Genomic Identification of Callose Synthase (CalS) Gene Family in Sorghum (Sorghum bicolor) and Comparative In Silico Expression Analysis under Aphid (Melanaphis sacchari) Infestation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Aphid Rearing
2.2. Identification of the SbCalS Gene Family
2.3. Physicochemical Characterization and Subcellular Localization Prediction of SbCalS Gene Family
2.4. Prediction of Gene Structure and Cis-Acting Elements in the SbCalS Gene Family
2.5. Chromosomal Mapping and Intraspecific Collinearity and Ka/Ks Value Analysis of SbCalS Gene Family
2.6. Interspecific Collinearity Analysis of SbCalS and Construction of Phylogenetic Tree
2.7. Analysis of the SbCalS Gene Family Using Published RNA-Seq Data
2.8. Expression Characteristics of SbCalS Genes under Aphid Infestation
2.9. Determination of Callose Content in Sorghum
3. Results
3.1. Identification of SbCalS Gene Family
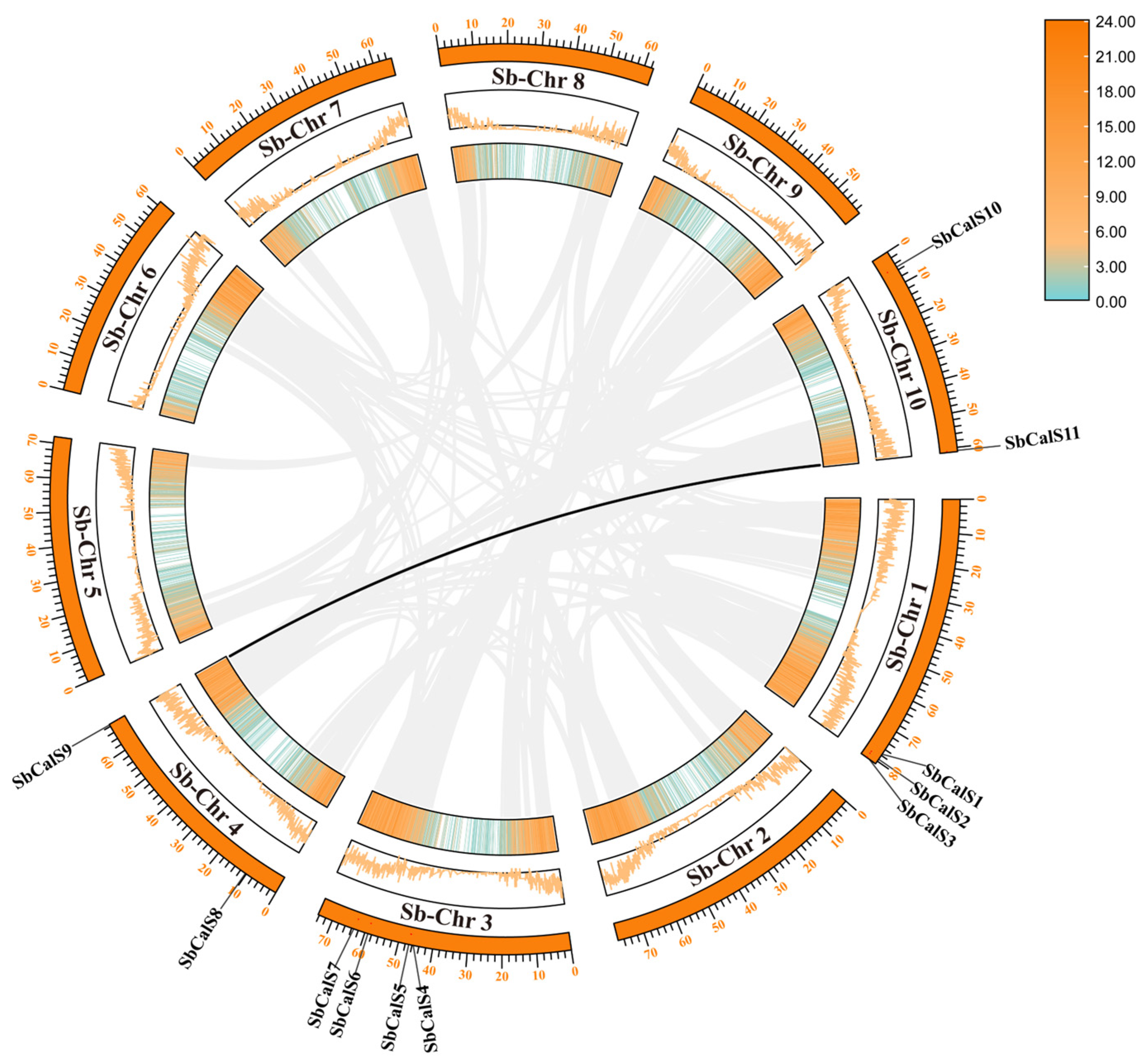
3.2. Chromosome Localization and Intraspecific Colinear and Ka/Ks Analysis of SbCalS Gene Family
3.3. Physiochemical Properties and Subcellular Localization Analysis of SbCalS Gene Family
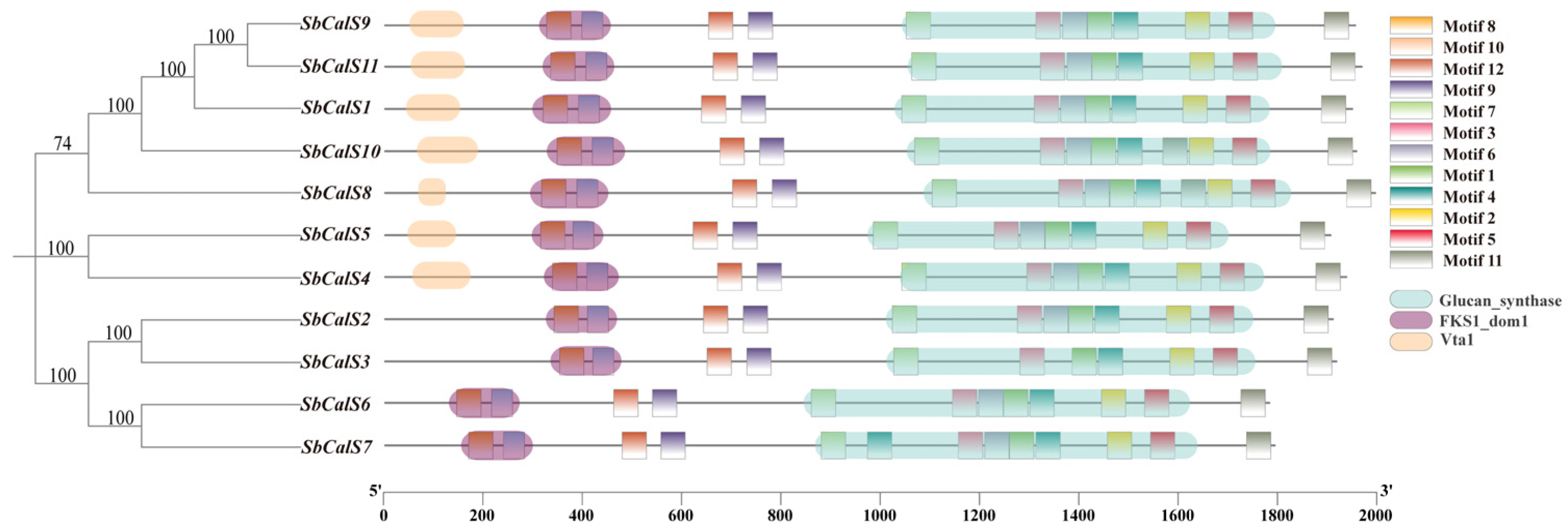
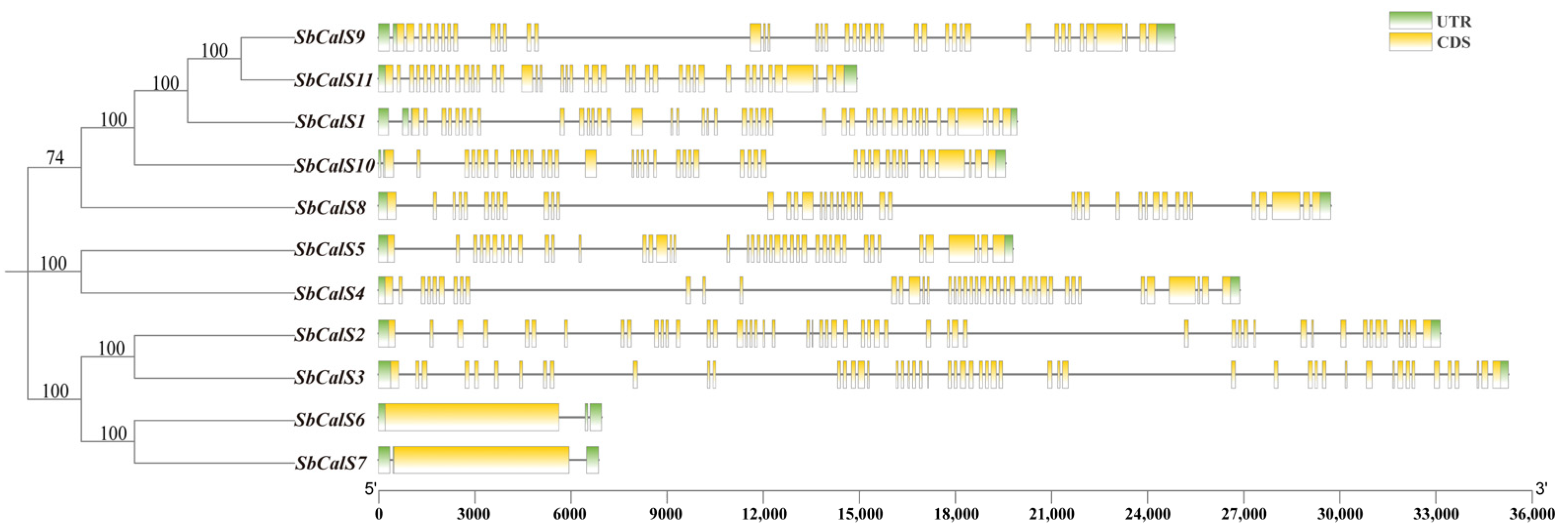
3.4. Structural Analysis of the SbCalS Gene Family
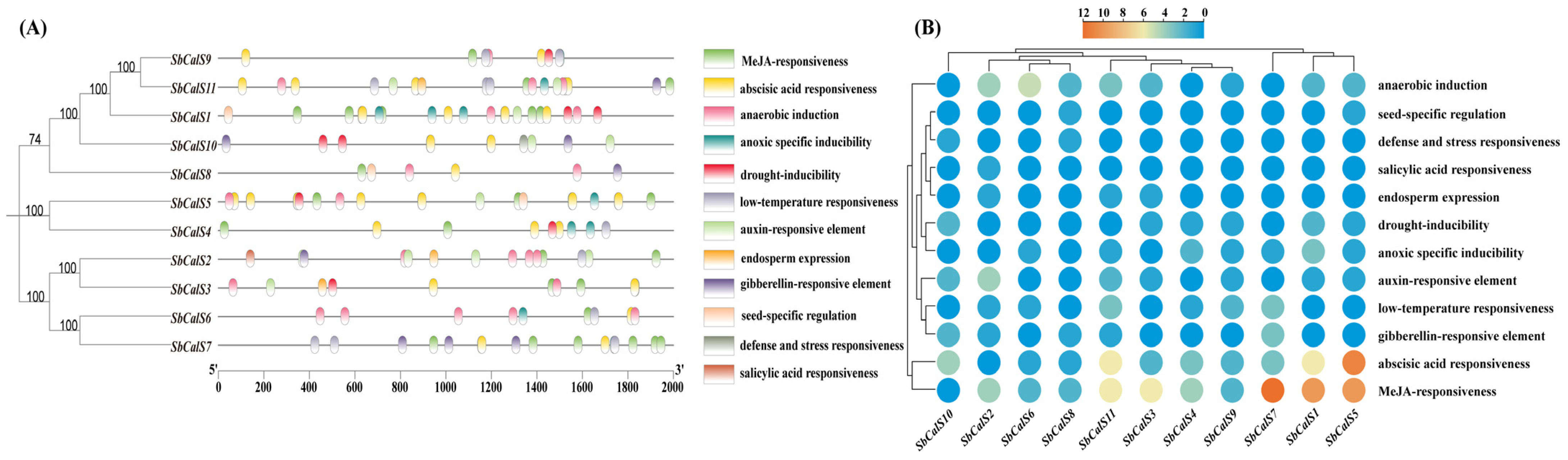
3.5. Cis-Acting Element Analysis of SbCalS Gene Family
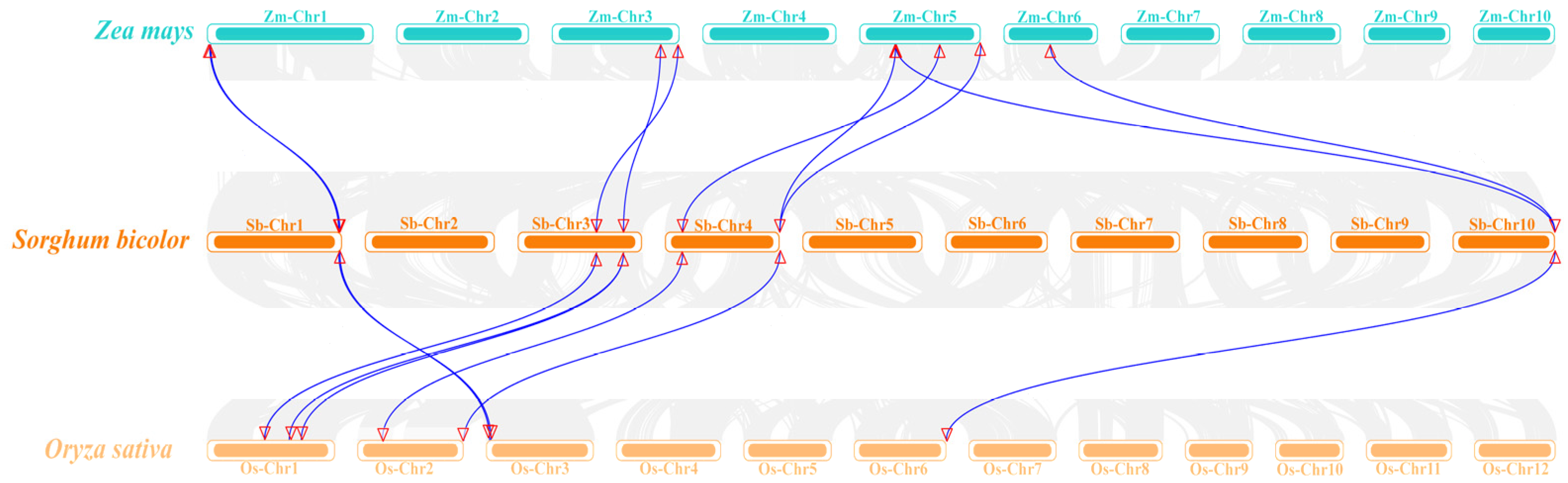
3.6. Analysis of SbCalS Interspecific Covariates and Phylogenetic Trees
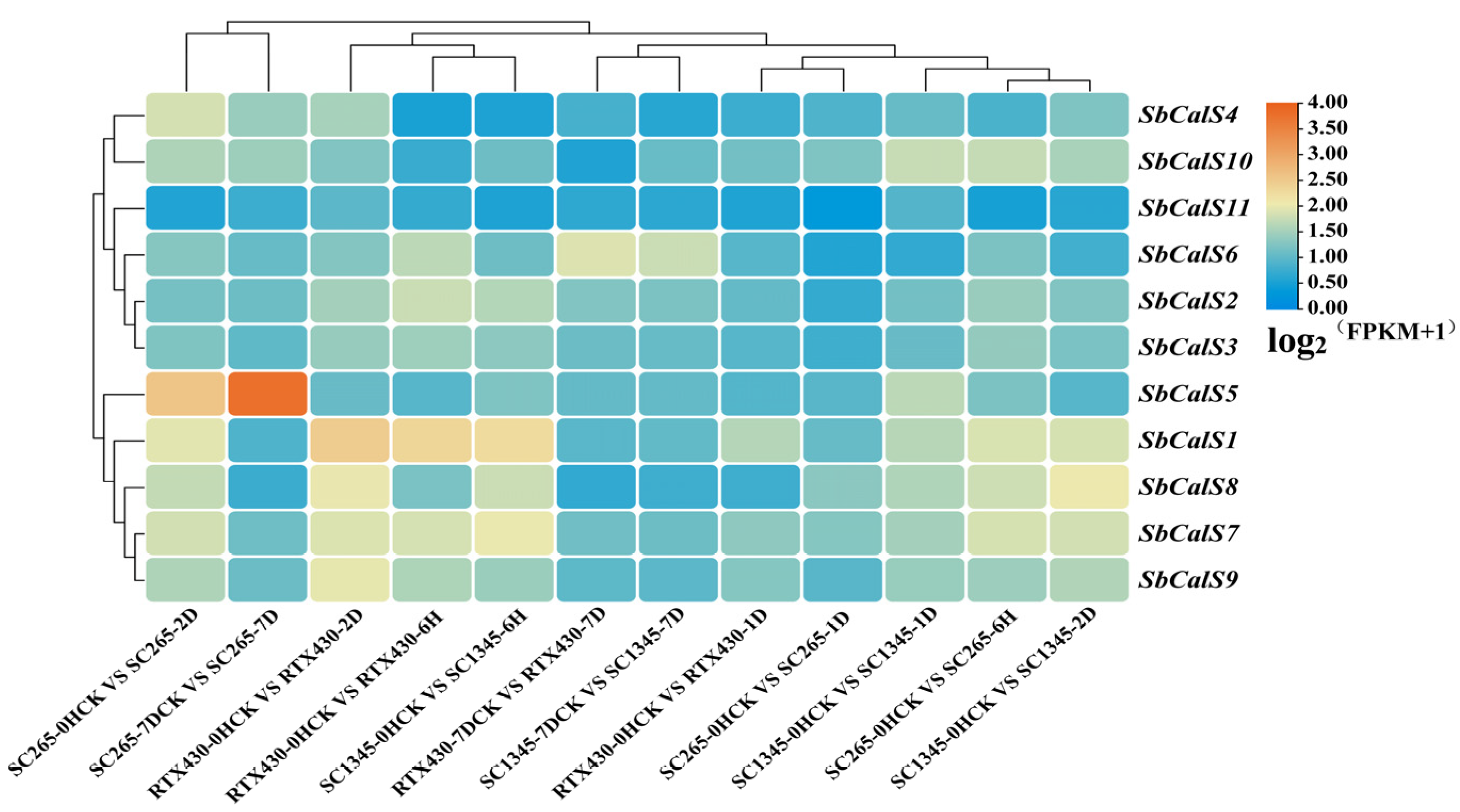
3.7. Transcriptome Analysis of the SbCalS Gene Family Following Aphid Infestation
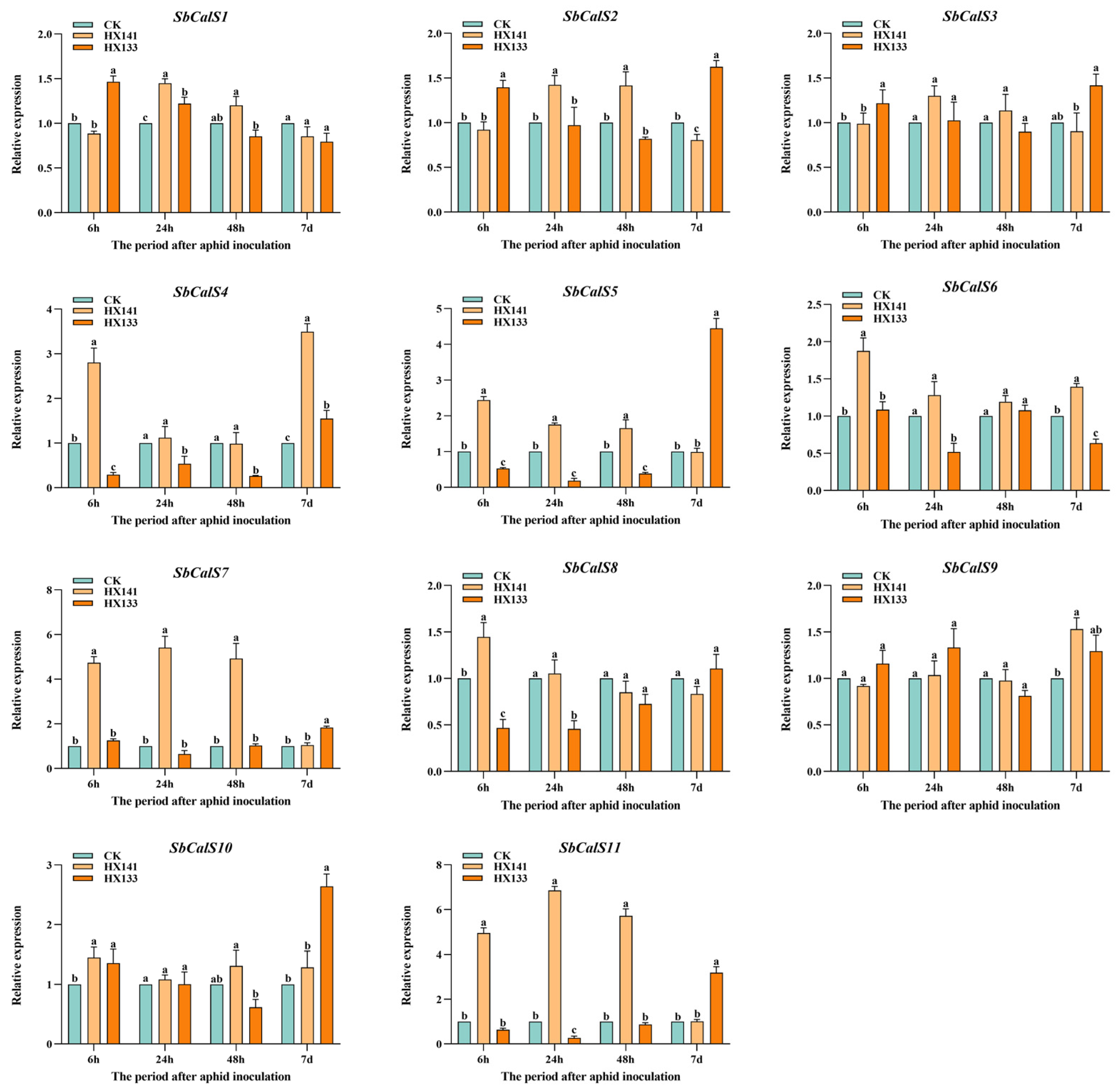
3.8. Expression Pattern Analysis of the SbCalS Gene Family Following Aphid Infestation
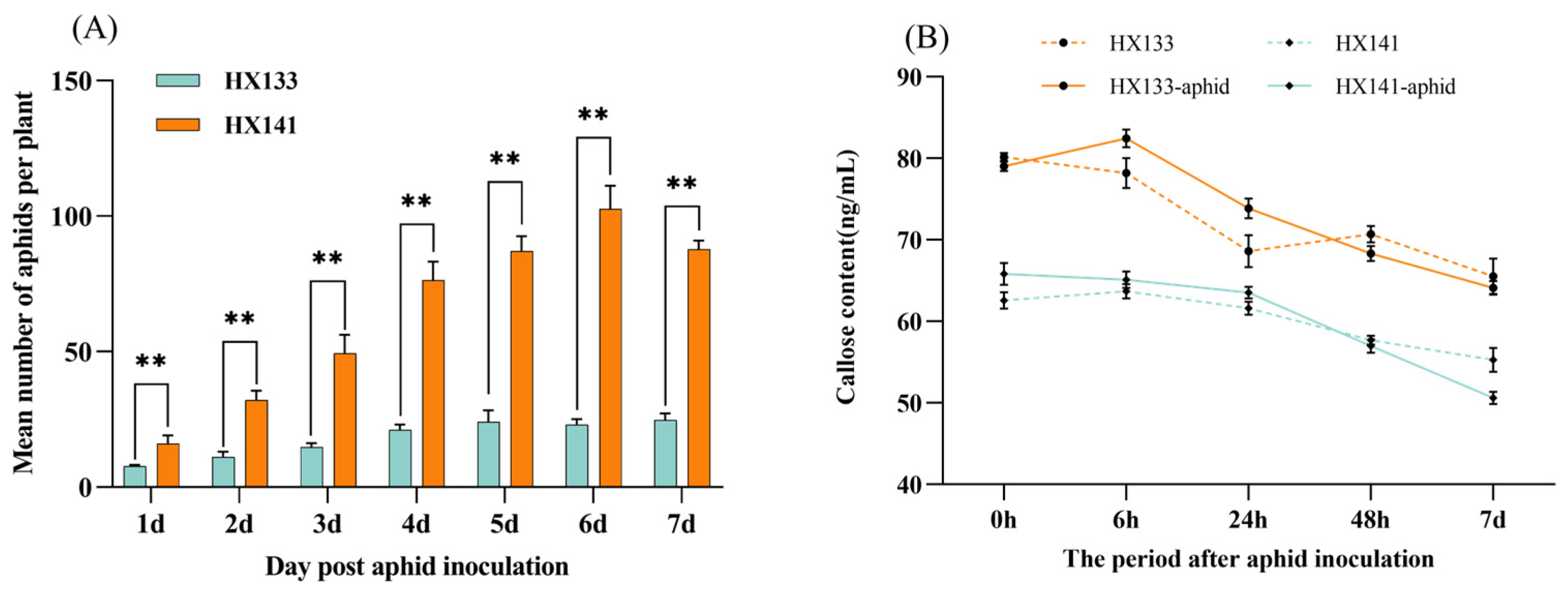
3.9. Analysis of Sorghum Aphid Population and Callose Content
4. Discussion
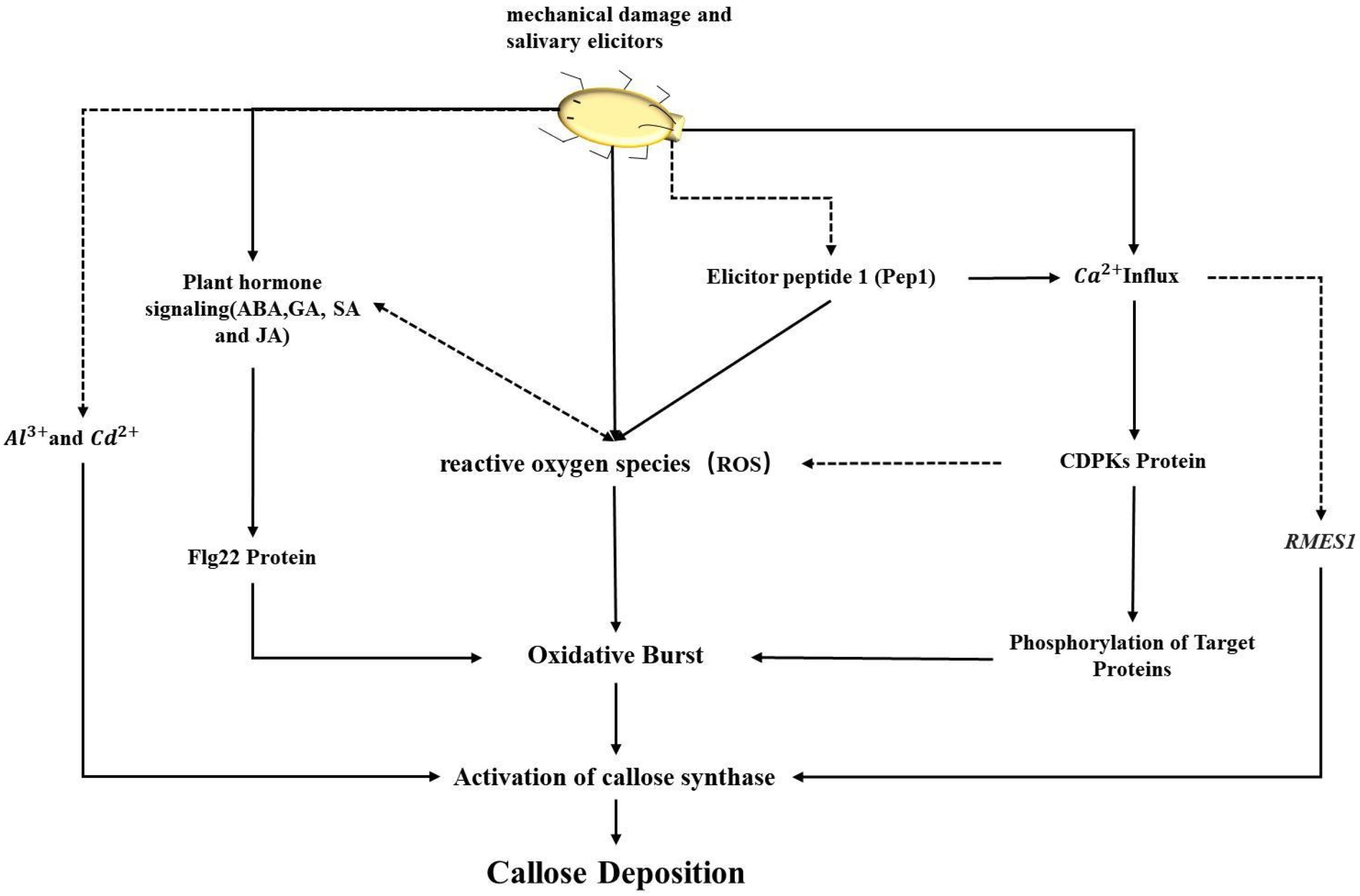
4.1. The Role and Mechanism of the CalS Gene Family in Plant Defense
4.2. The Multiple Roles and Regulation of the CalS Gene Family in Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Begna, T. Role of sorghum genetic diversity in tackling drought effect in Ethiopia. Int. J. Adv. Res. Biol. Sci. 2021, 8, 29–45. [Google Scholar]
- Afify, A.E.-M.M.; El-Beltagi, H.S.; Abd El-Salam, S.M.; Omran, A.A. Bioavailability of iron, zinc, phytate and phytase activity during soaking and germination of white sorghum varieties. PLoS ONE 2011, 6, e25512. [Google Scholar] [CrossRef] [PubMed]
- Taleon, V.; Dykes, L.; Rooney, W.; Rooney, L. Effect of genotype and environment on flavonoid concentration and profile of black sorghum grains. J. Cereal Sci. 2012, 56, 470–475. [Google Scholar] [CrossRef]
- Kim, J.-S.; Hyun, T.K.; Kim, M.-J. The inhibitory effects of ethanol extracts from sorghum, foxtail millet and proso millet on α-glucosidase and α-amylase activities. Food Chem. 2011, 124, 1647–1651. [Google Scholar] [CrossRef]
- Abd Elmoneim, O.E.; Schiffler, B.; Bernhardt, R. Effect of fermentation on the functional properties of sorghum flour. Food Chem. 2005, 92, 1–5. [Google Scholar]
- Gordy, J.W.; Brewer, M.J.; Bowling, R.D.; Buntin, G.D.; Seiter, N.J.; Kerns, D.L.; Reay-Jones, F.P.; Way, M. Development of economic thresholds for sugarcane aphid (Hemiptera: Aphididae) in susceptible grain sorghum hybrids. J. Econ. Entomol. 2019, 112, 1251–1259. [Google Scholar] [CrossRef]
- Bowling, R.D.; Brewer, M.J.; Kerns, D.L.; Gordy, J.; Seiter, N.; Elliott, N.E.; Buntin, G.D.; Way, M.; Royer, T.; Biles, S. Sugarcane aphid (Hemiptera: Aphididae): A new pest on sorghum in North America. J. Integr. Pest Manag. 2016, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Padmaja, P.; Seetharama, N. Biology and management of the sugarcane aphid, Melanaphis sacchari (Zehntner)(Homoptera: Aphididae), in sorghum: A review. Crop Prot. 2004, 23, 739–755. [Google Scholar] [CrossRef]
- Narayana, D. Screening for aphids and sooty molds in sorghum. Sorghum Newsl. 1975, 18, 21–22. [Google Scholar]
- Li, N.; Lin, Z.; Yu, P.; Zeng, Y.; Du, S.; Huang, L.-J. The multifarious role of callose and callose synthase in plant development and environment interactions. Front. Plant Sci. 2023, 14, 1183402. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Kim, J.-Y. Callose synthesis in higher plants. Plant Signal. Behav. 2009, 4, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, M.-C.; Wirthmueller, L.; Sklenar, J.; Findlay, K.; Piquerez, S.J.; Jones, A.M.; Robatzek, S.; Jones, J.D.; Faulkner, C. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 2014, 10, e1004496. [Google Scholar] [CrossRef] [PubMed]
- Blümke, A.; Somerville, S.C.; Voigt, C.A. Transient expression of the Arabidopsis thaliana callose synthase PMR4 increases penetration resistance to powdery mildew in barley. Adv. Biosci. Biotechnol. 2013, 2013, 35416. [Google Scholar]
- Sanmartín, N.; Pastor, V.; Pastor-Fernández, J.; Flors, V.; Pozo, M.J.; Sánchez-Bel, P. Role and mechanisms of callose priming in mycorrhiza-induced resistance. J. Exp. Bot. 2020, 71, 2769–2781. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Bengtsson, T.; Olsson, N.; Hot, V.; Zhu, L.-H.; Åhman, I. Mutations in two aphid-regulated β-1, 3-glucanase genes by CRISPR/Cas9 do not increase barley resistance to Rhopalosiphum padi L. Front. Plant Sci. 2020, 11, 538516. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Jander, G. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 2007, 49, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Botha, C.E.; Matsiliza, B. Reduction in transport in wheat (Triticum aestivum) is caused by sustained phloem feeding by the Russian wheat aphid (Diuraphis noxia). S. Afr. J. Bot. 2004, 70, 249–254. [Google Scholar] [CrossRef]
- Yao, L.; Zhong, Y.; Wang, B.; Yan, J.; Wu, T. BABA application improves soybean resistance to aphid through activation of phenylpropanoid metabolism and callose deposition. Pest Manag. Sci. 2020, 76, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Mbiza, N.I.T.; Hu, Z.; Zhang, H.; Zhang, Y.; Luo, X.; Wang, Y.; Wang, Y.; Liu, T.; Li, J.; Wang, X. GhCalS5 is involved in cotton response to aphid attack through mediating callose formation. Front. Plant Sci. 2022, 13, 892630. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Chen, A.; Huo, R.; Yan, H.; Zhang, Z.; Guo, H. Callose deposition regulates differences in cotton aphid resistance among six watermelon varieties. J. Pest Sci. 2024, 1–16. [Google Scholar] [CrossRef]
- Barratt, D.P.; Kölling, K.; Graf, A.; Pike, M.; Calder, G.; Findlay, K.; Zeeman, S.C.; Smith, A.M. Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol. 2011, 155, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.-Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.Y.; Shirasu, K.; Moon, J.S.; Lee, S.-G.; Kwon, S.-Y. The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS ONE 2014, 9, e88951. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, D.; Voigt, C.A. Callose biosynthesis in Arabidopsis with a focus on pathogen response: What we have learned within the last decade. Ann. Bot. 2014, 114, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Nalam, V.; Louis, J.; Shah, J. Plant defense against aphids, the pest extraordinaire. Plant Sci. 2019, 279, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sagar, S.; Biswas, D.K. Calcium dependent protein kinase, a versatile player in plant stress management and development. Crit. Rev. Plant Sci. 2017, 36, 336–352. [Google Scholar] [CrossRef]
- Molina-Moya, E.; Terrón-Camero, L.C.; Pescador-Azofra, L.; Sandalio, L.M.; Romero-Puertas, M.C. Reactive oxygen species and nitric oxide production, regulation and function during defense response. In Reactive Oxygen, Nitrogen and Sulfur Species in Plants: Production, Metabolism, Signaling and Defense Mechanisms; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2019; pp. 573–590. [Google Scholar]
- Kuśnierczyk, A.; Winge, P.; Jørstad, T.S.; Troczyńska, J.; Rossiter, J.T.; Bones, A.M. Towards global understanding of plant defence against aphids–timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 2008, 31, 1097–1115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Bao, Q.; Wang, H.; Hou, S. Plant elicitor peptide 1 fortifies root cell walls and triggers a systemic root-to-shoot immune signaling in Arabidopsis. Plant Signal. Behav. 2022, 17, 2034270. [Google Scholar] [CrossRef] [PubMed]
- O’Lexy, R.; Kasai, K.; Clark, N.; Fujiwara, T.; Sozzani, R.; Gallagher, K.L. Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. J. Exp. Bot. 2018, 69, 3715–3728. [Google Scholar] [CrossRef]
- Tetreault, H.M.; Grover, S.; Scully, E.D.; Gries, T.; Palmer, N.A.; Sarath, G.; Louis, J.; Sattler, S.E. Global responses of resistant and susceptible sorghum (Sorghum bicolor) to sugarcane aphid (Melanaphis sacchari). Front. Plant Sci. 2019, 10, 426693. [Google Scholar] [CrossRef]
- Hong, Z.; Delauney, A.J.; Verma, D.P.S. A cell plate–specific callose synthase and its interaction with phragmoplastin. Plant Cell 2001, 13, 755–768. [Google Scholar] [PubMed]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomic data. In Plant Genomics Databases: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–31. [Google Scholar]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Kb, N. GeneDoc: Analysis and visualization of genetic variation. EMBnet News 1997, 4, 1–4. [Google Scholar]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI biosystems database. Nucleic Acids Res. 2010, 38, D492–D496. [Google Scholar] [CrossRef]
- Yadav, C.B.; Bonthala, V.S.; Muthamilarasan, M.; Pandey, G.; Khan, Y.; Prasad, M. Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res. 2015, 22, 79–90. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Puri, H.; Grover, S.; Pingault, L.; Sattler, S.E.; Louis, J. Temporal transcriptomic profiling elucidates sorghum defense mechanisms against sugarcane aphids. BMC Genom. 2023, 24, 441. [Google Scholar] [CrossRef]
- Sayers, E.W.; O’Sullivan, C.; Karsch-Mizrachi, I. Using GenBank and SRA. In Plant Bioinformatics: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–25. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Logemann, J.; Schell, J.; Willmitzer, L. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 1987, 163, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, F.; Wang, L.; Zhan, Q.; Wu, P.; Du, J.; Yang, X.; Liu, Y. Cloning and expression analysis of cinnamoyl-CoA reductase (CCR) genes in sorghum. PeerJ 2016, 4, e2005. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, F.; Huang, W.; Sun, Q.; Huang, X. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci. Rep. 2019, 9, 8408. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H. Prism 5 statistics guide, 2007. GraphPad Softw. 2007, 31, 39–42. [Google Scholar]
- Ward, D.M.; Vaughn, M.B.; Shiflett, S.L.; White, P.L.; Pollock, A.L.; Hill, J.; Schnegelberger, R.; Sundquist, W.I.; Kaplan, J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 2005, 280, 10548–10555. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yang, P.; Chai, C.; Liu, J.; Sun, H.; Wu, Y.; Zhang, M.; Zhang, M.; Liu, X.; Yu, H. Structural and mechanistic insights into fungal β-1, 3-glucan synthase FKS1. Nature 2023, 616, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, J.; Lin, W.; Li, S.; Li, H.; Zhou, J.; Ni, P.; Dong, W.; Hu, S.; Zeng, C. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, e38. [Google Scholar] [CrossRef]
- Venkateswaran, K.; Elangovan, M.; Sivaraj, N. Origin, domestication and diffusion of Sorghum bicolor. In Breeding Sorghum for Diverse end Uses; Elsevier: Amsterdam, The Netherlands, 2019; pp. 15–31. [Google Scholar]
- Shen, Y.; Liu, M.; Wang, L.; Li, Z.; Taylor, D.C.; Li, Z.; Zhang, M. Identification, duplication, evolution and expression analyses of caleosins in Brassica plants and Arabidopsis subspecies. Mol. Genet. Genom. 2016, 291, 971–988. [Google Scholar] [CrossRef]
- Niu, Q.; Zhang, P.; Su, S.; Jiang, B.; Liu, X.; Li, C.; Yu, T.; Yi, H.; Tang, J.; Cao, M. Characterization and expression analyses of Callose synthase enzyme (Cals) family genes in maize (Zea mays L.). Biochem. Genet. 2022, 60, 351–369. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Regulation and function of defense-related callose deposition in plants. Int. J. Mol. Sci. 2021, 22, 2393. [Google Scholar] [CrossRef]
- Shikanai, Y.; Yoshida, R.; Hirano, T.; Enomoto, Y.; Li, B.; Asada, M.; Yamagami, M.; Yamaguchi, K.; Shigenobu, S.; Tabata, R. Callose synthesis suppresses cell death induced by low-calcium conditions in leaves. Plant Physiol. 2020, 182, 2199–2212. [Google Scholar] [CrossRef] [PubMed]
- Kauss, H.; Jeblick, W. Induced Ca2+ uptake and callose synthesis in suspension-cultured cells of Catharanthus roseus are decreased by the protein phosphatase inhibitor okadaic acid. Physiol. Plant. 1991, 81, 309–312. [Google Scholar] [CrossRef]
- Verma, D.P.S.; Hong, Z. Plant callose synthase complexes. Plant Mol. Biol. 2001, 47, 693–701. [Google Scholar] [CrossRef] [PubMed]
- He, D.; You, X.-L.; He, S.-L.; Zhang, M.-X.; Zhang, J.-R.; Hua, C.; Wang, Z.; Liu, Y.-P. Identification of callose synthetase gene family and functional analysis of PlCalS5 in Paeonia lactiflora. Sci. Agric. Sin. 2023, 56, 3183–3198. [Google Scholar]
- Dong, X.; Hong, Z.; Sivaramakrishnan, M.; Mahfouz, M.; Verma, D.P.S. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 2005, 42, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Enns, L.C.; Kanaoka, M.M.; Torii, K.U.; Comai, L.; Okada, K.; Cleland, R.E. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol. Biol. 2005, 58, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Tang, D.; Zhao, T.; Zhang, F.; Liu, C.; Xue, Z.; Shi, W.; Du, G.; Shen, Y.; Li, Y. Os SPL regulates meiotic fate acquisition in rice. New Phytol. 2018, 218, 789–803. [Google Scholar] [CrossRef]
- Zhang, Z.; Lee, Y.; Spetz, C.; Clarke, J.L.; Wang, Q.; Blystad, D.-R. Invasion of shoot apical meristems by Chrysanthemum stunt viroid differs among Argyranthemum cultivars. Front. Plant Sci. 2014, 6, 53. [Google Scholar] [CrossRef]
- Liu, F.; Zou, Z.; Fernando, W.D. Characterization of callose deposition and analysis of the callose synthase gene family of brassica napus in response to leptosphaeria maculans. Int. J. Mol. Sci. 2018, 19, 3769. [Google Scholar] [CrossRef]
- Huang, Q.; Lin, B.; Cao, Y.; Zhang, Y.; Song, H.; Huang, C.; Sun, T.; Long, C.; Liao, J.; Zhuo, K. CRISPR/Cas9-mediated mutagenesis of the susceptibility gene OsHPP04 in rice confers enhanced resistance to rice root-knot nematode. Front. Plant Sci. 2023, 14, 1134653. [Google Scholar] [CrossRef]
- Sun, M.; Voorrips, R.E.; Steenhuis-Broers, G.; van’t Westende, W.; Vosman, B. Reduced phloem uptake of Myzus persicae on an aphid resistant pepper accession. BMC Plant Biol. 2018, 18, 138. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Hyun, T.K.; Zhang, M.; Kumar, R.; Koh, E.-j.; Kang, B.-H.; Lucas, W.J.; Kim, J.-Y. Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell 2014, 28, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Du, H.; Ding, X.; Zhou, Y.; Xie, P.; Wu, J. Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stål (Hemiptera: Delphacidae). Pest Manag. Sci. 2017, 73, 2559–2568. [Google Scholar] [CrossRef]
- Wang, B.; Andargie, M.; Fang, R. The function and biosynthesis of callose in high plants. Heliyon 2022, 8, e09248. [Google Scholar] [CrossRef]
- Zhu-Salzman, K.; Salzman, R.A.; Ahn, J.-E.; Koiwa, H. Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol. 2004, 134, 420–431. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, X.-Y.; Sun, X.-Z.; Xu, B.-Y. Effect of methyl jasmonate on aphid resistance of chrysanthemum. Ying Yong Sheng tai xue bao J. Appl. Ecol. 2020, 31, 4197–4205. [Google Scholar]
- Underwood, W. The plant cell wall: A dynamic barrier against pathogen invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef]
- Repka, V.; Fischerova, I.; Šilhárová, K. Methyl jasmonate is a potent elicitor of multiple defense responses in grapevine leaves and cell-suspension cultures. Biol. Plant. 2004, 48, 273–283. [Google Scholar] [CrossRef]
- Yao, L.; Guo, X.; Su, J.; Zhang, Q.; Lian, M.; Xue, H.; Li, Q.; He, Y.; Zou, X.; Song, Z. ABA-CsABI5-CsCalS11 module upregulates Callose deposition of citrus infected with Candidatus Liberibacter asiaticus. Hortic. Res. 2024, 11, uhad276. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, L.; Baldwin, I.T. Methyl jasmonate-elicited herbivore resistance: Does MeJA function as a signal without being hydrolyzed to JA? Planta 2008, 227, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, S.; Guayazán-Palacios, N.; Steinbrenner, A.D. Molecular tug-of-war: Plant immune recognition of herbivory. Plant Cell 2022, 34, 1497–1513. [Google Scholar] [CrossRef] [PubMed]
- Farmer, E.E.; Gao, Y.Q.; Lenzoni, G.; Wolfender, J.L.; Wu, Q. Wound-and mechanostimulated electrical signals control hormone responses. New Phytol. 2020, 227, 1037–1050. [Google Scholar] [CrossRef]
- Wang, F.; Zhao, S.; Han, Y.; Shao, Y.; Dong, Z.; Gao, Y.; Zhang, K.; Liu, X.; Li, D.; Chang, J. Efficient and fine mapping of RMES1 conferring resistance to sorghum aphid Melanaphis sacchari. Mol. Breed. 2013, 31, 777–784. [Google Scholar] [CrossRef]
- Puri, H. Sorghum-Sugarcane Aphid Interactions: Multi-Omic Approaches to Elucidate Plant Defense Against Sap-Feeding Insects; The University of Nebraska-Lincoln: Lincoln, NE, USA, 2023. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, K.; Liu, Y.; Wang, T.; Guan, M.; Li, X.; Li, J.; Yu, H.; Wu, D.; Du, J. Genomic Identification of Callose Synthase (CalS) Gene Family in Sorghum (Sorghum bicolor) and Comparative In Silico Expression Analysis under Aphid (Melanaphis sacchari) Infestation. Agronomy 2024, 14, 1393. https://doi.org/10.3390/agronomy14071393
Zou K, Liu Y, Wang T, Guan M, Li X, Li J, Yu H, Wu D, Du J. Genomic Identification of Callose Synthase (CalS) Gene Family in Sorghum (Sorghum bicolor) and Comparative In Silico Expression Analysis under Aphid (Melanaphis sacchari) Infestation. Agronomy. 2024; 14(7):1393. https://doi.org/10.3390/agronomy14071393
Chicago/Turabian StyleZou, Kunliang, Yang Liu, Tonghan Wang, Minghui Guan, Xiaofei Li, Jieqin Li, Haibing Yu, Degong Wu, and Junli Du. 2024. "Genomic Identification of Callose Synthase (CalS) Gene Family in Sorghum (Sorghum bicolor) and Comparative In Silico Expression Analysis under Aphid (Melanaphis sacchari) Infestation" Agronomy 14, no. 7: 1393. https://doi.org/10.3390/agronomy14071393
APA StyleZou, K., Liu, Y., Wang, T., Guan, M., Li, X., Li, J., Yu, H., Wu, D., & Du, J. (2024). Genomic Identification of Callose Synthase (CalS) Gene Family in Sorghum (Sorghum bicolor) and Comparative In Silico Expression Analysis under Aphid (Melanaphis sacchari) Infestation. Agronomy, 14(7), 1393. https://doi.org/10.3390/agronomy14071393






