Engineered Rhizobia with Trehalose-Producing Genes Enhance Peanut Growth Under Salinity Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and the Rhizobial Strains
2.2. Construction of Engineerd Rhizobial Strains
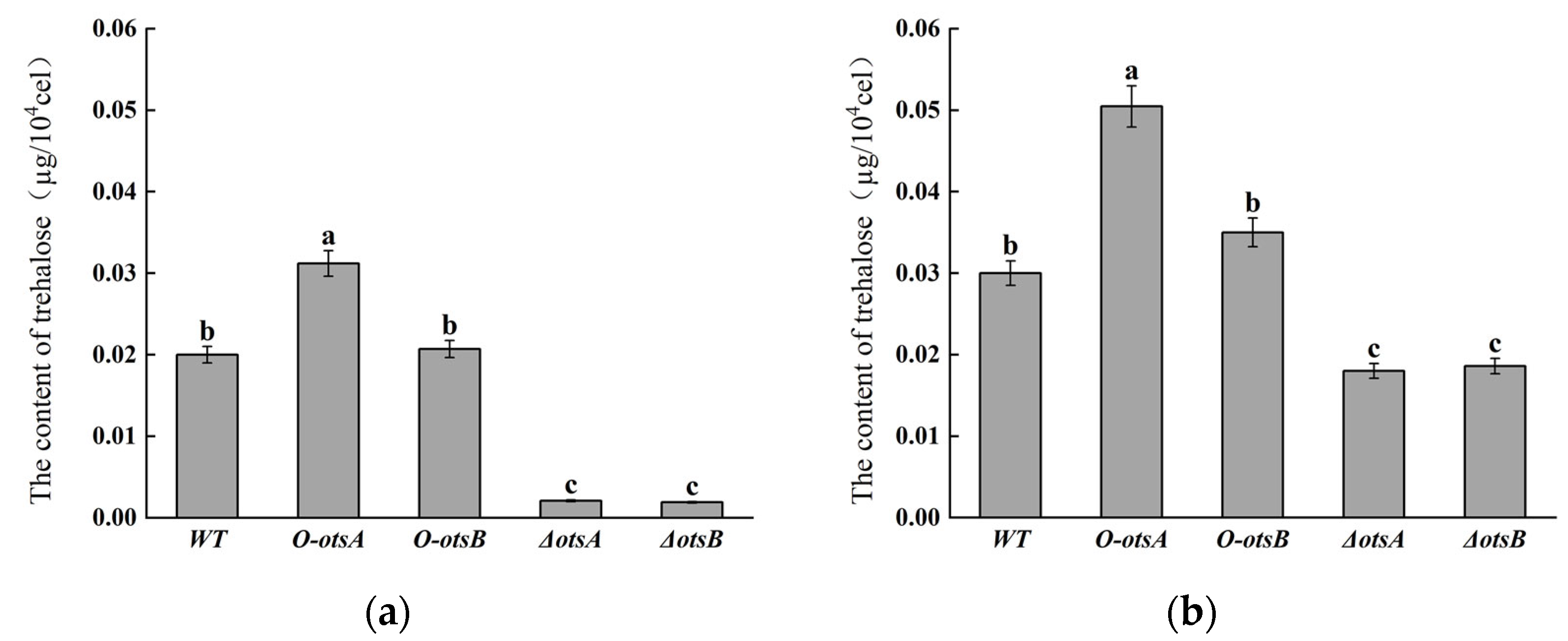
2.3. Determination of Trehalose Content and Salt Tolerance in Engineerd Rhizobial Strains
2.4. Methods of Pot Experiment, Rhizobial Inocualtion and Salt Treatment
2.5. qRT-PCR Analysis of Nodular Genes and Nitrogen-Fixing Genes in Peanut Root Nodules
2.6. Determination of Peanut Nitrogenase Activity
2.7. Determination of Peanut Leaf Photosynthesis and Agronomic Traits After Inoculation of Different Engineered Rhizobial Strains
2.8. Determination of Antioxidant Oxidase (SOD, POD) Activities, MDA and Proline Contents in Peanut
3. Results
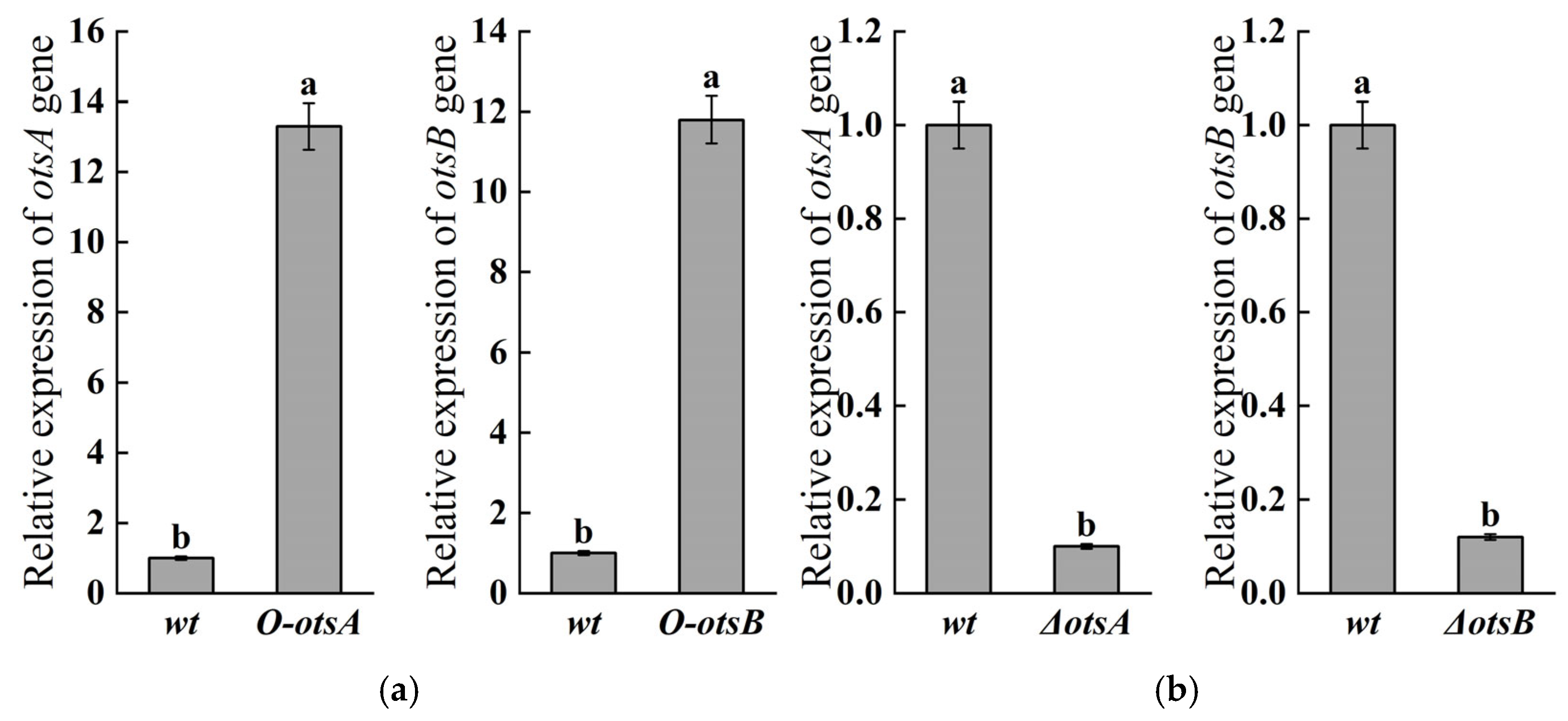
3.1. Construction of ostA and ostB Overexpression/Knockout Strains
3.2. Analysis of Agronomic Traits in Peanuts Inoculated with Engineered Rhizobial Strains
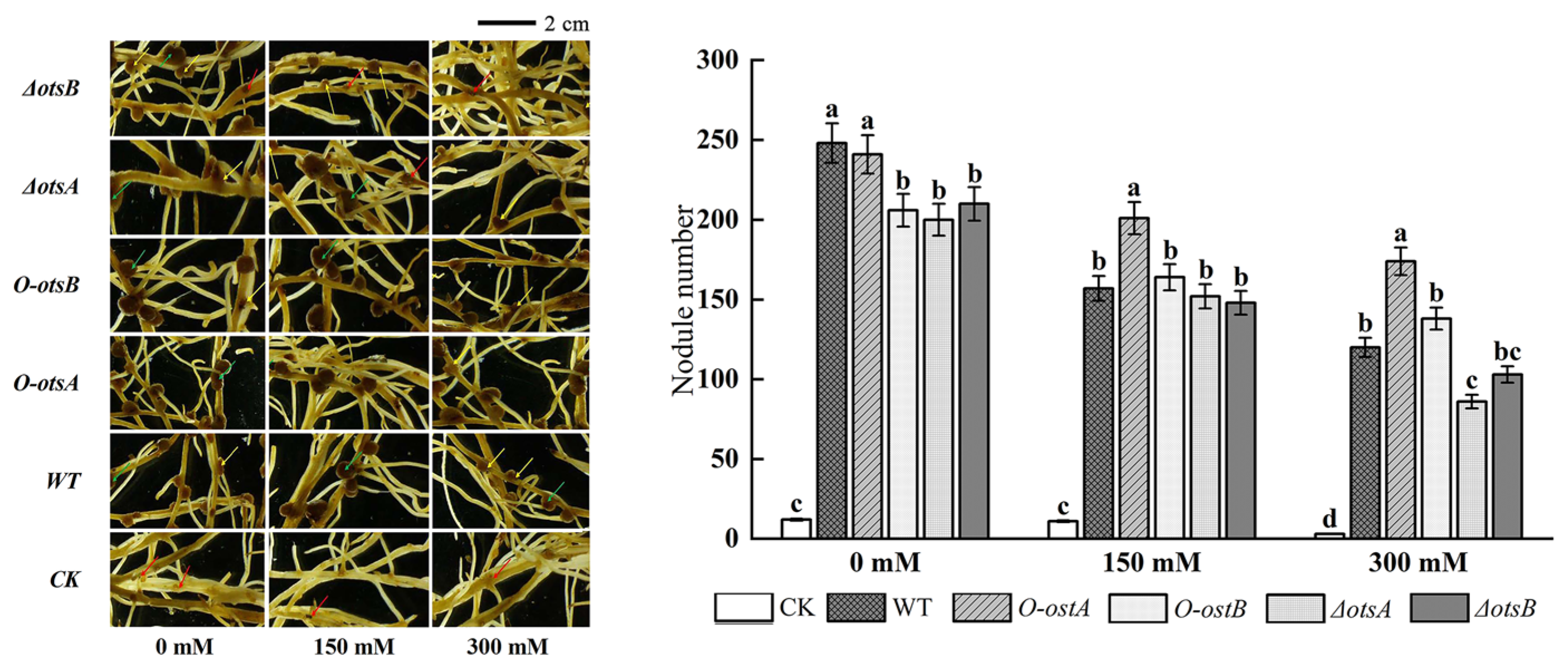
3.3. Analysis of the Growth of Peanut Root Nodules and Enzyme Activity
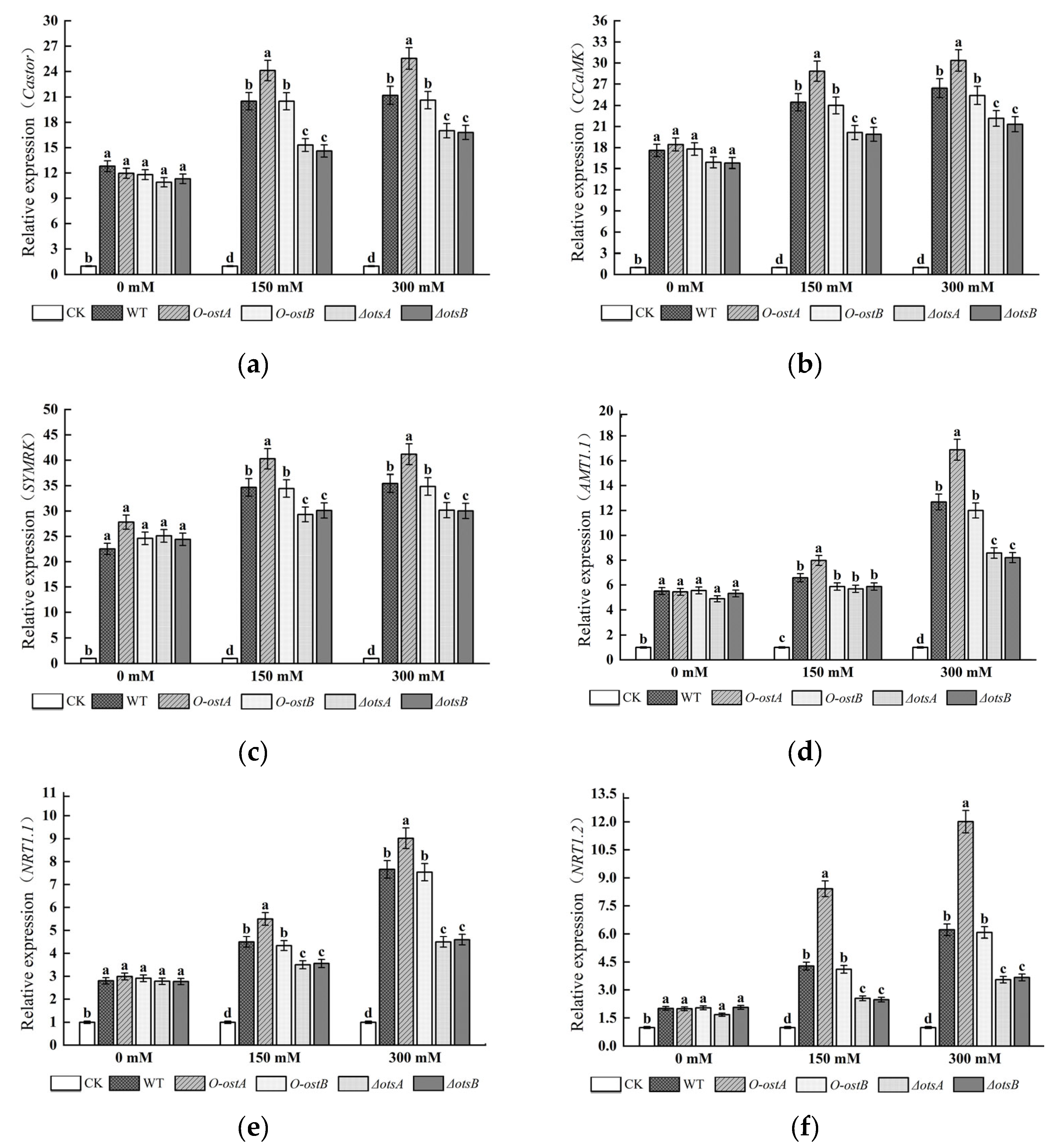
3.4. qRT-PCR Analysis of Nodulation and Nitrogen Fixation Genes in Peanut Roots
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Tre | Trehalose |
| DNA | Directory of open access journals |
| PCR | Polymerase chain reaction |
| Kan | Kanamycin |
| WT | Wide Type |
| YMA | Yeast-Mannitol Agar |
| SOD | Superoxide dismutase |
| POD | Peroxidase |
| MDA | Malondialdehyde |
References
- Wang, C.; Wang, Z.; Wei, Y.; Tang, Y.; Wang, F.; Han, H.; Sun, Y. Effect of variety and seed dressing on emergence of high-oleic peanut under low temperature and high soil humidity conditions. Oil Crop Sci. 2021, 6, 164–168. [Google Scholar] [CrossRef]
- Kong, Q.W. Analysis of peanut cultivation techniques and measures to improve planting benefits. Mod. Agric. 2022, 16, 13–14. [Google Scholar]
- Shao, C.L.; Wang, M.L. Some thoughts on the development of peanut industrialization. J. Peanut Sci. 2003, 32, 73–76. [Google Scholar]
- Raza, A.; Sharif, Y.; Chen, K.; Wang, L.; Fu, H.; Zhuang, Y.; Chitikineni, A.; Chen, H.; Zhang, C.; Varshney, R.K.; et al. Genome-wide characterization of ascorbate peroxidase gene family in peanut (Arachis hypogea L.) revealed their crucial role in growth and multiple stress tolerance. Front. Plant Sci. 2022, 13, 962182. [Google Scholar] [CrossRef]
- Wu, Y.F.; Liu, Q.M.; Liu, W.H.; Meng, A.P.; Chen, Z.X.; Liu, L. Effects of AMF and rhizobia on photosynthesis and respiratory metabolism of intercropping soybean. J. Guangxi Norm. Univ. Nat. Sci. Ed. 2022, 40, 231–241. [Google Scholar]
- Xu, Y.; Zhang, Z.; Ding, H.; Wen, S.; Zhang, G.; Qin, F.; Dai, L. Comprehensive effects of salt stress and peanut cultivars on the rhizosphere bacterial community diversity of peanut. Arch. Microbiol. 2021, 204, 15. [Google Scholar] [CrossRef]
- Rao, D.L.; Giller, K.E.; Yeo, A.R.; Flowers, T.J. The effects of salinity and sodicity upon nodulation and nitrogen fixation in chickpea (Cicer arietinum). Ann. Bot. 2002, 89, 563–570. [Google Scholar] [CrossRef]
- Fan, P.; Feng, J.; Jiang, P.; Chen, X.; Bao, H.; Nie, L.; Jiang, D.; Lv, S.; Kuang, T.; Li, Y. Coordination of carbon fixation and nitrogen metabolism in Salicornia europaea under salinity: Comparative proteomic analysis on chloroplast proteins. Proteomics 2011, 11, 4346–4367. [Google Scholar] [CrossRef]
- Singer, M.A.; Lindquist, S. Thermotolerance in Saccharomyces cerevisiae: The Yin and Yang of trehalose. Trends Biotechnol. 1998, 16, 460–468. [Google Scholar] [CrossRef]
- Goddijn, O.J.M.; van Dun, K. Trehalose metabolism in plants. Trends Plant Sci. 1999, 4, 315–319. [Google Scholar] [CrossRef]
- Bledsoe, S.W.; Henry, C.; Griffiths, C.A.; Paul, M.J.; Feil, R.; Lunn, J.E.; Stitt, M.; Lagrimini, L.M. The role of Tre6P and SnRK1 in maize early kernel development and events leading to stress-induced kernel abortion. BMC Plant Biol. 2017, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Duan, X.C.; Zhang, L.; Li, P.F. Effect of Lactobacillus plantarum ATCC14917 combined with trehalose on the formation of atherosclerosis in APOE-/- mice. J. Qingdao Univ. Nat. Sci. Ed. 2024. Available online: https://link.cnki.net/urlid/37.1245.N.20241213.0845.002 (accessed on 1 April 2025).
- Li, X.; Zheng, Z.J.; Yue, T.W.; Ouyang, J. Biosynthesis of trehalose from cellobiose by recombinant Escherichia coli. Food Sci. 2019, 40, 180–186. [Google Scholar]
- Karim, S.; Aronsson, H.; Ericson, H.; Pirhonen, M.; Leyman, B.; Welin, B.; Mäntylä, E.; Palva, E.T.; Van Dijck, P.; Holmström, K.O. Improved drought tolerance without undesired side effects in transgenic plants producing trehalose. Plant Mol. Biol. 2007, 64, 371–386. [Google Scholar] [CrossRef]
- Strøm, A.R.; Kaasen, I. Trehalose metabolism in Escherichia coli: Stress protection and stress regulation of gene expression. Mol. Microbiol. 1993, 8, 205–210. [Google Scholar] [CrossRef]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 1990, 56, 2–4. [Google Scholar] [CrossRef]
- Kovach, M.E.; Phillips, R.W.; Elzer, P.H.; Roop, R.M., 2nd; Peterson, K.M. pBBR1MCS: A broad-host-range cloning vector. BioTechniques 1994, 16, 800–802. [Google Scholar]
- Quandt, J.; Hynes, M.F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 1993, 127, 15–21. [Google Scholar] [CrossRef]
- Origoni, M.; Cristoforoni, P.; Carminati, G.; Stefani, C.; Costa, S.; Sandri, M.T.; Mariani, L.; Preti, M. E6/E7 mRNA testing for 487 human papilloma virus-induced high-grade cervical intraepithelial disease (CIN2/CIN3): A promising perspective. eCancerMedicalScience 2015, 9, 533. [Google Scholar] [CrossRef]
- Miao, L.; St Clair, D.K. Regulation of superoxide dismutase genes: Implications in disease. Free Radic. Biol. Med. 2009, 47, 344–356. [Google Scholar] [CrossRef]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef]
- Magné, C.; Larher, F. High sugar content of extracts interferes with colorimetric determination of amino acids and free proline. Anal. Biochem. 1992, 200, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.; Wang, L.; Poudel, R.; Acharya, J.P.; Victores, S.; Souza, C.H.L.; Rios, E.; Wang, J. Elucidating the effects of organic vs. conventional cropping practice and rhizobia inoculation on rhizosphere microbial diversity and yield of peanut. Environ. Microbiome 2003, 18, 60. [Google Scholar] [CrossRef] [PubMed]
- ElAkhal, M.R.; Rincon, A.; El Mourabit, N.; Pueyo, J.; Barrijal, S. Phenotypic and genotypic characterizations of rhizobia isolated from root nodules of peanut (Arachis hypogaea L.) grown in Moroccan soils. J. Basic Microbiol. 2009, 49, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Mustapha, M.; Sanjay, J.K.; Elias, S.N.K.; Ahiabor, D.K.B.; Dakora, D.F. Symbiotic N2 fixation and grain yield of endangered kerstings groundnut landraces in response to soil and plant associated bradyrhizobium inoculation to promote ecological resource-use efficiency. Front. Microbiol. 2018, 9, 2105. [Google Scholar]
- Goyal, R.K.; Schmidt, M.A.; Hynes, M.F. Molecular biology in the improvement of biological nitrogen fixation by rhizobia and extending the scope to cereals. Microorganisms 2021, 9, 125. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, Q.; Song, X. Research advances on arachidonic acid production by fermentation and genetic modification of Mortierella alpina. World J. Microbiol. Biotechnol. 2021, 37, 4. [Google Scholar] [CrossRef]
- Ambrosio, R.; Ortiz-Marquez, J.C.F.; Curatti, L. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metab. Eng. 2017, 40, 59–68. [Google Scholar] [CrossRef]
- Cui, J.; Sun, T.; Li, S.; Xie, Y.; Song, X.; Wang, F.; Chen, L.; Zhang, W. Improved salt tolerance and metabolomics analysis of Synechococcus elongatus UTEX 2973 by overexpressing Mrp antiporters. Front. Bioeng. Biotechnol. 2020, 8, 500. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; Al-Harrasi, A. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Zeng, F.; Zhu, Y.; Zhang, D.; Zhao, Z.; Li, Q.; Ma, P.; Zhang, G.; Wang, Y.; Wu, S.; Guo, S.; et al. Metagenomic analysis of the soil microbial composition and salt tolerance mechanism in Yuncheng Salt Lake, Shanxi Province. Front. Microbiol. 2022, 13, 1004556. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wang, S.; Dao, Y.; Wang, J.; Wang, K. Arabidopsis thaliana trehalose-6-phosphate phosphatase gene TPPI enhances drought tolerance by regulating stomatal apertures. J. Exp. Bot. 2020, 71, 4285–4297. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Broyart, C.; Rabanal, F.A.; Jonak, C. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid. Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef] [PubMed]
- Li, H.W.; Zang, B.S.; Deng, X.W.; Wang, X.P. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018. [Google Scholar] [CrossRef]
- Yu, W.; Zhao, R.; Wang, L.; Zhang, S.; Li, R.; Sheng, J.; Shen, L. ABA signaling rather than ABA metabolism is involved in trehalose-induced drought tolerance in tomato plants. Planta 2019, 250, 643–655. [Google Scholar] [CrossRef]
- Cai, X.; Seitl, I.; Mu, W.; Zhang, T.; Stressler, T.; Fischer, L.; Jiang, B. Biotechnical production of trehalose through the trehalose synthase pathway: Current status and future prospects. Appl. Microbiol. Biotechnol. 2018, 102, 2965–2976. [Google Scholar] [CrossRef]
- Li, H.; Su, H.; Kim, S.B.; Chang, Y.K.; Hong, S.K.; Seo, Y.G.; Kim, C.J. Enhanced production of trehalose in Escherichia coli by homologous expression of otsBA in the presence of the trehalase inhibitor, validamycin A, at high osmolarity. J. Biosci. Bioeng. 2012, 113, 224–232. [Google Scholar] [CrossRef]
- Chen, X.; An, L.; Fan, X.; Ju, F.; Zhang, B.; Sun, H.; Xiao, J.; Hu, W.; Qu, T.; Guan, L.; et al. A trehalose biosynthetic enzyme doubles as an osmotic stress sensor to regulate bacterial morphogenesis. PLoS Genet. 2017, 13, e1007062. [Google Scholar] [CrossRef]
- Lee, J.; Jeong, B.; Bae, H.R.; Jang, H.A.; Kim, J.K. Trehalose biosynthesis gene otsA protects against stress in the initial infection stage of Burkholderia-Bean bug symbiosis. Microbiol. Spectr. 2023, 11, e0351022. [Google Scholar] [CrossRef]
- Chen, J.; Gou, J.Y.; Zhao, Q.; Han, Q.Q.; Li, H.P.; Yao, D.; Zhang, J.L. Effects of rhizobium of haloxylon on growth and salt tolerance of alfalfa. J. Bioresour. Bioprod. 2021, 45, 99–110. [Google Scholar]
- Kunau, W.H.; Hartig, A. Peroxisome biogenesis in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 1992, 62, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kloepper, J.W.; Ryu, C.M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; He, Y.; Zhang, Z.; Wu, H.; He, L.; Jiang, J.; Meng, W.; Huang, Z.; Xiong, F.; Liu, J.; et al. Beneficial shift of rhizosphere soil nutrients and metabolites under a sugarcane/peanut intercropping system. Front. Plant Sci. 2022, 13, 1018727. [Google Scholar] [CrossRef]


| Id of Primers | Sequence 5′→3′ |
|---|---|
| AMT1.1-FP | GTTGGCGGCAAAGGTGAAG |
| AMT1.1-RP | TAAGGCCTCTCCGATCCGTA |
| NRT1.1-FP | AGGTCTGTGGATGCTCCTA |
| NRT1.1-RP | GATGGAAATGAGAAGCAGC |
| NRT1.2-FP | AGGTTTTGTACCGTAGACT |
| NRT1.2-RP | CTTCAATCCGTCGATAGCTC |
| Castor-FP | CGCACTCGCGACGTTGA |
| Castor-RP | TCGCCCAGTAATGTGGAACTC |
| SYMRK-FP | CCTGGTGCCTCTTCTTGGTT |
| SYMRK-RP | TTCTCTTTGCAGGTTCTCCATA |
| CCaMK-FP | CCTCTTGGAAGTGATGCGGT |
| CCaMK-RP | CCGGATCTGTCCCTTCCTGA |
| LOC-FP | CAGGATTTGCCGGTGATGATG |
| LOC-RP | TCTGTTGGCCTTCGGGTTGAG |
| Salt Concentration | Strain | Net Photosynthetic Rate (μmol/m2/s) | Average Plant Height (cm) | Average Fresh Weight (g) | Average Dry Weight (g) |
|---|---|---|---|---|---|
| 0 mM | CK | 16.81 b | 13.31 b | 12.13 a | 4.52 b |
| WT | 22.39 a | 16.90 a | 17.01 ab | 6.81 a | |
| O-otsA | 23.63 a | 16.52 a | 18.10 a | 7.50 a | |
| O-otsB | 23.81 a | 16.01 a | 15.89 bc | 7.34 a | |
| ΔotsA | 24.04 a | 16.60 a | 14.34 c | 6.90 a | |
| ΔotsB | 22.41 a | 15.90 a | 14.77 c | 6.77 a | |
| 150 mM | CK | 13.66 b | 9.50 e | 7.17 c | 3.78 b |
| WT | 19.14 a | 15.60 b | 11.09 a | 5.49 ab | |
| O-otsA | 19.64 a | 17.80 a | 12.05 a | 6.03 a | |
| O-otsB | 18.88 a | 15.00 bc | 10.88 ab | 5.57 ab | |
| ΔotsA | 18.82 a | 12.00 d | 10.59 ab | 5.10 ab | |
| ΔotsB | 18.59 a | 13.70 c | 9.29 b | 4.89 ab | |
| 300 mM | CK | 8.91 c | 6.15 d | 4.59 c | 1.53 b |
| WT | 11.36 b | 10.00 bc | 6.79 b | 3.24 ab | |
| O-otsA | 14.50 a | 11.87 a | 8.37 a | 4.57 a | |
| O-otsB | 11.32 b | 10.98 ab | 6.18 bc | 3.53 a | |
| ΔotsA | 10.93 b | 9.12 c | 5.36 bc | 2.91 ab | |
| ΔotsB | 11.87 b | 8.98 c | 5.17 bc | 2.98 ab |
| Salt Concentration | Strain | Nitrogenase Activity (U/g) | SOD Activity (U/g) | POD Activity (U/g) | MDA Content (mol/g) | Proline Content(μg/g) |
|---|---|---|---|---|---|---|
| 0 mM | CK | 15.79 c | 72.03 a | 4.82 b | 2.70 a | 69.80 a |
| WT | 25.52 a | 78.92 a | 5.01 b | 2.55 a | 64.50 bc | |
| O-otsA | 25.03 a | 68.13 a | 7.94 a | 2.47 a | 65.10 b | |
| O-otsB | 24.90 a | 74.78 a | 5.63 b | 2.44 a | 62.80 c | |
| ΔotsA | 23.00 b | 75.66 a | 5.68 b | 2.52 a | 64.10 bc | |
| ΔotsB | 22.61 b | 70.24 a | 7.69 a | 2.51 a | 63.20 c | |
| 150 mM | CK | 15.78 b | 148.12 b | 10.20 c | 10.00 a | 80.30 a |
| WT | 15.69 b | 151.98 b | 14.80 b | 7.30 cd | 75.80 b | |
| O-otsA | 18.61 a | 161.67 a | 17.69 a | 5.63 e | 69.80 d | |
| O-otsB | 16.54 b | 138.44 c | 15.65 b | 6.11 de | 72.00 c | |
| ΔotsA | 16.02 b | 125.39 d | 14.34 b | 9.22 ab | 72.50 c | |
| ΔotsB | 16.00 b | 134.61 cd | 14.90 b | 8.20 bc | 71.90 c | |
| 300 mM | CK | 7.52 a | 180.10 d | 15.21 e | 25.21 a | 104.80 a |
| WT | 14.68 a | 266.39 b | 23.08 b | 15.18 c | 95.70 a | |
| O-otsA | 16.28 a | 281.69 a | 28.37 a | 9.27 e | 84.30 b | |
| O-otsB | 14.73 a | 250.22 c | 20.32 c | 11.80 d | 94.00 a | |
| ΔotsA | 11.83 b | 178.01 d | 19.71 c | 20.01 b | 97.50 a | |
| ΔotsB | 11.90 b | 186.65 d | 17.20 d | 19.68 b | 100.90 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Wang, D.; Tong, R.; Ye, S.; Zhao, Y.; Wu, J.; Gan, Y. Engineered Rhizobia with Trehalose-Producing Genes Enhance Peanut Growth Under Salinity Stress. Agronomy 2025, 15, 974. https://doi.org/10.3390/agronomy15040974
Liu J, Wang D, Tong R, Ye S, Zhao Y, Wu J, Gan Y. Engineered Rhizobia with Trehalose-Producing Genes Enhance Peanut Growth Under Salinity Stress. Agronomy. 2025; 15(4):974. https://doi.org/10.3390/agronomy15040974
Chicago/Turabian StyleLiu, Jialin, Dong Wang, Ruiqi Tong, Shengyue Ye, Yanhao Zhao, Jiangwen Wu, and Yi Gan. 2025. "Engineered Rhizobia with Trehalose-Producing Genes Enhance Peanut Growth Under Salinity Stress" Agronomy 15, no. 4: 974. https://doi.org/10.3390/agronomy15040974
APA StyleLiu, J., Wang, D., Tong, R., Ye, S., Zhao, Y., Wu, J., & Gan, Y. (2025). Engineered Rhizobia with Trehalose-Producing Genes Enhance Peanut Growth Under Salinity Stress. Agronomy, 15(4), 974. https://doi.org/10.3390/agronomy15040974





