Abstract
Bananas, a staple food globally and a key agricultural commodity, face a severe threat from the fungus Fusarium oxysporum f. sp. cubense (Foc), significantly impacting production. Genetic improvement to develop resistant cultivars stands out as a crucial strategy to mitigate this disease. This study focused on assessing and recommending enhanced diploid banana varieties for resistance against Foc subtropical race 4 (ST4) and Foc race 1 (R1). Twenty-four improved diploids developed by Embrapa, Brazil, underwent evaluation. Utilizing a scale for internal symptoms at 90 days after inoculation, genotypes were categorized from highly resistant to highly susceptible based on the internal symptom index. The diploid M53 exhibited high resistance to Foc R1 and resistance to Foc ST4, while only the diploid CNPMF 0534 demonstrated complete resistance to both R1 and ST4, with resistance to the latter likely associated with penetration, primarily due to the presence of callose. These findings provide valuable insights for banana and plantain breeding programs, offering selected diploids for crossbreeding with commercial cultivars to develop new, resistant genotypes against Foc.
1. Introduction
Bananas and plantains (Musa spp.) are cultivated in over 150 tropical and subtropical countries, playing a crucial role in the economies of developing nations [1]. Banana cultivation stands as a leading sector in global agricultural production, with India as the largest producer, followed by China, Indonesia, Nigeria, and Brazil [1]. Fusarium wilt, caused by Fusarium oxysporum f. sp. cubense (Foc), stands out as a primary limitation to banana production, ranking among the top five diseases impacting the economic sector and fruit-producing regions [2,3]. Fusarium wilt is caused by Foc, a soil-inhabiting fungus with the ability to colonize plants, induce their demise, and employ intricate pathways to suppress plant defenses [4,5].
In infested soils, the structures of Foc, such as macroconidia, microconidia, and chlamydospores, remain dormant until they come into direct contact with susceptible root tissues or are stimulated to germinate by root exudates. Foc conidia and hyphae can adhere to root surfaces after inoculation, with infection occurring through secondary or tertiary roots, and penetration can happen directly or through wounds [4,6]. Most infection attempts appear to be blocked by the host, whose cell wall acts as a barrier to infection, being composed of cellulose, hemicellulose, pectin, or callose [6].
Isolates of Foc pathogenic to bananas are categorized within the forma specialis (f. sp.) cubense and classified into three races based on their infectivity to different cultivars within the species [3,5]. Race 1 (R1) affects the Gros Michel (AAA) and Manzano/Silk/Latundan (AAB) sub-groups. Race 2 (R2) targets cultivars susceptible to R1 and those in the Bluggoe subgroup. Race 4 (R4) impacts all cultivars in the Cavendish subgroup (AAA), and those susceptible to R1 and R2 [4,6]. R4 has further subdivisions into tropical race 4 (TR4) and subtropical race 4 (ST4). TR4 affects Cavendish in both tropical and subtropical conditions, while Foc ST4 isolates cause disease in Cavendish cultivars in subtropical regions under specific conditions, such as temperature extremes or water deficits [7,8]. The ST4 and TR4 variants can be differentiated by their vegetative compatibility group (VCG). Among the 24 known Foc VCGs, VCGs 0120, 01201, 01202, 01209, 01210, 01211, 01215, and 0120/15 are associated with Foc ST4; for Foc TR4, only VCG 01213/16 has been identified [6,8].
Foc TR4 poses a significant threat to banana and plantain cultivation across various regions globally. Apart from its extensive presence in Asia, Africa, and Australia, it has also encroached upon cultivation areas in Latin America and the Caribbean, including Colombia, Peru, and Venezuela [3,9,10]. The escalating concern regarding the damage inflicted on banana cultivation by this threat has redirected research efforts, prompting a worldwide search for new insights based on epidemiological, genetic, and management data. Notably, the absence of a cultivar with sufficient resistance to replace those of the Cavendish subgroup intensifies the urgency of such research [3,6,11,12].
Consequently, genetic improvement programs are actively exploring solutions to mitigate the impacts of Fusarium wilt, as well as addressing pest and abiotic stresses affecting banana crops [13]. Research institutions worldwide, including the Honduran Foundation for Agricultural Research (FHIA) in Honduras, the African Center for Research on Bananas and Plantains (CARBAP) in Cameroon, the International Institute of Tropical Agriculture (IITA) in Nigeria and Uganda, the Brazilian Agricultural Research Corporation (Embrapa) in Brazil, the National Banana Research Center (NCRB) in India, the National Agricultural Research Organization (NARO) in Uganda, and the International Cooperation Center for Agricultural Research and Development (CIRAD) in France, are collaboratively working towards identifying resistant cultivars [12].
In the exploration of Foc TR4, it was observed that the prevalence of resistant genotypes is linked to diploid genomes. These findings underscore that, currently, resistance sources to Foc TR4 mainly consist of wild or improved diploids that have not yet been integrated into the development of commercial cultivars through hybridization, unlike Foc R1, which benefits from a broad array of available resistant cultivars [14]. To enhance the genetic diversity of banana cultivars and achieve resistance to Fusarium wilt, researchers globally have been amassing germplasms, including from wild relatives. Despite numerous evaluations conducted in both greenhouse and field conditions, the assessment of resistance to Foc TR4 across a large number of banana genotypes remains notably limited [15,16].
Embrapa employs improved diploids as male parents in crosses with commercial cultivars to develop hybrids. These diploids result from the crossing of different wild diploids, showcasing resistance to key diseases affecting banana cultivation, including Yellow Sigatoka, Black Sigatoka, and Fusarium wilt. Additionally, they possess other agronomic characteristics of interest for breeding [12]. Notably, the genealogy of some parents of Embrapa’s improved diploids, such as Malaccensis, Tjau Lagada, Calcutta 4, and Tuu Gia, includes wild diploids resistant to Foc R1 and Black Sigatoka [17]. In Foc R4T research, the parents Calcutta 4 and Tuu Gia emerged as valuable resources for resistance genes, positioning them as promising options for banana breeding and for the study of mechanisms resisting Fusarium wilt [5,14,16]. Histochemical and histological evaluations were conducted to identify defense responses in the plant–pathogen interaction; however, new microscopic analyses can be performed at the early stages of infection, making it possible to quantify fungal germination, hyphal penetration and colonization, and host defense responses in future studies. Given the pivotal role of improved diploids in the development of commercial banana hybrids, this study aimed to assess the behavior of 24 improved diploids. The evaluation involves symptomatology analyses post-inoculation with Foc R1 and ST4 in the greenhouse. Additionally, histochemical and histological evaluations were conducted to identify defense responses in the plant–pathogen interaction.
2. Materials and Methods
2.1. Fusarium Strains
The isolates belonging to R1 and ST4, identified in the biological collection of the Phytopathology Laboratory of Embrapa and designated as Foc 0801 and Foc 218A, were selected for this study. Foc 0801 serves as a standard for R1, providing a foundational basis for studies [12]. Isolate 218A, collected in the state of São Paulo, induces disease in Nanica (Cavendish), and was characterized in previous studies as part of VCG 0120, being included as an ST4 isolate [14].
The isolates of Foc R1 and ST4 were multiplied from cultures on plates containing potato dextrose agar and incubated in biochemical oxygen demands (BODs) at a temperature of 25 °C with a 12 h photoperiod. After colony growth, the culture medium was prepared, comprising 20 mL of spore suspension and 500 g of autoclaved rice. Incubation occurred in a BOD at a temperature of 25 °C with a 12 h photoperiod. After 20 days, a serial dilution of the infected rice was prepared, and the colony-forming units (CFUs) were counted to adjust the concentration and verify spore viability. CFUs were counted using the Neubauer chamber, and the concentration was adjusted to 106 CFU/g of substrate [18].
2.2. Plant Materials and Growth
Twenty-one improved diploids developed by Embrapa, one diploid developed in Jamaica (M53), and two diploids (SH3263 and SH3362) developed by the Honduran Foundation for Agricultural Research (FHIA) were used. M53 is an improved diploid utilized by Embrapa’s genetic improvement program and is part of the lineage of various hybrids developed in Brazil. The SH hybrids are resistant to Black Sigatoka and are utilized in breeding schemes in Brazil.
The rooted plants were transplanted into plastic trays containing a substrate composed of coconut fiber and acclimatized in a greenhouse for 30 to 45 days until they reached a height of approximately 15 cm. Twenty-four genotypes were employed, along with the cultivar Maçã (Silk type), which served as the control. Detailed information on each genotype can be found in Table 1.

Table 1.
Banana genotypes used for the evaluation of resistance to Foc R1 and ST4 in a greenhouse.
2.3. Plant Inoculation
The acclimatized plants were individually inoculated with Foc R1 and ST4 in two independent greenhouse experiments, both in a completely randomized design (CRD) with 10 replications, where the experimental unit was one plant. For the experiment with Foc R1, pots with a capacity of 3 L of soil were used, where seedlings of each genotype, approximately 15 cm tall, were planted. Inoculation of the seedlings with Foc R1 was performed with infected rice at a concentration of 106 CFU/g, distributing a total of 40 g of inoculum in four holes around the plants. For the experiment with Foc ST4, polyethylene boxes with a capacity of 310 L, filled with soil, were used. To infect the soil, 1 kg of inoculum per box was used at a concentration of 106 CFU/g. Thus, plants of each genotype were planted with a density of 25 seedlings per box. In both experiments, 10 plants of the Silk cultivar were included as positive controls, given their known susceptibility.
2.4. Assessment of Symptoms
For both experiments with Foc R1 and ST4, internal symptoms were assessed 90 days after inoculation (DAI). Measurements associated with internal symptoms were made by crosswise cutting the rhizome and observing discoloration based on the scale proposed by Dita: 1: No symptoms; 2: Initial rhizome discoloration; 3: Slight rhizome discoloration along the whole vascular system; 4: Rhizome with most of the internal tissues showing necrosis; 5: Rhizome totally necrotic [19].
Using the obtained data, the disease severity index (DSI) was estimated following the formula by [20]: DSI = [∑ (disease score × number of plants with the score) × 100] (Number of plants evaluated per genotype × highest score adopted in the scale). Nine plants per treatment (genotype) were used, comprising three replicates for calculating the DSI.
Based on the mean DSI values and the standard deviation of three replicates, the following disease categories were established: highly resistant (HR); resistant (R); moderately resistant (MR); moderately susceptible (MS); susceptible (S); and highly susceptible (HS). Using the internal symptom indices, a hierarchical grouping analysis of the genotypes was conducted based on a heatmap, producing a graphical interpretation in which each genotype’s data is represented by colors. Shades of green are associated with levels of resistance, while shades of red indicate susceptibility. The statistical package used for the analysis was gplots in the R software v.4.4 [21].
Utilizing the “Performance Analytics” package in R, a Pearson correlation analysis was performed between the DSIs related to Foc R1, Foc ST4, and between the average DSIs of both breeds.
2.5. Histological and Histochemical Analysis
2.5.1. Whitening and Staining of Fungal Structures in Roots
At 90 DAI, plants were evaluated, and root fragments were collected and immediately immersed in formaldehyde–acetic acid–alcohol solution. Root clarification and staining of fungal structures were carried out according to a method described by Phillips [22] with modifications. For clarification, roots were immersed in a 10% potassium hydroxide solution at room temperature for 48 h and then in a 1% HCl solution for 30 min. Trypan blue dye in a 0.05% solution (lactic acid 2:1:1: glycerol: water) was applied for 1 h to stain the structures. After staining, slides were prepared, and fragments were microphotographed under a light microscope (Olympus Latin America Inc., Tokyo, Japan).
2.5.2. Histochemical Analysis
At 90 DAI, small root fragments were collected and immersed in Karnovsky solution [23]. Fragments remained in the Karnovsky solution for 48 h; they were then dehydrated in an increasing ethanol series at intervals of 3 h each (30–100%). They were infiltrated and embedded with historesin (hydroxyethyl methacrylate, Leica Heidelberg, Germany). After the historesin polymerization process, histological sections (8 μm) were obtained using a Leitz 1516 microtome. The slides were then assembled with the cuts and stained with aniline blue [24] to identify the presence of callose. The histological sections were analyzed and photographed using a B x S1 fluorescence microscope (Olympus Latin America Inc., Tokyo, Japan).
2.5.3. Scanning Electron Microscopy
Root samples were collected at 90 DAI. In the initial steps, the samples underwent dehydration in a series of ethanol and were dried in a critical point apparatus (Leica EM CPD 030) using liquid CO2. Following this, they were affixed to a metal support (stubs) with double-sided carbon adhesive tape and metallized with gold in a JEOL Smart Coater DII-29010SCTR device. After metallization, micrographs were captured using a JEOL JSM-6390LV scanning electron microscope in the electron microscopy laboratory of the Gonçalo Moniz Institute, Fiocruz, Salvador-BA, Brazil.
3. Results
3.1. Screening for Resistance to Foc R1
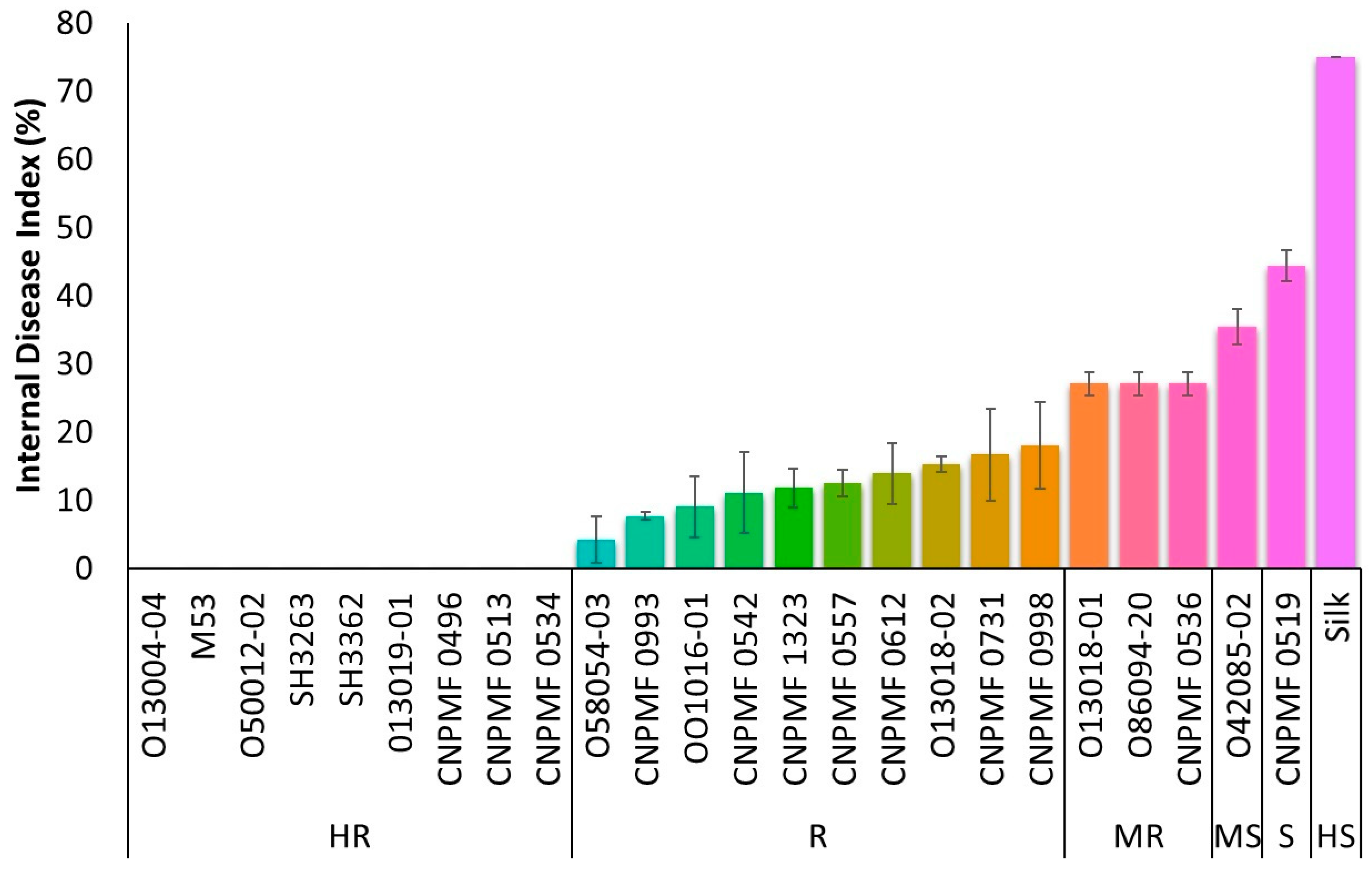
After 90 DAI, based on cutting the pseudostem close to the rhizome, genotypes were classified regarding the presence of typical symptoms of Foc R1. Improved diploids 013004-04, M53, 050012-02, SH3263, SH3362, 013019-01, CNPMF 0496, CNPMF 0513, and CNPMF 0534 showed complete resistance to Foc, exhibiting no symptoms of the pathogen (Figure 1). Improved diploids 058054-03, CNPMF 0993, 001016-01, CNPMF 0542, CNPMF 0557, CNPMF 1323, 013018-02, CNPMF 0612, CNPMF 0731, and CNPF 0998 displayed spots or discoloration around the xylem and were classified as resistant. Genotypes with slight discoloration of the rhizome in the vascular system, namely CNPMF 0536, 086094-20, and 013018-01, were classified as moderately resistant. Genotype 042085-02, with discoloration around the rhizome in the vascular system, was characterized as moderately susceptible while diploid CNPMF 0519 was characterized as susceptible. The control cultivar, Maçã (Silk type), exhibited over 50% discoloration of the rhizome and was categorized as highly susceptible to Foc R1.
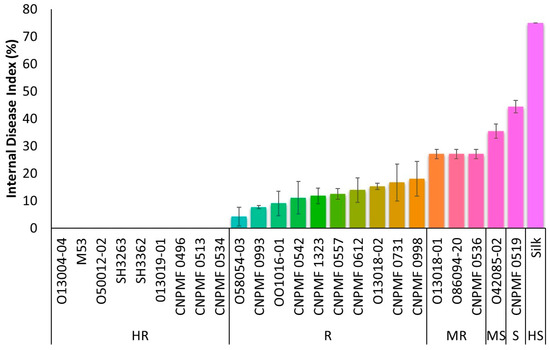
Figure 1.
Responses of different improved banana diploids to Fusarium oxysporum f. sp. cubense race 1 in a greenhouse. The data are presented as the mean ± SD (standard deviation) of three replicates. HR: highly resistant; R: resistant; MR: moderately resistant; MS: moderately susceptible; S: susceptible; HS: highly susceptible.
3.2. Screening for Resistance to Foc ST4
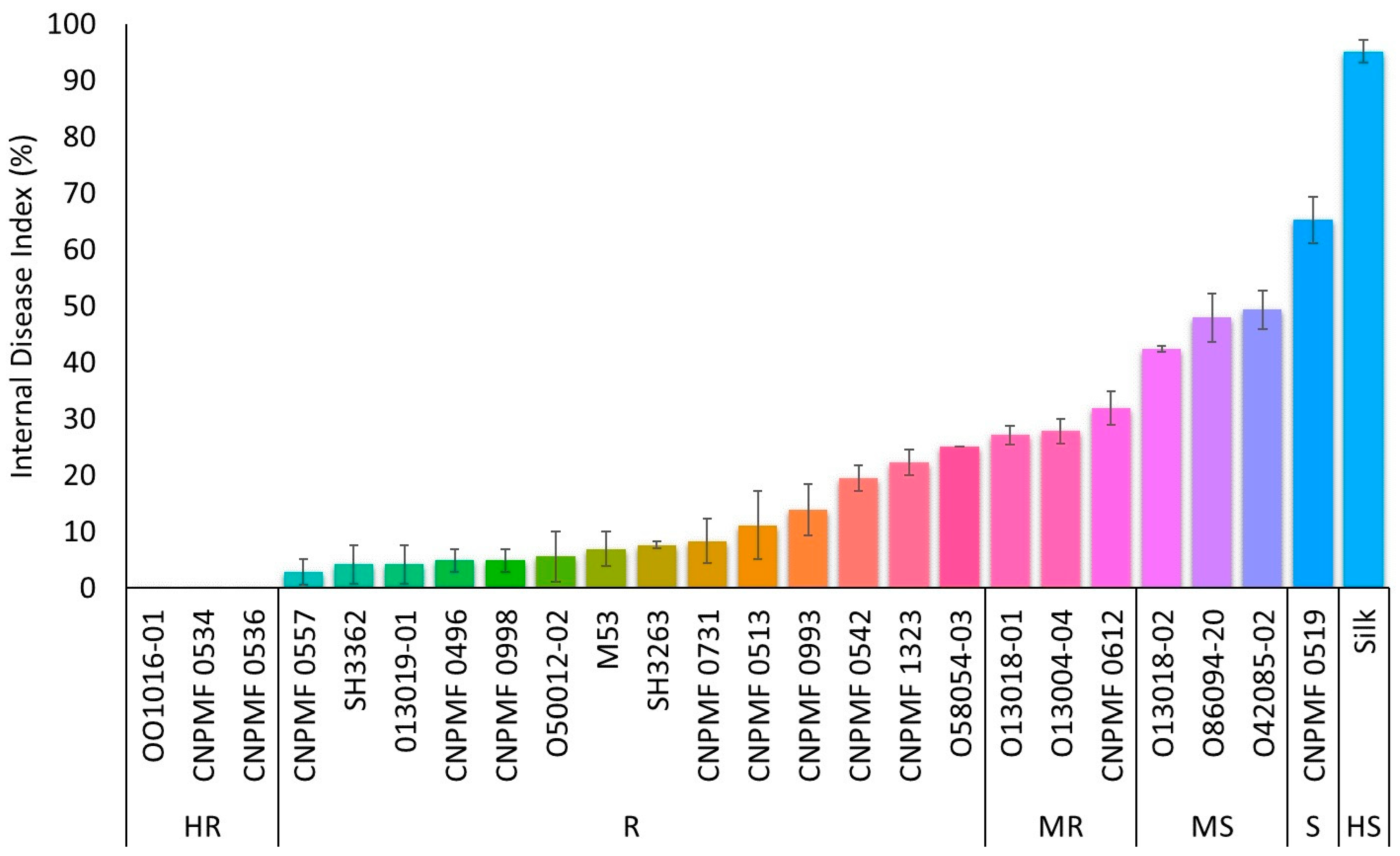
Three improved diploids, 001016-01, CNPMF 0534, and CNPMF 0536, demonstrated complete resistance to Foc ST4, displaying no internal symptoms of the disease (Figure 2). Fourteen other genotypes were classified as resistant due to some spots or discoloration around the xylem. Diploids 013004-04, 013018-01, and CNPMF 0612 exhibited slight discoloration of the rhizome along with the entire vascular system and were categorized as moderately resistant. Regarding susceptibility to Foc ST4, diploids 013018-02, 086094-20, and 042085-02 were moderately susceptible, diploid CNPMF 0519 was classified as susceptible, and the cultivar Silk was highly susceptible, following the grading scale used in the evaluation.
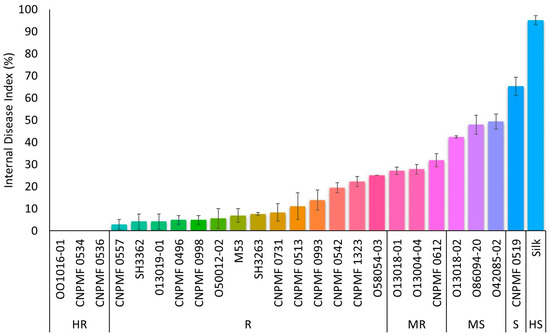
Figure 2.
Response of different improved banana diploids to Fusarium oxysporum f. sp. cubense subtropical race 4 in a greenhouse. The data are presented as the mean ± SD (standard deviation) of three replicates. HR: highly resistant; R: resistant; MR: moderately resistant; MS: moderately susceptible; S: susceptible; HS: highly susceptible.
3.3. Cluster Analysis for Responses to Foc R1 and Foc ST4
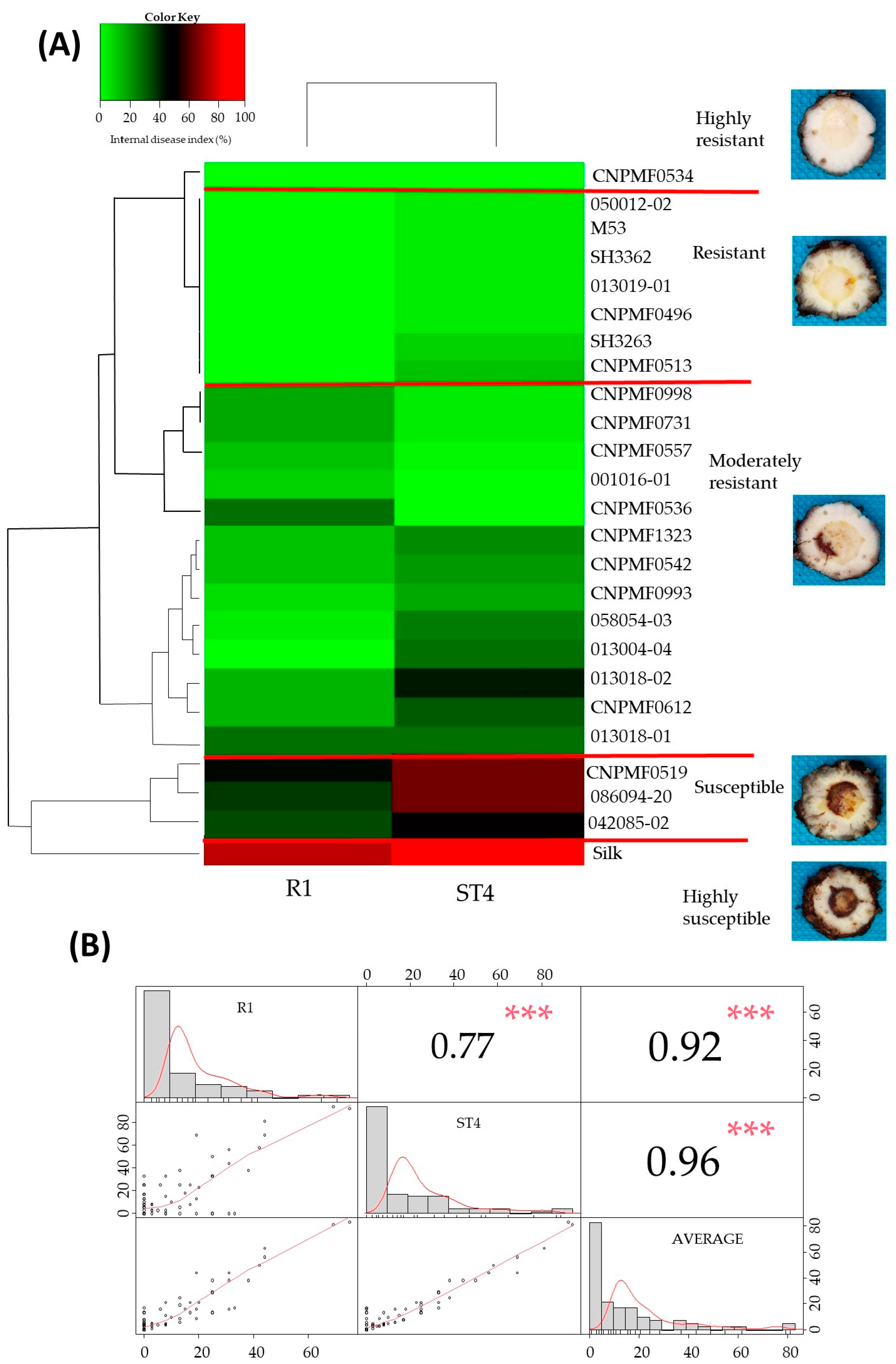
The cluster analysis depicted in the heat map (Figure 3A) considers disease indices per genotype for evaluations of resistance to Foc R1 and Foc ST4. CNPMF 0534 exhibited high resistance to both Foc R1 and ST4, while M53, 050012-02, SH3362, 013019-01, CNPMF 0496, SH3263, CNPMF 0513, and 042085-02 were classified as resistant. Thirteen other diploids fell into the moderately resistant category. Diploids 086094-20 and CNPMF 0519 were grouped as susceptible, and the cultivar Maçã (Silk type) formed an exclusive group, highly susceptible, aligning with expectations. Correlation analysis (Figure 3B) showed a high correlation between evaluations of Foc R1 and ST4 (0.77), Foc R1 and the average of IDs (0.92), and Foc ST4 and the average (0.96), indicating similar responses in the two races.
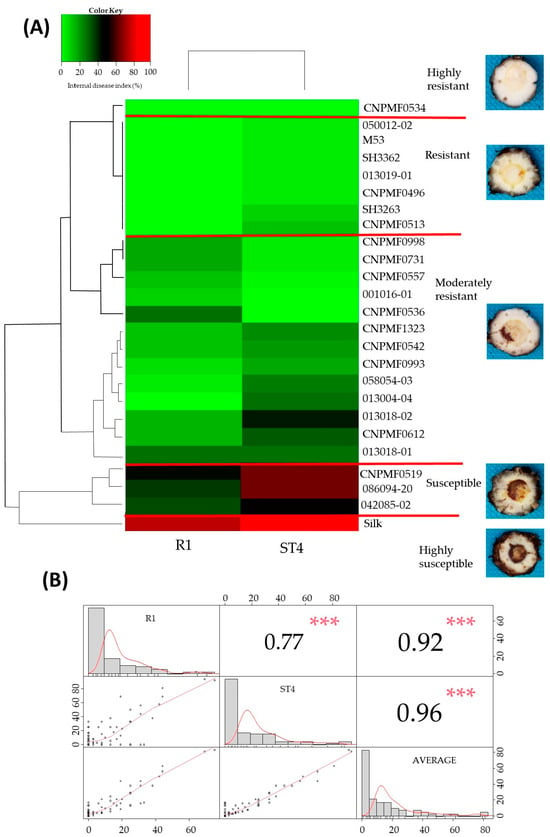
Figure 3.
Cluster analysis depicting the reaction of banana diploids to Fusarium oxysporum f. sp. cubense races 1 and subtropical 4 in a greenhouse (A). Pearson correlation matrix analysis between internal symptom indices of Foc race 1 (R1), Foc subtropical 4 (ST4), and race averages (mean): scatterplots, values, and statistical significance of each correlation coefficient (Pearson correlation coefficients [r]) (B). Signification codes: *** 0.001.
3.4. Histological and Histochemical Analyses
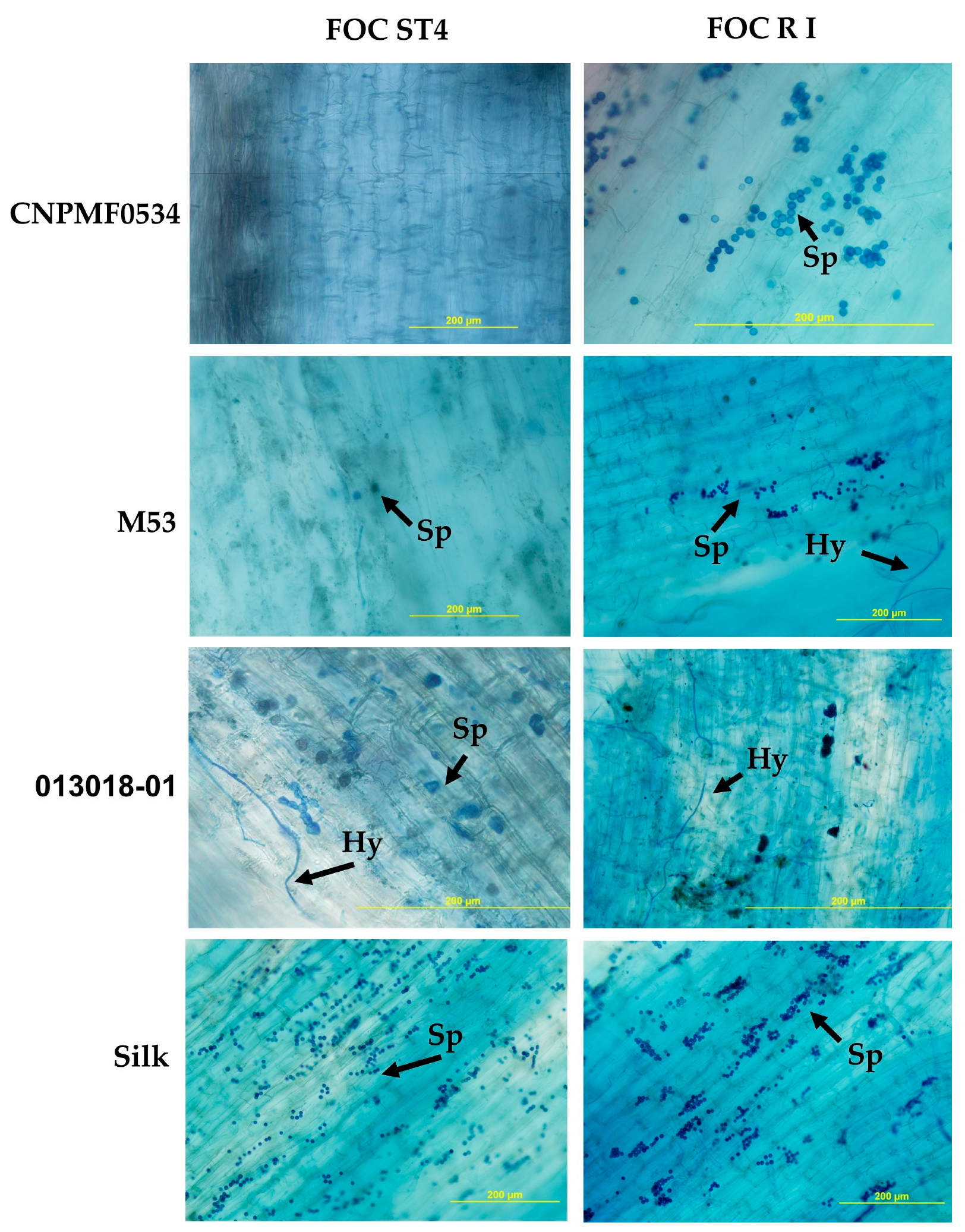
Irrespective of diploid behavior, whether resistant or susceptible, pathogen structures were observed in the roots of most evaluated genotypes. Notably, the highly resistant CNPMF 0534 exhibited Foc R1 microconidia in the roots, similar to observations in the cultivar Maçã (Silk type). Conversely, for Foc ST4, no fungal structures were detected in CNPMF 0534, possibly due to either fungus non-penetration or incapacity to colonize tissues post-penetration (Figure 4). In diploid M53, fungal hyphae and spores were observed in interaction with Foc R1, while only spores were seen with Foc ST4. Diploid 013018-01, classified as moderately resistant, displayed the presence of spores and hyphae in the tissues. The cultivar Maçã (Silk type) exhibited an abundance of spores in interaction with both isolates (Figure 4).
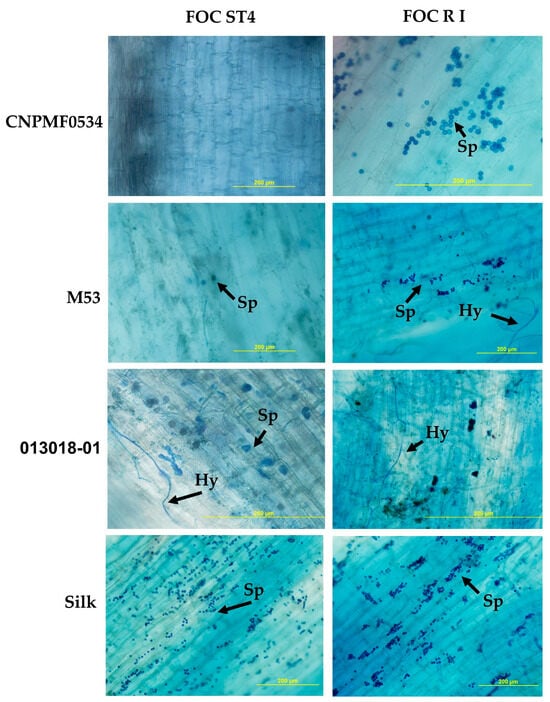
Figure 4.
Whitening and staining of fungal structures with Trypan blue dye in roots of banana genotypes after infection by two isolates of Fusarium oxysporum f. sp. cubense (Foc). Black arrows indicate spores (Sp) and hyphae (Hy). R1: race 1; ST4: subtropical race 4.
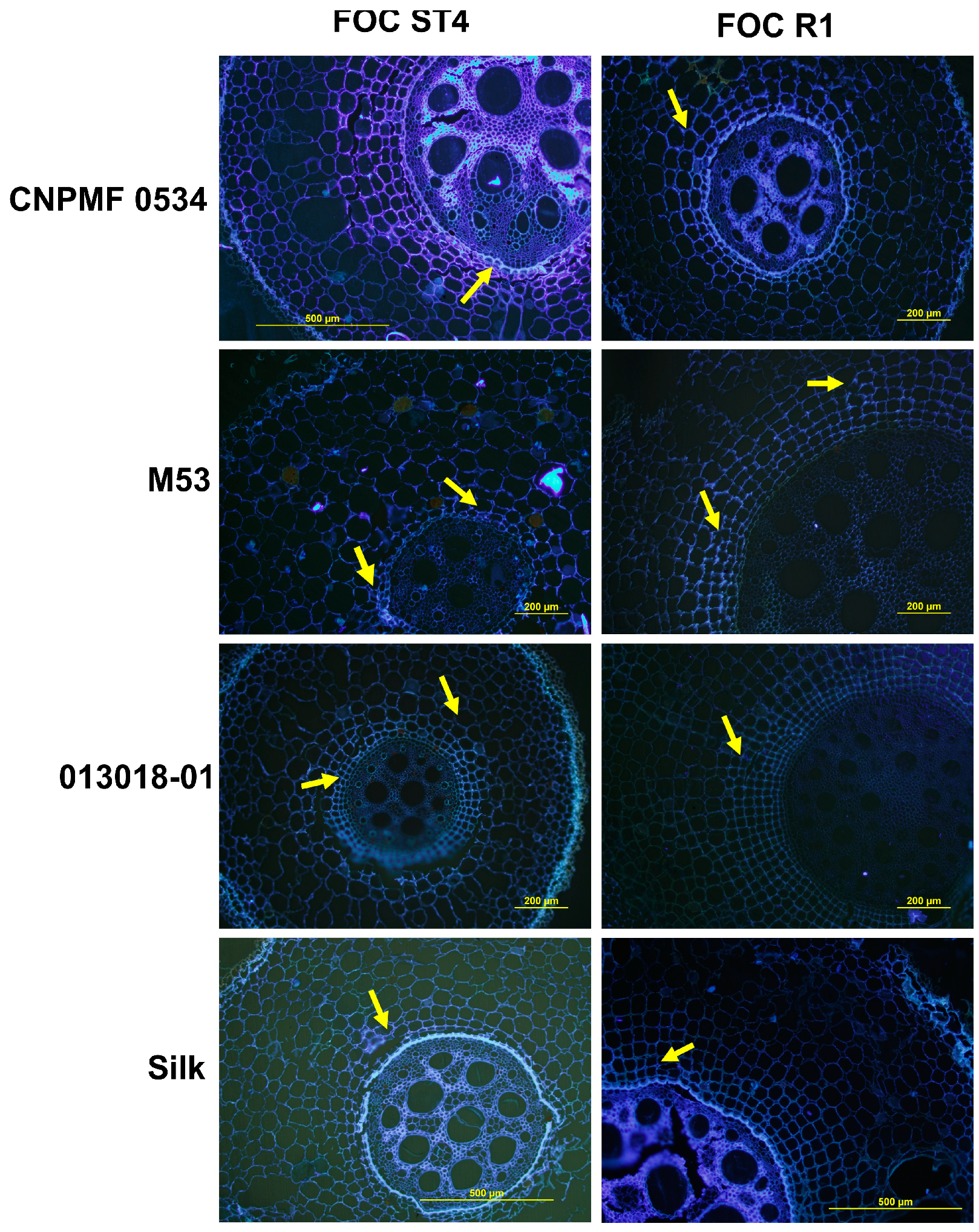
Upon staining root tissues with aniline blue dye to assess callose production, blue–white fluorescence indicated the presence of this mechanism. At 90 DAI, CNPMF 0534 exhibited pronounced fluorescence when inoculated with isolates Foc R1 and ST4 (Figure 5). The resistant diploid M53 also displayed fluorescence, indicating callose presence (Figure 5). Diploid 013018-01, moderately resistant, showed less callose emission. The cultivar Maçã (Silk type), when inoculated with isolates R1 and ST4, displayed slight fluorescence, indicating low levels of callose.
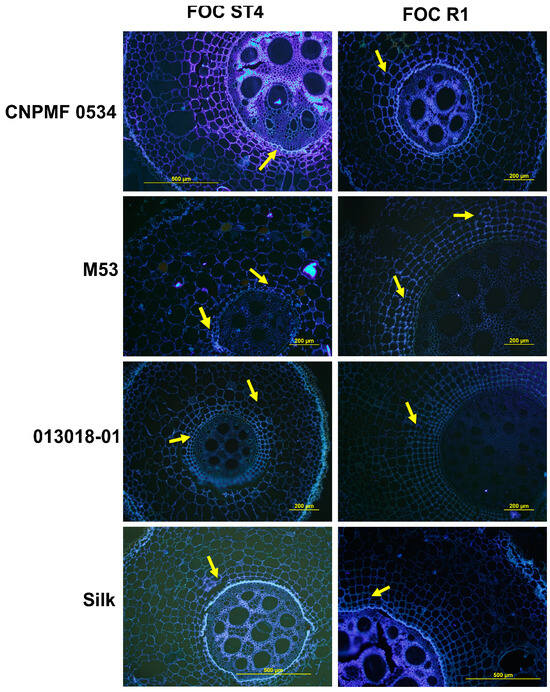
Figure 5.
Transverse section of the rhizome of banana genotypes infected by two races of Fusarium oxysporum f. sp. cubense in fluorescence micrographs. White–blue fluorescence indicates the presence of callose in the tissues stained with aniline blue. Foc: F. oxysporum f. sp. cubense; R1: race 1; ST4: subtropical race 4.
The scanning electron microscopy (SEM) study examined the interaction between genotypes contrasting in susceptibility or resistance to Foc R1 and ST4 at 90 DAI. In both cases, variability in structures and tissue reactions were observed, including abundant calcium oxalate crystals, varying in quantity and shape, and signs of tissue obstruction by gums and fungal structures, such as mycelium, hyphae, and spores.
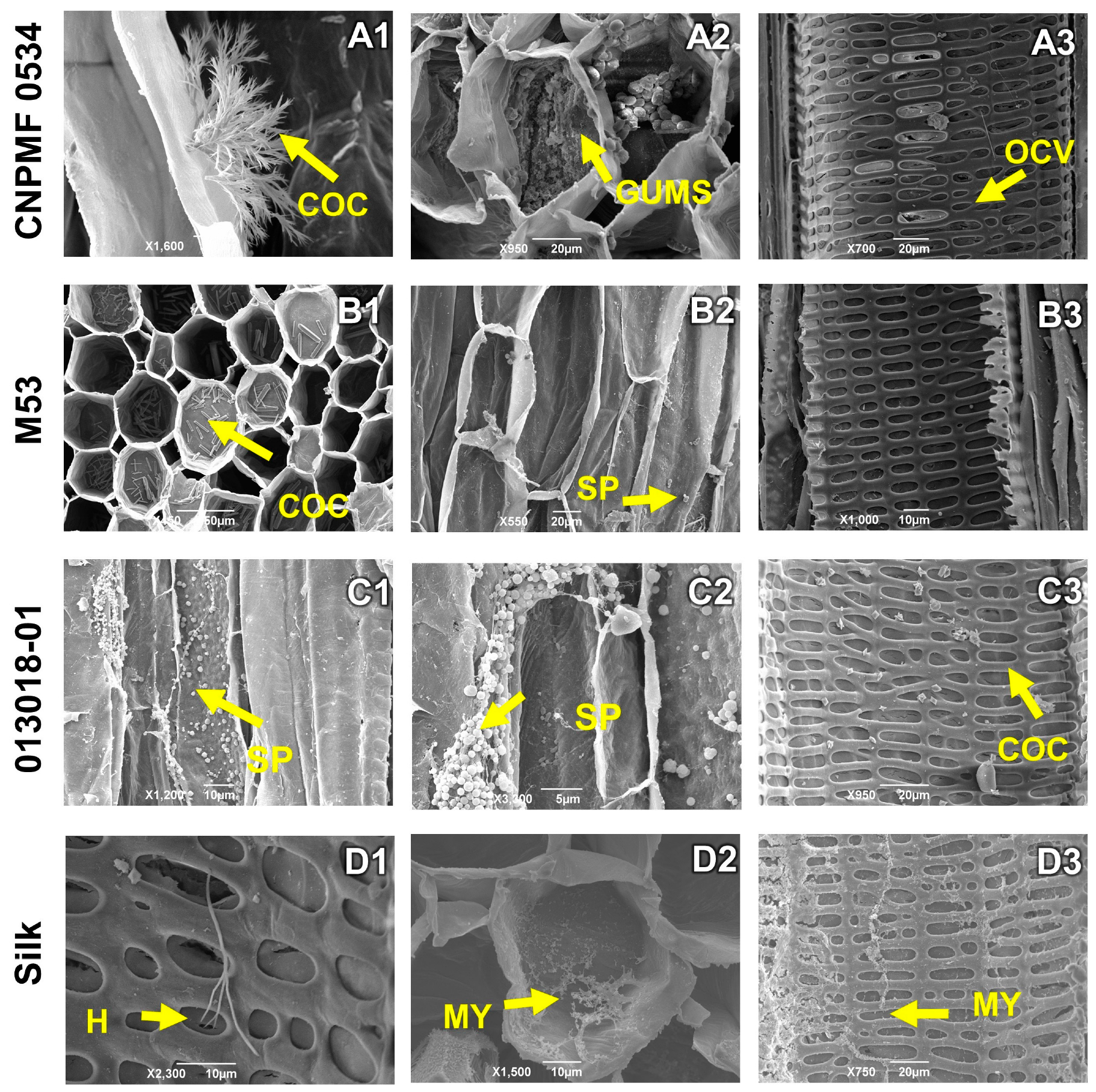
The highly resistant CNPMF 0534 diploid produced calcium oxalate crystals in the tissue (Figure 6(A1)), as well as gums (Figure 6(A2)) and vessel occlusion (Figure 6(A3)). Resistant diploid M53 showed a significant deposition of calcium oxalate crystals on the vessels and spores of the pathogen on the tissue (Figure 6(B1,B2)). Moderately resistant diploid 013018-01 exhibited intense infection by Foc R1, with concentrated spores in the tissues and the presence of some calcium oxalate crystals (Figure 6(C1–C3)). In the cultivar Maçã, the study revealed the growth of hyphae, fungal mycelium, and obstruction of conducting vessels (Figure 6(D1–D3)).
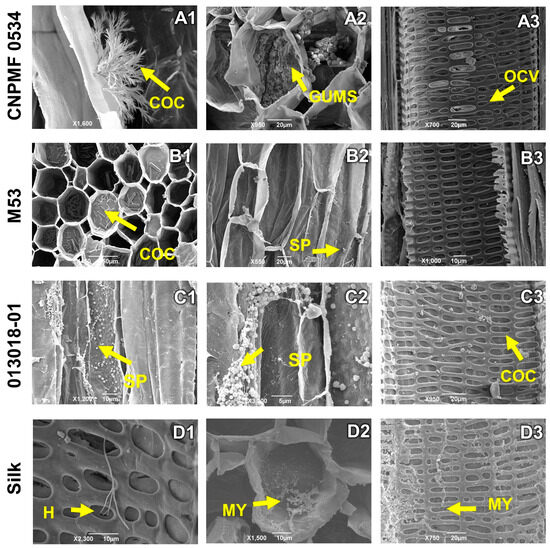
Figure 6.
Longitudinal and transverse sections of root sections of banana genotypes infected by Fusarium oxysporum f. sp. cubense race 1 (R1). H: hyphae; COC: calcium oxalate crystals; MY: mycelium; SP: spores, GUMS: gums, OCV: vessel occlusion. As grouped subfigures (A1–D3) represent photographs of different sections of the roots of the same genotype.
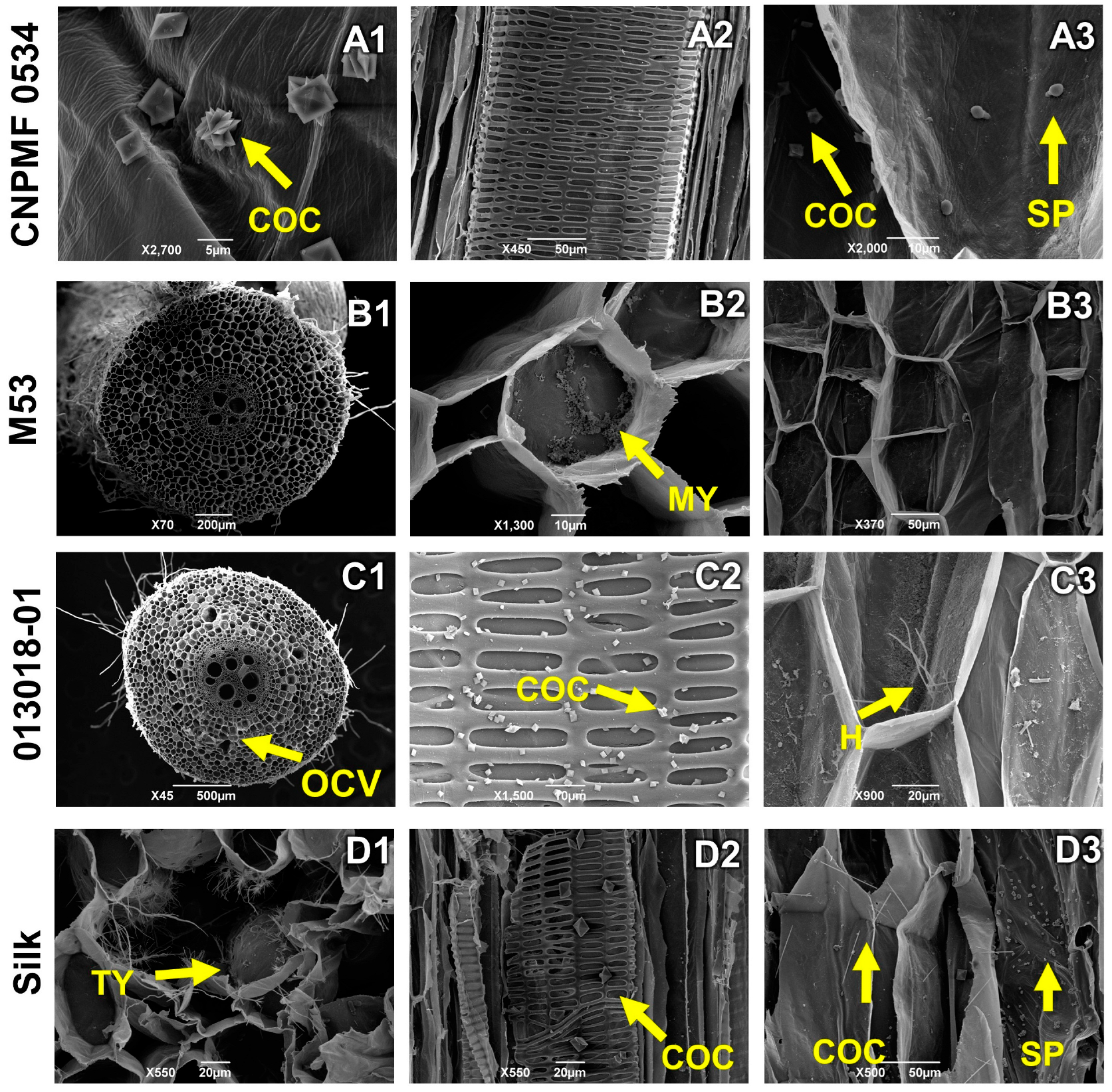
In the interaction between genotypes and Foc ST4, diploid CNPMF 0534 exhibited intense production of calcium oxalate crystals in the form of a druse (Figure 7(A1)). No vessel occlusions were observed; instead, scattered spores throughout the tissue were seen, still in the process of germination, indicating delayed fungal growth, considering the analysis at 90 DAI (Figure 7(A2,A3)). Resistant diploid M53 showed minimal vessel occlusion; however, mycelial growth was observed in some spaces where occlusion occurred (Figure 7(B2)). Moderately resistant diploid 013018-01 displayed vessel occlusion, hyphae growth (Figure 7(C2)), and the presence of calcium oxalate crystals. The cultivar Maçã (Silk type) showed spores, calcium oxalate crystal production in the form of a druse similar to a needle (Figure 7(D3)), and thylose deposition (Figure 7(D1)), characterized as the physiological process of xylem occlusion.
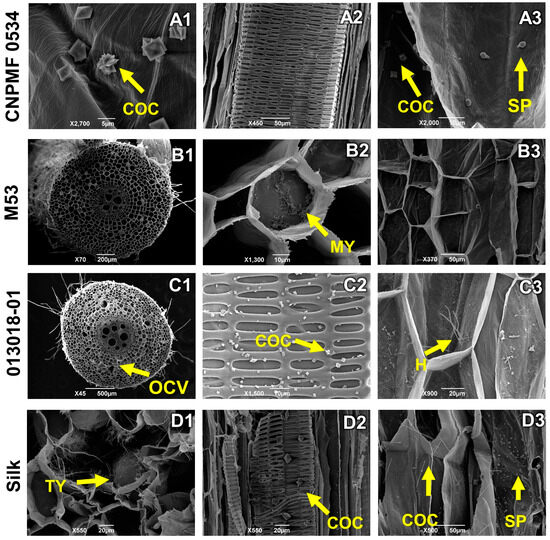
Figure 7.
Longitudinal and transverse sections of root sections of banana genotypes infected by Fusarium oxysporum f. sp. cubense subtropical race 4 (ST4). H: hyphae; COC: calcium oxalate crystals; MY: mycelium; SP: spores, GUMS: gums, OCV: vessel occlusion. As grouped subfigures (A1–D3) represent photographs of different sections of the roots of the same genotype.
4. Discussion
4.1. Selection of Improved DIPLOIDS Resistant to Foc R1 and Foc ST4
In this study, we assessed 24 improved banana diploid hybrids for resistance to Foc R1 and ST4 in a greenhouse setting. The diploids were categorized as highly resistant, resistant, moderately resistant, moderately susceptible, susceptible, and highly susceptible based on the mean values of the rhizome discoloration index. Selected for further analysis were CNPMF0534 (highly resistant), M53 (resistant), 013018-01 (moderately resistant), and the cultivar Maçã (Silk type) (highly susceptible) as a control. Among these, only the diploid hybrid CNPMF0534 exhibited resistance to both races and displayed no characteristic vascular bundle discoloration in the rhizome, with all plants scoring 1.
By averaging the scores and calculating the standard deviation from 10 repetitions for the diploid response profiles to both races, we observed that 10 genotypes were grouped in the highly resistant category against Foc R1. In contrast, only three were grouped in this category when evaluated against Foc ST4 (Figure 1 and Figure 2). Notably, differences between categories emerged, as two diploids were classified in the interaction with Foc ST4, whereas none received this classification in the interaction with Foc R1.
These findings are linked to the known virulence and aggressiveness of ST4 isolates in environmental conditions featuring temperature extremes, such as the intense cold prevalent in certain regions of Brazil [7,14]. The data underscore the importance of using appropriate Foc isolates and methods that exert the necessary selection pressure to draw accurate conclusions about resistance, both in the greenhouse and subsequently in the field, as practiced in other studies [5,16,25].
In a cluster and correlation analysis utilizing estimated Foc R1 and Foc ST4 IDs, we demonstrated that hybrids are classified in the same categories for both races. There is a positive correlation between evaluations of Foc R1 and ST4 IDs, as well as the average of both (Figure 3B). To our knowledge, this is the first report correlating resistance profiles to Foc R1 and ST4 based on disease index estimates. These data reinforce the accuracy of the selection method adopted by the Embrapa breeding program in Brazil for over 20 years and suggest that all plant materials that have been selected and indicated as resistant to Foc R1 over these years may also be resistant to Foc ST4 and possibly to Foc TR4, such as the hybrids BRS Platina (Prata type, AAAB) and BRS Princesa (Silk type, AAAB) already widespread in the Brazilian market.
Previous studies by Gonçalves et al. [17] described the resistance capacity of the improved diploids developed by Embrapa to Foc R1 under field conditions. The diploids CNPMF 1323, CNPMF 0612, CNPMF 0534, CNPMF 0998, CNPMF 0731, and CNPMF 0542 exhibited resistance to Foc R1, which aligns with the results of the present work. These improved diploids have parents that include Calcutta 4, M53, Malaccensis, Tjau Lagada, M61, and Tuu Gia, all confirmed to be resistant to R1 and ST4 [14,16,17].
4.2. Histological and Histochemical Responses of Improved Diploids Inoculated with Foc R1 and Foc ST4
From our histological analyses, we have observed no distinction between highly resistant and susceptible genotypes concerning the presence of Foc R1 within tissues post-bleaching and staining with Trypan blue. This reaffirms that penetration occurs in both cases, suggesting that defense strategies distinguishing resistant from susceptible genotypes manifest after the penetration process. This aligns with findings by Li et al. [15], Rocha et al. [12], and Rocha et al. [14]. Conversely, in the interaction with Foc ST4, a notable difference was observed in the CNPMF 0534 diploid. This diploid, considered highly resistant, did not exhibit pathogen structures inside the tissue, possibly associated with the absence of penetration or advancement in colonization. SEM analysis confirmed this observation, revealing only calcium oxalate crystals inside the tissue, and the spores were still in the process of germination, indicating a delay in infection (Figure 7(A1,A3)).
These results suggest that the direct penetration of roots and the vascular colonization of the rhizome by Foc are key steps for successful infection, as has been reported in previous studies involving Cavendish cultivars [26]. In the aforementioned study, it was clear that moderately resistant cultivars reduced spore germination and hyphal growth in the tissue, while susceptible cultivars did not affect fungal germination and growth. According to previous studies, the inoculation of Foc triggers a series of coordinated responses in banana roots that directly contribute to host resistance. Furthermore, the banana activates defense mechanisms mediated by salicylic acid and jasmonic acid/ethylene, possibly involving systemic acquired resistance and regulation of the expression of pathogenesis-related genes and cell wall biosynthesis [6,7,26]. This suggests that the ability to limit the initial colonization of the fungus may be an important resistance mechanism. Differential expression of genes related to cell wall modification has been observed in resistant plants compared to susceptible plants. The upregulation of these genes in resistant plants may strengthen the cell wall, making fungal penetration more difficult and thus limiting its ability to colonize tissues [14,15,26].
These findings parallel those obtained in a study of genotypic interaction with Foc ST4, aiming to select and evaluate resistant banana somaclones. The absence of pathogen structures in root tissues suggests that resistant somaclones may have developed physical and/or chemical barriers that impede pathogen penetration [27].
In SEM analyses, we consistently observed the presence of calcium oxalate crystals, with quantity and distribution in the tissue seemingly associated with genotypes classified as resistant. While specific data on the direct influence of these crystals on resistance are lacking, studies have documented their varied forms and functions across more than 215 plant families. Other research suggests that these crystals play a crucial role, as their degradation can produce reactive oxygen species linked to the response to pathogen infection and the inhibition of such infections [28,29].
Our results align with previous findings indicating abundant calcium oxalate crystals in the cultivar BRS Platina, a tetraploid banana hybrid of the Prata subgroup, inoculated with Foc ST4. These crystals may play vital roles in resistance [14]. Furthermore, we emphasize the importance of conducting additional studies to investigate the role of calcium oxalate crystals not only in the resistance of banana genotypes to Fusarium wilt, but also their role and differences in wild diploid genotypes. These wild diploids form the basis for genetic improvement and are the primary source of genetic variability in Musa spp.
In the histochemical analyses, evaluating plants’ response to fungal attack through the production of chemical compounds, callose was assessed. All genotypes exhibited fluorescence, indicating the presence of this compound. However, in the diploid CNPMF 0534, identified as highly resistant, notably high levels of fluorescence were observed, suggesting a potential increase in production during interactions with both Foc R1 and Foc ST4 (Figure 5). This could be linked to the fact that callose deposition is induced by various factors, particularly biotic and abiotic stresses, such as pathogen attacks, exposure to heavy metals, and wounds [30]. The formation of callose, gels, and tyloses in infected vessels has been reported in resistant banana plants as a mechanism to immobilize spores, preventing their invasion during pathogen–host interaction [31,32].
4.3. Associations Based on Improved Diploid Genealogy and Future Perspectives
The improved diploids M53, 013004-04, 013018-01, 013018-02, 013019-01, CNPMF0542, CNPMF0612, and CNPMF1323 result from crosses with wild diploids like Malaccensis, Pahang, Calcutta-4, Pisang Lilin, and Tuu Gia, which are part of their genealogy. These diploids have been previously reported for their resistance to Foc TR4 [14].
The Malaccensis diploid stands out as a promising candidate for crosses with elite cultivars to transfer resistance alleles to Foc, given its relevant breeding characteristics. The resistance of this diploid can be further explored by isolating a resistance gene (R) of the putative nucleotide-binding and leucine-rich repeat type from Musa acuminata ssp. Malaccensis, which has been utilized in transgenic experiments [33,34].
Besides the Malaccensis diploid, 10 genotypes, including four wild diploids (Jaran, Birmanie, Pipit) and a tetraploid hybrid of the Silk type developed by Embrapa named BRS Princesa (from the cross between Yangambi Nº2—AAB and the M53 diploid—AA), were classified as resistant to Fusarium wilt. The Pahang diploid, extensively studied, has demonstrated resistance to Foc TR4 in both greenhouse and field experiments [11,16,35]. In a study by Zuo et al. [16], 129 accessions from the germplasm bank were evaluated for resistance to Foc TR4 in both greenhouse and field conditions, identifying DH Phang, Tuu Gia, Calcutta 4, and Borneo as highly resistant. These genotypes also contribute to the genealogy of some of the improved diploids assessed in this study, such as 013019-01, CNPMF 0557, CNPMF 0496, and CNPMF 0731, all characterized as resistant to both Foc R1 and ST4. Based on the genealogy of the diploids under study, a potential resistance to TR4 in these hybrids can be hypothesized, warranting confirmation under natural or artificially induced pathogen infection conditions.
The comprehensive analyses conducted in this study suggest that the resistance observed in some of the evaluated improved diploids, particularly to Foc ST4, may be inherited from their parent genotypes. Notably, diploids CNPMF 0534 and CNPMF 0536, classified as highly resistant to Foc ST4, have Calcutta 4 in their genealogy, while those grouped as resistant have Malaccensis and Tuu Gia. These wild relatives of edible bananas are recognized as valuable sources of resistance genes to Foc TR4 [14,15,26]. Furthermore, diploid hybrids 001016-01, CNPMF 0534, and CNPMF 0536, displaying total resistance to Black Sigatoka, hold the potential for crosses with susceptible commercial cultivars to transfer resistance alleles to commercial germplasm [17]. The diploid M53, used as a parent in crosses for generating commercially relevant cultivars, has been identified as resistant to Foc R1 [17], with previous reports confirming its resistance to Foc TR4 in a heavily infected field in Australia [36].
Consequently, the results discussed herein can significantly contribute to banana breeding programs. The improved diploids identified as resistant have the potential for use in crosses with commercial cultivars, particularly 001016-01, CNPMF 0534, and CNPMF 0536, exhibiting high resistance to Foc ST4, and 013004-04, M53, 050012-02, SH3263, SH3362, 013019-01, CNPMF 0496, CNPMF 0513, and CNPMF 0534, displaying high resistance to Foc R1. Moreover, the diploid hybrids evaluated as resistant or moderately resistant for both races, with low Fusarium wilt symptom scores, can be considered quantitatively resistant. These hybrids are valuable for transferring genes with minor effects, are easily selectable, and are efficient in cultivation systems based on integrated management.
5. Conclusions
The screening conducted in this study, under the specified conditions, facilitated the selection of diploid hybrids 001016-01, CNPMF 0534, and CNPMF 0536, highly resistant to Foc ST4, and 013004-04, M53, 050012-02, SH3263, SH3362, 013019-01, CNPMF 0496, CNPMF 0513, and CNPMF 0534, highly resistant to Foc R1, exhibiting no symptoms of Fusarium wilt in the greenhouse. Remarkably, diploid CNPMF0534 demonstrated complete resistance to both evaluated races, with its resistance to Foc ST4 possibly attributable to impeding fungal penetration.
Author Contributions
Conceptualization, W.S.S., A.d.J.R. and E.P.A.; methodology, W.S.S., A.d.J.R., V.B.O.d.A., A.P.d.S.R. and E.P.A.; software, W.S.S. and A.d.J.R.; validation, W.S.S., A.d.J.R., V.B.O.d.A., W.B.d.S. and E.P.A.; formal analysis, A.d.J.R.; investigation, W.S.S., A.d.J.R., F.H. and E.P.A.; resources, E.P.A.; data curation, W.S.S., A.d.J.R. and V.B.O.d.A.; writing—original draft preparation, W.S.S., A.d.J.R. and E.P.A.; writing—review and editing, W.S.S., A.d.J.R., A.P.d.S.R., V.B.O.d.A. and E.P.A.; visualization, W.S.S., A.d.J.R., V.B.O.d.A., A.P.d.S.R., W.B.d.S., F.H. and E.P.A.; supervision, E.P.A.; project administration, E.P.A.; funding acquisition, E.P.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by IITA/The Bill and Melinda Gates Foundation—Accelerated Breeding of Better Bananas, ID OPP1093845.
Data Availability Statement
The data are contained within the article.
Acknowledgments
The authors thank the Postgraduate Program in Genetic Resources of the Federal University of Recôncavo da Bahia (UFRB), as well as the CNPq (National Council for Scientific and Technological Development) for the research productivity grants for E.P.A. and C.F.F., and Fapesb (Bahia Research Foundation) for granting MSc scholarships to W.S.S.
Conflicts of Interest
The authors Anelita de Jesus Rocha and Vanusia Batista de Oliveira Amorim receive research grants from Embrapa; Andresa Priscila de Souza Ramos, Fernando Haddad, and Edson Perito Amorim are employees of Embrapa. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- FAOSTAT. 2024. Available online: https://www.fao.org/faostat/en/#data (accessed on 19 March 2024).
- Mintoff, S.J.L.; Nguyen, T.V.; Kelly, C.; Cullen, S.; Hearnden, M.; Williams, R.; Daniells, J.W.; Tran-Nguyen, L.T.T. Banana Cultivar Field Screening for Resistance to Fusarium oxysporum f. sp. cubense Tropical Race 4 in the Northern Territory. J. Fungi 2021, 7, 627. [Google Scholar] [CrossRef] [PubMed]
- Ploetz, R.C. Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop Prot. 2015, 73, 7–15. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 2006, 96, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Sun, J.; Matthews, A.; Armas-Egas, L.; Chen, N.; Hamill, S.; Mintoff, S.; Tran-Nguyen, L.T.; Batley, J.; Aitken, E.A. Assessing Variations in Host Resistance to Fusarium oxysporum f. sp. cubense Race 4 in Musa Species, with a Focus on the Subtropical Race 4. Front. Microbiol. 2019, 10, 1062. [Google Scholar]
- Dita, M.; Barquero, M.; Heck, D.; Mizubuti, E.S.G.; Staver, C.P. Fusarium wilt of banana: Current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 2018, 9, 1468. [Google Scholar] [CrossRef] [PubMed]
- Batista, I.C.; Heck, D.W.; Santos, A.; Alves, G.; Ferro, C.G.; Dita, M.; Mizubuti, E.S. The Brazilian population of Fusarium oxysporum f. sp. cubense is not structured by VCG or by geographic origin. bioRxiv 2022, 112, 11. [Google Scholar] [CrossRef]
- Buddenhagen, I. Understanding Strain Diversity in Fusarium oxysporum f. sp. cubense and History of Introduction of ‘Tropical Race 4’ to Better Manage Banana Production. Acta Hortic. 2009, 828, 193–204. [Google Scholar]
- García-Bastidas, F.A.; Quintero-Vargas, J.C.; Ayala-Vasquez, M.; Schermer, T.; Seidl, M.F.; Santos-Paiva, M.; Noguera, A.M.; Aguilera-Galvez, C.; Wittenberg, A.; Hofstede, R.; et al. First report of Fusarium wilt Tropical race 4 in Cavendish bananas caused by Fusarium odoratissimum in Colombia. Plant Dis. 2020, 104, 994. [Google Scholar] [CrossRef]
- Martínez, G.; Olivares, B.O.; Rey, J.C.; Rojas, J.; Cardenas, J.; Muentes, C.; Dawson, C. The Advance of Fusarium Wilt Tropical Race 4 in Musaceae of Latin America and the Caribbean: Current Situation. Pathogens 2023, 12, 277. [Google Scholar] [CrossRef]
- Zheng, S.J.; García-Bastidas, F.A.; Li, X.D.; Zeng, L.; Bai, T.T.; Xu, S.T.; Yin, K.S.; Li, H.; Fu, G.; Yu, Y.C.; et al. New incursions of Fusarium oxysporum f. sp. cubense tropical Race 4 across the Greater Me kong Subregion. Front. Plant Sci. 2018, 4, 208–218. [Google Scholar]
- Rocha, A.D.J.; Soares, J.M.D.S.; Nascimento, F.D.S.; Santos, A.S.; Amorim, V.B.D.O.; Ferreira, C.F.; Haddad, F.; Santos-Serejo, J.A.D.; Amorim, E.P. Improvements in the Resistance of the Banana Species to Fusarium Wilt: A Systematic Review of Methods and Perspectives. J. Fungi 2022, 7, 249. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ge, X.; Wu, W.; Wang, W.; Hu, Y.; Mo, Y.; Xie, J. Identification of defense-related genes in banana roots infected by Fusarium oxysporum f. sp. cubense tropical race 4. Euphytica 2015, 205, 837–849. [Google Scholar]
- Rocha, A.D.J.; Soares, J.M.D.S.; Nascimento, F.D.S.; Rocha, A.d.S.; Amorim, V.B.O.D.; Ramos, E.S.T.E.; Ferreira, C.F.; Haddad, F.; Amorim, E.P. Molecular, histological and histochemical responses of banana cultivars challenged with Fusarium oxysporum f. sp. cubense with different levels of virulence. Plants 2022, 11, 2339. [Google Scholar] [CrossRef] [PubMed]
- Li, W.M.; Dita, M.; Rouard, M.; Wu, W.; Roux, N.; Xie, J.H.; Ge, X.J. Deep RNA-seq analysis reveals key responding aspects of wild banana relative resistance to Fusarium oxysporum f. sp. cubense tropical race 4. Funct. Integr. Genom. 2020, 20, 551–562. [Google Scholar]
- Zuo, C.; Deng, G.; Li, B.; Huo, H.; Li, C.; Hu, C.; Kuang, R.; Yang, Q.; Dong, T.; Sheng, O.; et al. Germplasm screening of Musa spp. for resistance to Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4). Eur. J. Plant Pathol. 2018, 151, 723–734. [Google Scholar]
- Gonçalves, Z.S.; Rocha, A.D.J.; Haddad, F.; Amorim, V.B.O.; Ferreira, C.F.; Amorim, E.P. Selection of Diploid and Tetraploid Banana Hybrids Resistant to Pseudocercospora fijiensis. Agronomy 2021, 11, 2483. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Bender, K.S.; Buckley, D.H.; Stahl, D.A. Microbiologia de Brock, 14th ed.; Artmed Editora: Porto Alegre, Brazil, 2016. [Google Scholar]
- Dita, M.A.; Pérez Vicente, L.; Martínez, E. Inoculation of Fusarium oxysporum f. sp. cubense causal agent of fusarium wilt in banana. In Technical Manual: Prevention and Diagnostic of Fusarium Wilt of Banana Caused by Fusarium Oxysporum f. sp. cubense Tropical Race 4 (TR4); Pérez Vicente, L., Dita, M.A., de la Parte, E.M., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014; pp. 55–58. [Google Scholar]
- Mckinney, H.H. Influence of soil, temperature and on infection of wheat seedlings by Helminthosporium sativum. J. Agric. Res. 1923, 26, 195–217. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2020; Volume 1. [Google Scholar]
- Phillips, J.M.; Hayman, D.S. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Karnovsk, M.J. A formaldehydeglutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 1965, 27, 137. [Google Scholar]
- Foster, A.S. Practical Plant Anatomy, 2nd ed.; Van Nostrand: Torronto, ON, Canada, 1949; p. 228. [Google Scholar]
- Ndayihanzamaso, P.; Mostert, D.; Matthews, M.C.; Mahuku, G.; Jomanga, K.; Mpanda, H.J.; Mduma, H.; Brown, A.; Uwimana, B.; Swennen, R.; et al. Evaluation of Mchare and Matooke Bananas for Resistance to Fusarium oxysporum f. sp. cubense Race 1. Plants 2020, 9, 1082. [Google Scholar] [CrossRef]
- Li, C.Q.; Yang, J.H.; Li, W.B.; Sun, J.B.; Peng, M. Direct Root Penetration and Rhizome Vascular Colonization by Fusarium oxysporum f. sp. cubense are the Key Steps in the Successful Infection of Brazil Cavendish. Plant Dis. 2017, 101, 2073–2078. [Google Scholar] [PubMed]
- Rebouças, T.A.; Haddad, F.; Ferreira, C.F.; de Oliveira, S.A.S.; Ledo, C.A.D.S.; Amorim, E.P. Identification of banana genotypes resistant to Fusarium wilt race 1 under field and greenhouse conditions. Sci. Hortic. 2018, 239, 308–313. [Google Scholar] [CrossRef]
- Nakata, P.A. Advances in our understanding of calcium oxalate crystal formation and function in plants. Plant Sci. 2003, 164, 901–909. [Google Scholar] [CrossRef]
- Ceita, G.D.O.; Macedo, J.N.A.; Santos, T.B.; Alemanno, L.; Gesteira, A.D.; Micheli, F.; Mariano, A.C.; Gramacho, K.P.; Silva, D.D.C.; Meinhardt, L.; et al. Involvement of calcium oxalate degradation during programmed cell death in Theobroma cacao tissues triggered by the hemibiotrophic fungus Moniliophthora pemiciosa. Plant Sci. 2007, 173, 106–117. [Google Scholar] [CrossRef]
- Zavaliev, R.; Ueki, S.; Epel, B.L.; Citovsky, V. Biology of callose (β-1, 3-glucan) turnover at plasmodesmata. Protoplasma 2011, 248, 117–130. [Google Scholar] [CrossRef]
- Dong, H.; Ye, Y.; Guo, Y.; Li, H. Comparison transcriptome analysis revealed resistance differences of Cavendish bananas to Fusarium oxysporum f. sp. cubense race1 and race 4. BMC Genom. 2020, 21, 122. [Google Scholar]
- García-Velasco, R.; Portal-González, N.; Santos-Bermúdez, R.; Rodríguez-García, A.; Companioni-González, B. Genetic improvement for resistance to Fusarium wilt in banana. Rev. Mex. Fitopatol. 2021, 39, 122–146. [Google Scholar] [CrossRef]
- Peraza-Echeverria, S.; Dale, J.L.; Harding, R.M.; Smith, M.K.; Collet, C. Characterization of disease resistance gene candidates of the nucleotide binding site (NBS) type from banana and correlation of a transcriptional polymorphism with resistance to Fusarium oxysporum f. sp. cubense race 4. Mol. Breed. 2008, 22, 565–579. [Google Scholar]
- Dale, J.; James, A.; Paul, J.Y.; Khanna, H.; Smith, M.; Peraza-Echeverria, S.; Harding, R. Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 2017, 8, 1496. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 21, 213–217. [Google Scholar] [CrossRef]
- Better Bananas. Panama TR4 Variety Screening Trial (December 2018) Sub-Trial Results (Plant and First Ratoon). Available online: https://betterbananas.com.au/2022/03/04/panama-tr4-variety-screening-trial-december-2018-sub-trial-results-plant-and-first-ratoon/ (accessed on 3 January 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).