramanian, S. Could fall armyworm, Spodoptera frugiperda (J. E. Smith) invasion in Africa contribute to the displacement of cereal stemborers in maize and sorghum cropping systems. Int. J. Trop. Insect Sci. 2021, 41, 1753–1762. [Google Scholar] [CrossRef]
- Godt, M.J.W.; Caplow, F.; Hamrick, J.L. Allozyme diversity in the federally threatened golden paintbrush, Castilleja levisecta (Scrophulariaceae). Conserv. Genet. 2005, 6, 87–99. [Google Scholar] [CrossRef]
- Martin, S.H.; Möst, M.; Palmer, W.J.; Salazar, C.; McMillan, W.O.; Jiggins, F.M.; Jiggins, C.D. Natural Selection and Genetic Diversity in the Butterfly Heliconius melpomene. Genetics 2016, 203, 525–541. [Google Scholar] [CrossRef]
- Xiao, H.; Ye, X.; Xu, H.; Mei, Y.; Yang, Y.; Chen, X.; Yang, Y.; Liu, T.; Yu, Y.; Yang, W.; et al. The genetic adaptations of fall armyworm Spodoptera frugiperda facilitated its rapid global dispersal and invasion. Mol. Ecol. Resour. 2020, 20, 1050–1068. [Google Scholar] [CrossRef] [PubMed]
- Nayyar, N.; Gracy, R.G.; Ashika, T.R.; Mohan, G.; Swathi, R.S.; Mohan, M.; Chaudhary, M.; Bakthavatsalam, N.; Venkatesan, T. Population structure and genetic diversity of invasive Fall Armyworm after 2 years of introduction in India. Sci. Rep. 2021, 11, 7760. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, M.; Ramesh, R. Occurrence, damage pattern and biology of fall armyworm, Spodoptera frugiperda (J.E. smith) (Lepidoptera: Noctuidae) on fodder crops and green amaranth in Goa, India. Phytoparasitica 2020, 48, 15–23. [Google Scholar] [CrossRef]
- Chormule, A.; Shejawal, N.; Sharanbasasapa; Kalleshwaraswamy, C.; Asokan, R.; Swamy, H. First report of the fall Armyworm, Spodoptera frugiperda (J. E. Smith) (Lepidoptera, Noctuidae) on sugarcane and other crops from Maharashtra, India. J. Entomol. Zool. Stud. 2019, 7, 114–117. [Google Scholar]
- Dharmayanthi, A.; Subagyo, V.N.O.; Nugraha, R.T.P.; Rahmini, R.; Rahmadi, C.; Darmawan, D.; Sutrisno, H. Genetic characteristics and strain types of the invasive fall armyworm Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) in Indonesia. Biodiversitas J. Biol. Divers. 2022, 23, 3928–3935. [Google Scholar] [CrossRef]
- Prowell, D.; McMichael, M.; Silvain, J.-F. Multilocus Genetic Analysis of Host Use, Introgression, and Speciation in Host Strains of Fall Armyworm (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 2009, 97, 1034–1044. [Google Scholar] [CrossRef]
- Cañas-Hoyos, N.; Márquez, E.; Saldamando-Benjumea, C. Differentiation of Spodoptera frugiperda (Lepidoptera: Noctuidae) Corn and Rice Strains From Central Colombia: A Wing Morphometric Approach. Ann. Entomol. Soc. Am. 2014, 107, 575–581. [Google Scholar] [CrossRef]
- Nagoshi, R.; Meagher, R. Review of Fall Armyworm (Lepidoptera: Noctuidae) Genetic Complexity and Migration. Fla. Entomol. 2008, 91, 546–554. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Meagher, R.L.; Flanders, K.; Gore, J.; Jackson, R.; Lopez, J.; Armstrong, J.S.; David Buntin, G.; Sansone, C.; Rogers Leonard, B. Using haplotypes to monitor the migration of fall armyworm (Lepidoptera: Noctuidae) corn-strain populations from Texas and Florida. J. Econ. Entomol. 2008, 101, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Murúa, M.G.; Nagoshi, R.N.; Dos Santos, D.A.; Hay-Roe, M.M.; Meagher, R.L.; Vilardi, J.C. Demonstration Using Field Collections that Argentina Fall Armyworm Populations Exhibit Strain-specific Host Plant Preferences. J. Econ. Entomol. 2015, 108, 2305–2315. [Google Scholar] [CrossRef] [PubMed]
- Levy, H.C.; Garcia-Maruniak, A.; Maruniak, J.E. Strain identification of Spodoptera frugiperda (Lepidoptera: Noctuidae) insects and cell line: Pcr-rflp of cytochrome oxidase c subunit i gene. Fla. Entomol. 2002, 85, 186–190. [Google Scholar] [CrossRef]
- Lewter, J.A.; Szalanski, A.L. Molecular Identification of the Fall Armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) Using PCR-RFLP. J. Agric. Urban Entomol. 2007, 24, 51–57. [Google Scholar] [CrossRef]
- McMichael, M.; Prowell, D.P. Differences in Amplified Fragment-Length Polymorphisms in Fall Armyworm (Lepidoptera: Noctuidae) Host Strains. Ann. Entomol. Soc. Am. 1999, 92, 175–181. [Google Scholar] [CrossRef]
- Mariela, L.-H. Molecular Characterization and Genetic Differentiation of Spodoptera frugiperda J. E. Smith (Lepidoptera: Noctuidae) in Maize, Rice, and Cotton Fields of Colombia with AFLP. Southwest. Entomol. 2012, 37, 193–207. [Google Scholar]
- Cano-Calle, D.; Arango Isaza, R.; Saldamando-Benjumea, C. Molecular Identification of Spodoptera frugiperda (Lepidoptera: Noctuidae) Corn and Rice Strains in Colombia by Using a PCR-RFLP of the Mitochondrial Gene Cytochrome Oxydase I (COI) and a PCR of the Gene FR (For Rice). Ann. Entomol. Soc. Am. 2015, 108, 172–180. [Google Scholar] [CrossRef]


| Country | COXI Accession Numbers | Continent |
|---|---|---|
| Bangladesh | MT933063.1 | Asia |
| Bhutan | MT324066.1 | Asia |
| Brazil | JF854740.1 | South America |
| Burkina Faso | MT152769.1 | Africa |
| Cape Verde | MT152770.1 | Africa |
| Congo | MT933053.1 | Africa |
| Guinea | MT152763.1 | Africa |
| India | MH639004.1 | Asia |
| Indonesia | OQ891323.1 | Asia |
| Japan | LC546859.1 | Asia |
| Korea | MT641268.1 | Asia |
| Mali | MT152759.1 | Africa |
| Nepal | MT103345.1 | Asia |
| Niger | MT152811.1 | Africa |
| Senegal | MT152792.1 | Africa |
| South Africa | MF593245.1 | Africa |
| Tanzania | MT103348.1 | Africa |
| Taiwan | LC508690.1 | Asia |
| Togo | MT152775.1 | Africa |
| Uganda | MT933059.1 | Africa |
| USA | MK790611.1 | North America |
| Vietnam | MT103337.1 | Asia |
| Zimbabwe | MT103346.1 | Africa |
| Sites District/City | Samples | ||
|---|---|---|---|
| ID | Coordinate | Accession Number | |
| Shebeen El-koam, Menufia | Shebeen El-koam | 30°33′37.6416″ N, 31°0′28.6128″ E | OQ925919.1 |
| Qillin, Kafr El-sheikh | egy-kafr el-sheikh1 | 31°02′47″ N 30°51′16″ E/31.04639° N 30.854581° E/31.04639; 30.854581 | OP647509.1 |
| Qillin, Kafr El-sheikh | egy-kafr el-sheikh 2 | 26°36′0″ N, 31°39′0″ E | OP649580.1 |
| Qillin, Kafr El-sheikh | Kafr Elsheikh | 26°36′0″ N, 31°39′0″ E | OQ920981.1 |
| Shandawil, Sohag | egy-Sohag | 26°37′11″ N 31°38′46″ E | OP649582.1 |
| Benha, Qalyubia | egy-kaliopia | 30°27′57.5748″ N, 31°11′5.3916″ E | OP648094.1 |
| Kom ombo, Aswan | Aswan1 | 24°28′ N 32°57′ E/24.467° N 32.950° E/24.467 | OQ920901.1 |
| Kom ombo, Aswan | Aswan2 | 24°28′ N 32°57′ E/24.467° N 32.950° E/24.467 | OQ150895.1 |
| Shandawil, Sohag | egy-KRHOSSAM | 26°37′11″ N 31°38′46″ E | OP648105.1 |
| Accession Numbers, Strains | Nucleotide Position | ||||
|---|---|---|---|---|---|
| 105 | 195 | 246 | 477 | 558 | |
| OP648105, egy-KRHOSSAM (laboratory strain—this study) | G | T | C | T | C |
| MT152769, S. frugiperda, Burkina Faso (Corn strain) | G | T | C | T | C |
| OP647509, egy-kafr el-sheikh1 (this study) | A | A | T | C | T |
| OP649580, egy-kafr el-sheikh 2 (this study) | A | A | T | C | T |
| OQ920981, Kafr Elshaikh (this study) | A | A | T | T | C |
| OQ925919, Shebeen El-koam (this study) | G | T | T | T | C |
| OP649582, egy-Sohag (this study) | G | T | C | T | C |
| OP648094, egy-kaliopia (this study) | G | T | C | T | C |
| OQ920901, Aswan1 (this study) | G | A | C | T | C |
| OQ150895, Aswan2 (this study) | G | A | T | T | C |
| MT152811- S. frugiperda, Niger (Rice strain) | A | A | T | C | T |
| Number of Polymorphic Sites | Nucleotide Diversity | Haplotype Diversity | Variance of Haplotype Diversity | Fu’s Fs | Fu and Li’s D | Tajima’s D |
|---|---|---|---|---|---|---|
| 5 | 0.00258 | 0.392 | 0.01759 | 2.074 | 1.30779 | −0.95225 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebody, K.A.E.; Salim, R.G.; El-Sayed, G.M.; Mahmoud, S.H. Identification and Genetic Diversity of Spodoptera frugiperda J. E. Smith (Lepidoptera: Noctuidae) in Egypt. Agronomy 2024, 14, 809. https://doi.org/10.3390/agronomy14040809
Lebody KAE, Salim RG, El-Sayed GM, Mahmoud SH. Identification and Genetic Diversity of Spodoptera frugiperda J. E. Smith (Lepidoptera: Noctuidae) in Egypt. Agronomy. 2024; 14(4):809. https://doi.org/10.3390/agronomy14040809
Chicago/Turabian StyleLebody, Kreema A. El, Rasha G. Salim, Ghada M. El-Sayed, and Shaymaa H. Mahmoud. 2024. "Identification and Genetic Diversity of Spodoptera frugiperda J. E. Smith (Lepidoptera: Noctuidae) in Egypt" Agronomy 14, no. 4: 809. https://doi.org/10.3390/agronomy14040809
APA StyleLebody, K. A. E., Salim, R. G., El-Sayed, G. M., & Mahmoud, S. H. (2024). Identification and Genetic Diversity of Spodoptera frugiperda J. E. Smith (Lepidoptera: Noctuidae) in Egypt. Agronomy, 14(4), 809. https://doi.org/10.3390/agronomy14040809




