Characterization and RNA-Seq Analysis of Yellow-Green Leaf Mutants in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Cultivation
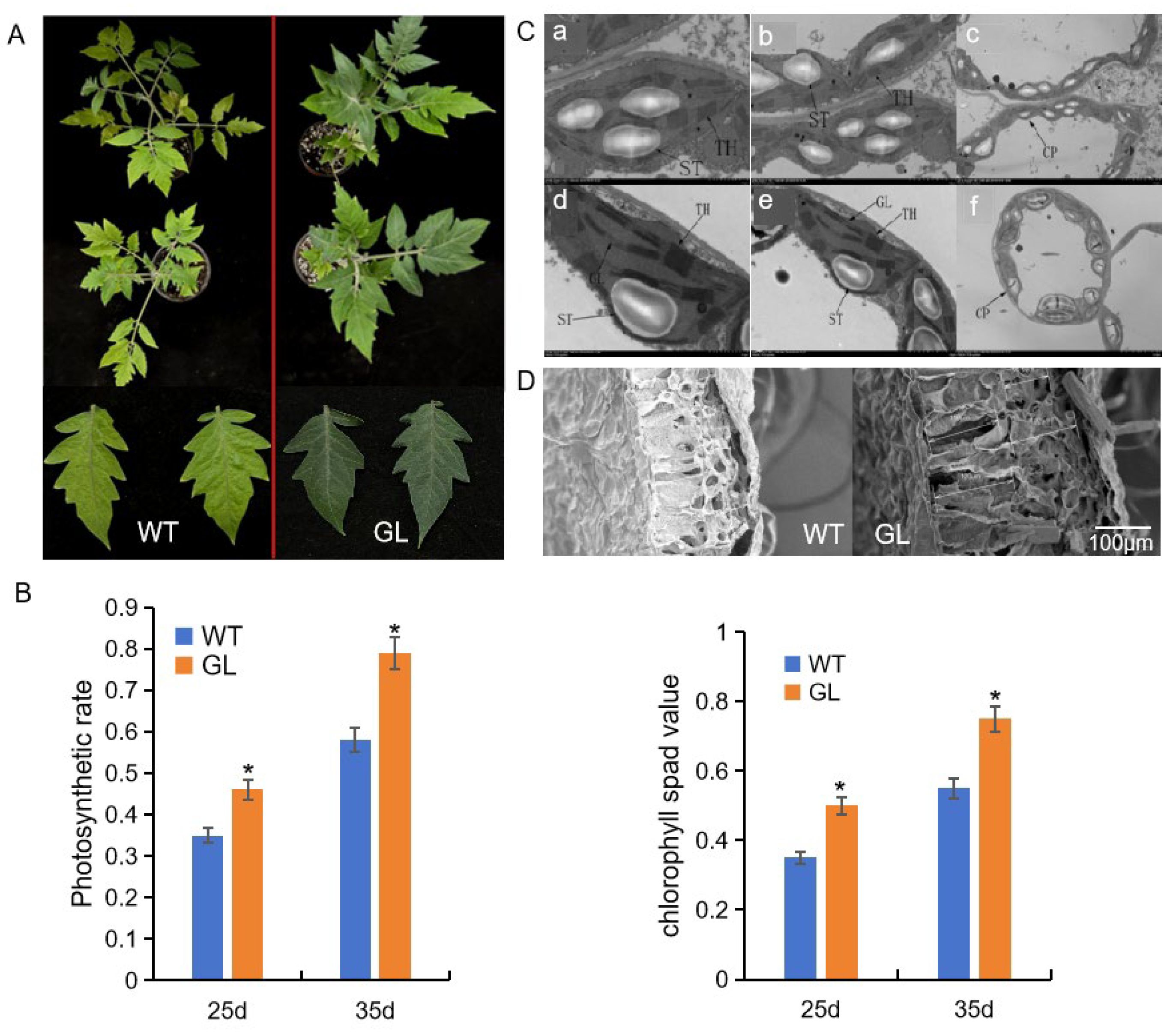
2.2. Measurement of Phenotypic and Physiological Trait
2.3. Sample Preparation for Sequencing
2.4. Transcriptome Library Construction
2.5. RNA-Seq Data Analysis
2.6. qRT-PCR Verification of Differentially Expressed Genes
3. Results
3.1. Phenotype, Physiological, and Microstructural Analyses
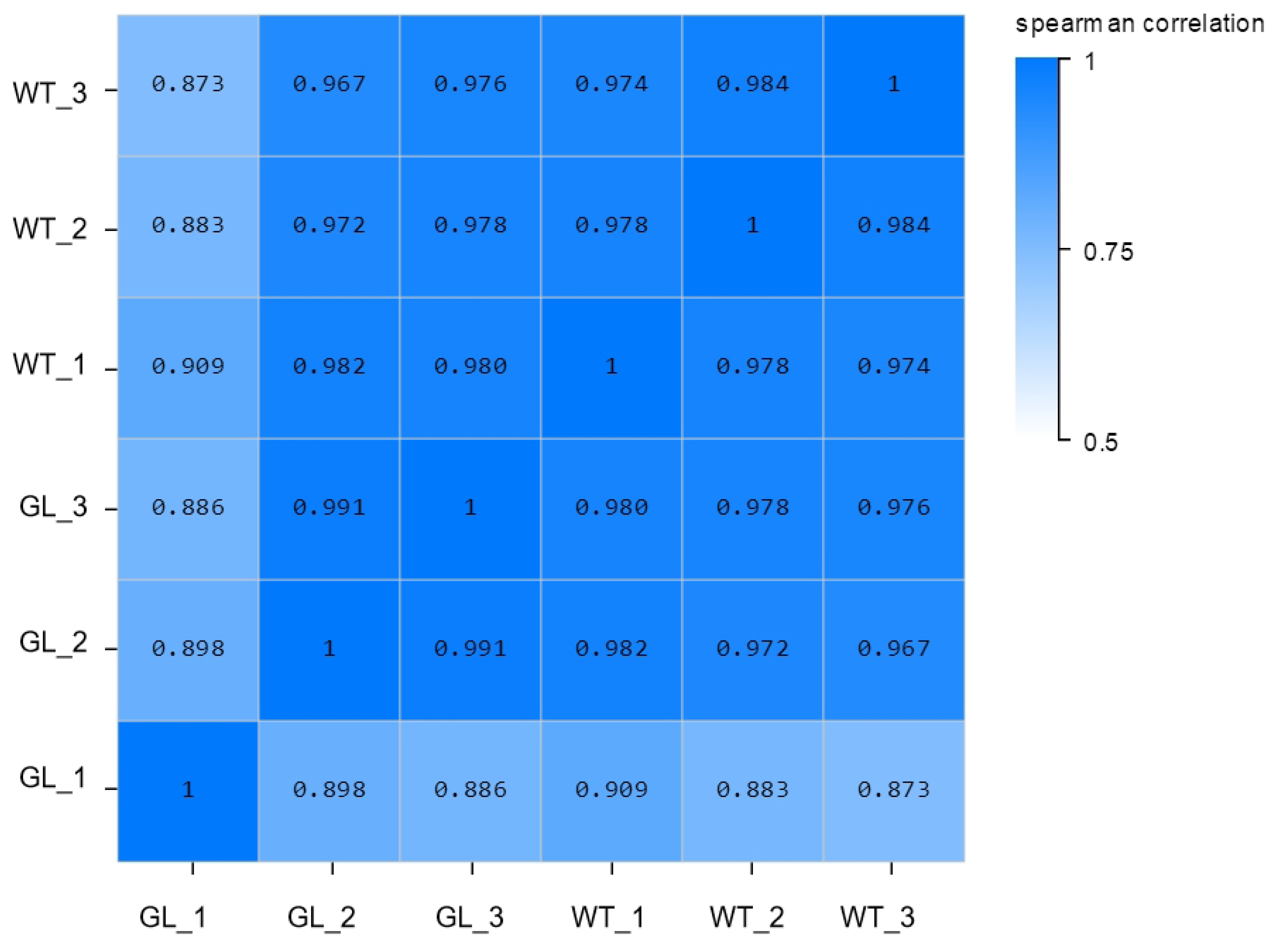
3.2. Total RNA Extraction and Quality Analysis
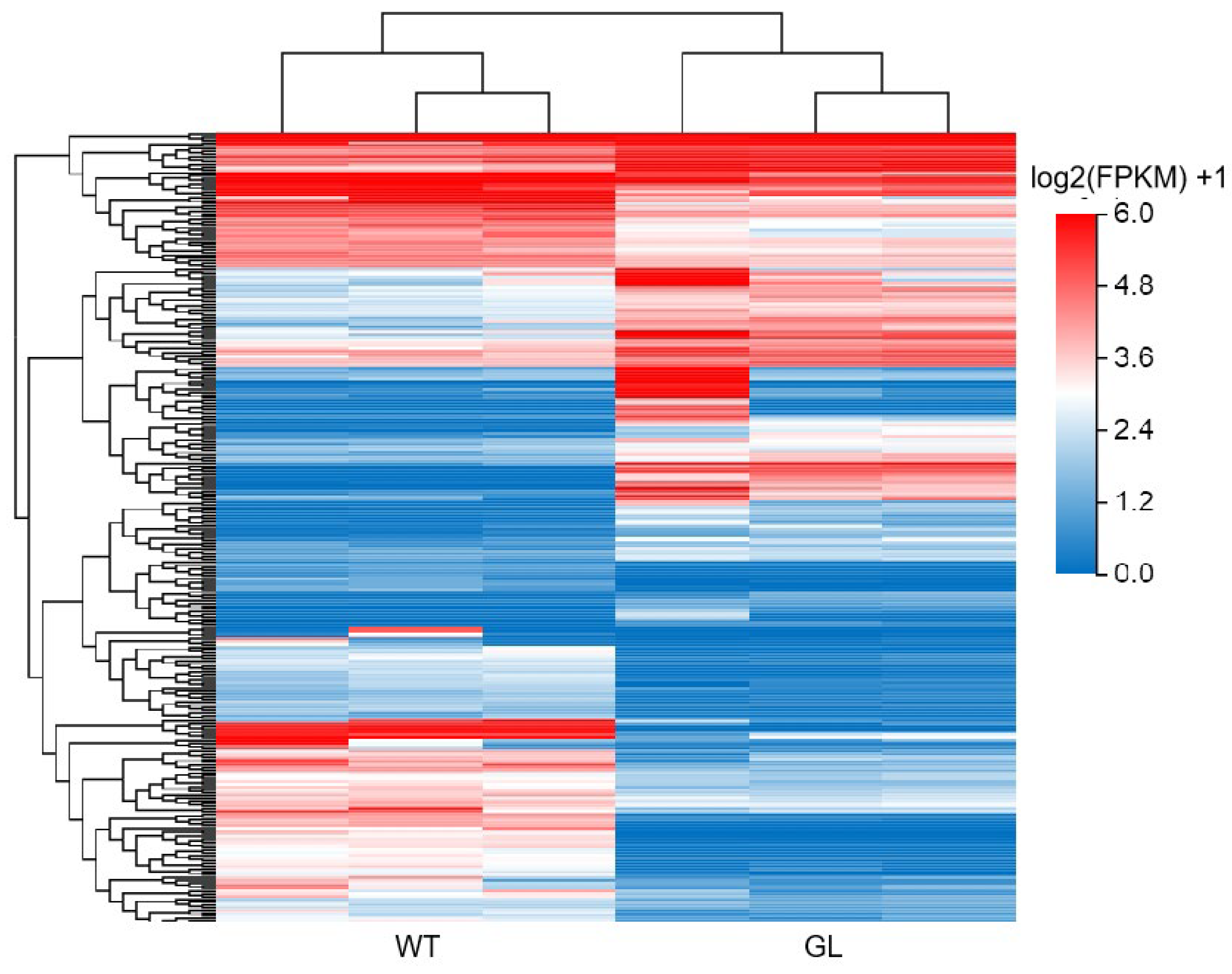
3.3. Gene Expression Analysis
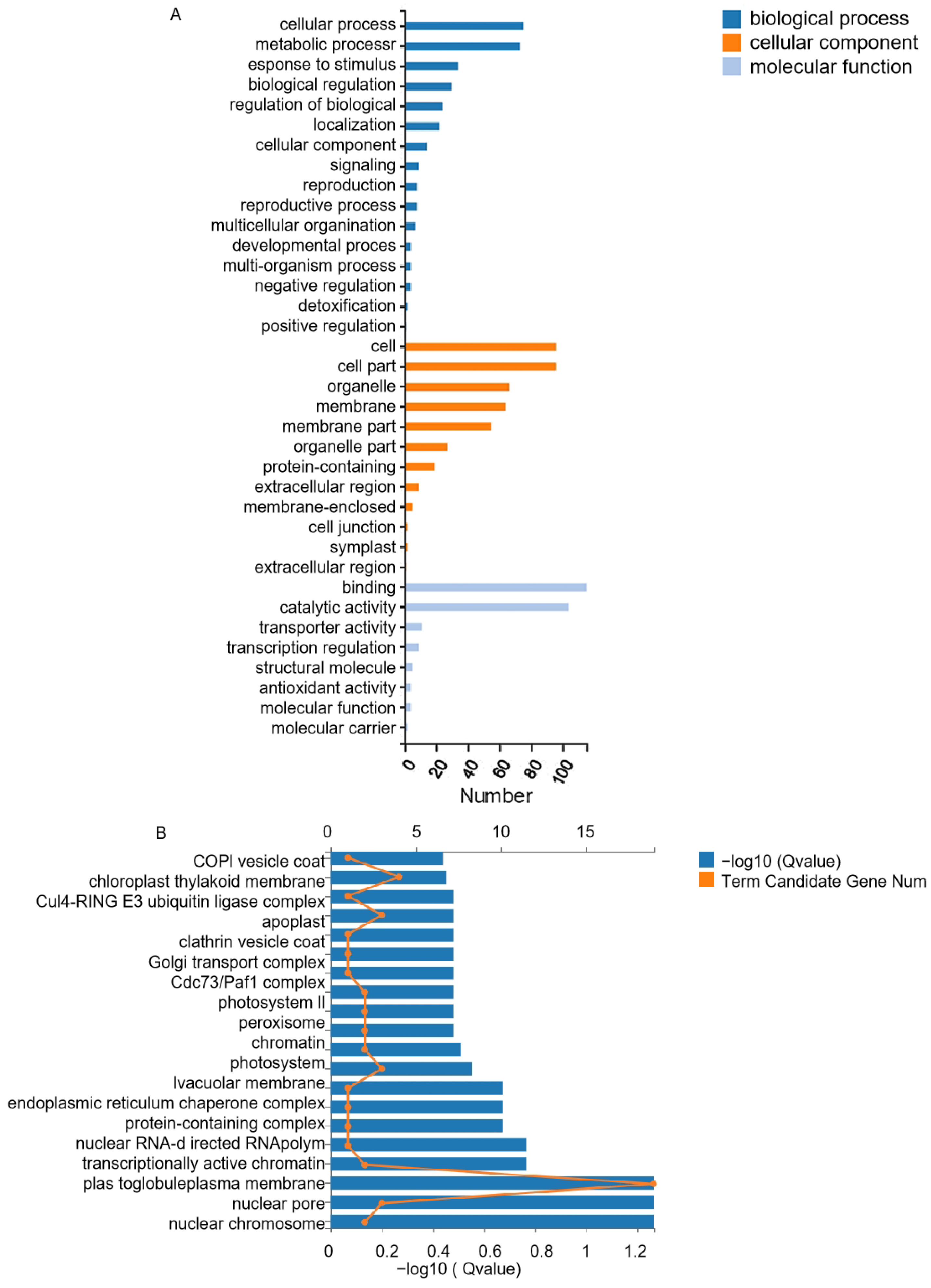
3.4. GO Analysis of DEGs
3.5. KEGG Analysis of DEGs
3.6. Weighted Gene Coexpression Network Analysis of (WGCNA) DEGs
3.7. Network Analysis of Photosynthesis Antenna Proteins
3.8. qRT-PCR Analysis of Genes Involved in Photosynthesis
4. Discussion
4.1. Characterization of Photosynthesis between WT and GL Plants
4.2. DEGs in Photosynthesis Antenna Proteins Pathways
4.3. DEGs in Diterpenoid Biosynthesis Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gao, Y.; Zhao, Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for Crispr-mediated genome editing. J. Integr. Plant Biol. 2014, 56, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zou, Y.; Li, X.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D.; Long, Y.; et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of m RNA silencing. Plant Cell 2006, 18, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Hur, J.; Ryu, C.H.; Choi, Y.; Chung, Y.Y.; Miyao, A.; Hirochika, H.; An, G. Characterization of a Rice Chlorophyll-Deficient Mutant Using the T-DNA Gene-Trap System. Plant Cell Physiol. 2003, 44, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, R.; Ito, T.; Kobayashi, M.; Taji, T.; Nagata, N.; Asami, T.; Yoshida, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 2003, 34, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Kusaba, M.; Ito, H.; Morita, R.; Lida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 Is Involved in Light-Harvesting Complex II and Grana Degradation during Leaf Senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The Senescence-Induced Staygreen Protein Regulates Chlorophyll Degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, R.E.; Terry, M.J. The aurea and yellow-green-2 Mutants of Tomato Are Deficient in Phytochrome Chromophore Synthesis. J. Biol. Chem. 1996, 271, 21681–21686. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, A.; Yang, W.; Jia, X.; Fu, Q.; Zhao, T.; Jiang, J.; Li, J.; Yang, H.; Xu, X. Transcriptome Analysis and Screening of Genes Associated with Flower Size in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2022, 23, 15624. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.M.; Wang, L.; Wang, Q.; Zhu, J.; Jiang, J.G. High-value bioproducts from microalgae: Strategies and progress. Crit. Rev. Food Sci. Nutr. 2019, 59, 2423–2441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, D.; Xu, J.W.; Zhao, P.; Li, T.; Ma, H.; Yu, X. Melatonin enhances lipid production in Monoraphidium sp. QLY-1 under nitrogen deficiency conditions via a multi-level mechanism. Bioresour. Technol. 2018, 259, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Xu, J.; Rao, Y.C.; Zeng, Y.J.; Liu, H.J.; Zheng, T.T.; Zhang, G.H.; Hu, J.; Guo, L.B.; Qian, Q.; et al. Cloning and functional analysis of pale-green leaf (PGL10) in rice (Oryza sativa L.). Plant Growth Regul. 2016, 78, 69–77. [Google Scholar] [CrossRef]
- Zuo, L.; Zhang, S.; Liu, Y.; Huang, Y.; Yang, M.; Wang, J. The Reason for Growth Inhibition of Ulmus pumila ‘Jinye’: Lower Resistance and Abnormal Development of Chloroplasts Slow Down the Accumulation of Energy. Int. J. Mol. Sci. 2019, 20, 4227. [Google Scholar] [CrossRef] [PubMed]
- Klimmek, F.; Sjödin, A.; Noutsos, C.; Leister, D.; Jansson, S. Abundantly and Rarely Expressed Lhc Protein Genes Exhibit Distinct Regulation Patterns in Plants. Plant Physiol. 2006, 140, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, H.; Ono, T.A.; Minagawa, J. Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 2001, 42, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Luciński, R.; Jackowski, G. The structure, functions and degradation of pigment binding proteins of photosystem II. Acta Biochim. Pol. 2006, 53, 693–708. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; Cao, D.; He, Y.; Kuang, T.; Yang, C. Structural and functional analysis of the antiparallel strands in the lumenal loop of the major light-harvesting chlorophyll a/b complex of photosystem II (LHCIIb) by site-directed mutagenesis. J. Biol. Chem. 2008, 283, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Dou, F.F.; Zhang, L.P.; Wang, Y.K.; Yu, K.; Liu, H.F. Effects of high-temperature stress on photosynthesis and gene expression of different grape cultivars. J. Fruit. Sci. 2021, 38, 871–883. [Google Scholar] [CrossRef]
- Yang, H.; Song, X.Z.; Zheng, G.L.; Shen, J. Effects of High-temperature stress on Physiological Indexes and Growth of Rhododendron championae. Acta Agric. Univ. Jiangxiensis 2020, 42, 259–265. [Google Scholar]
- Zhang, L.; Yang, Z.Q.; Sun, J.H.; Hong, J.W.; Li, S.G.; Wang, J.B. Genetic and transcriptome analysis of albino mutants of litchi (Litchi chinensis Sonn.) leaves. Mol. Plant Breed. 2020, 18, 5943–5950. [Google Scholar] [CrossRef]
- Cutolo, E.A.; Caferri, R.; Guardini, Z.; Dall’Osto, L.; Bassi, R. Analysis of state 1-state 2 transitions by genome editing and complementation reveals a quenching component independent from the formation of PSI-LHCI-LHCII supercomplex in Arabidopsis thaliana. Biol. Direct 2023, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Terry, N.; Abadia, J. Function of iron in chloroplasts. J. Plant Nutr. 1986, 9, 609–646. [Google Scholar] [CrossRef]
- Landsberg, E.C. Regulation of iron stress response by whole plant activity. J. Plant Nutr. 1984, 7, 609–621. [Google Scholar] [CrossRef]
- Bughio, N.; Takahashi, M.; Yoshimura, E.; Nishizawa, N.; Mori, S. Light-dependent iron transport into isolated barley chloroplasts. Plant Cell Physiol. 1997, 38, 101–105. [Google Scholar] [CrossRef]
- Shikanai, T.; Muller, P.; Munekaga, Y.; Niyogi, K.; Pilon, M. PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 2003, 15, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Moseley, J.; Quinn, J.; Eriksson, M.; Merchant, S. The Crdl gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 2014, 19, 2139–2151. [Google Scholar] [CrossRef] [PubMed]
- Tottey, S.; Mlock, M.A.; Allen, M.; Westergren, T.; Albrieux, C.; Scheller, H.V.; Merchant, S.; Jense, P.E. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 2003, 100, 16119–16124. [Google Scholar] [CrossRef] [PubMed]
- Murgia, I.; Tarantino, D.; Soave, C.; Morandini, P. Arabidopsis CYP82C4 expression is dependent on Fe availability and circadian rhythm, and correlates with genes involved in the early Fe deficiency response. J. Plant Physiol. 2011, 168, 894–902. [Google Scholar] [CrossRef]





| Sample | Total Raw Reads (M) | Total Clean Reads (M) | Total Clean Bases (Gb) | Q20 (%) | Q30 (%) | Ratio (%) |
|---|---|---|---|---|---|---|
| GL_1 | 47.19 | 44.89 | 6.73 | 97.84 | 93.27 | 95.13 |
| GL_2 | 47.19 | 45.03 | 6.76 | 97.86 | 93.19 | 95.44 |
| GL_3 | 47.19 | 45.27 | 6.79 | 97.94 | 93.52 | 95.93 |
| WT_1 | 47.19 | 45.16 | 6.77 | 98.08 | 93.9 | 95.71 |
| WT_2 | 47.19 | 45.13 | 6.77 | 97.93 | 93.39 | 95.64 |
| WT_3 | 47.19 | 45.33 | 6.8 | 98.02 | 93.7 | 96.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Zhang, P.; Fan, X.; Yang, H. Characterization and RNA-Seq Analysis of Yellow-Green Leaf Mutants in Tomato. Agronomy 2024, 14, 828. https://doi.org/10.3390/agronomy14040828
Guo X, Zhang P, Fan X, Yang H. Characterization and RNA-Seq Analysis of Yellow-Green Leaf Mutants in Tomato. Agronomy. 2024; 14(4):828. https://doi.org/10.3390/agronomy14040828
Chicago/Turabian StyleGuo, Xiao, Ping Zhang, Xing Fan, and Huanhuan Yang. 2024. "Characterization and RNA-Seq Analysis of Yellow-Green Leaf Mutants in Tomato" Agronomy 14, no. 4: 828. https://doi.org/10.3390/agronomy14040828
APA StyleGuo, X., Zhang, P., Fan, X., & Yang, H. (2024). Characterization and RNA-Seq Analysis of Yellow-Green Leaf Mutants in Tomato. Agronomy, 14(4), 828. https://doi.org/10.3390/agronomy14040828




