Dysregulation of Immature Sertoli Cell Functions by Exposure to Acetaminophen and Genistein in Rodent Cell Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. TM4 Mouse Sertoli Cell Line Culture
2.3. Primary Immature Rat Sertoli Cell Isolation
2.4. Sertoli Cell Treatments
2.5. Cell Viability/Proliferation with MTT Assay
2.6. Cell Proliferation with EdU Assay
2.7. Gene Expression Measured by qRT-PCR
2.8. PGD2 & PGE2 ELISA
2.9. Immunofluorescent (IF) Staining
2.10. Whole Transcriptome Sequencing (Total RNA-Seq)
2.11. Statistical Analysis
3. Results
3.1. Sertoli Cell Viability and Proliferation Are Dysregulated by Exposure to APAP and GEN
3.2. Eicosanoid Pathway Is Dysregulated in Immature Sertoli Cells Exposed to APAP, GEN, and Their Mixtures
3.3. Estrogen Receptors Dysregulation in Immature Sertoli Cells by APAP and GEN
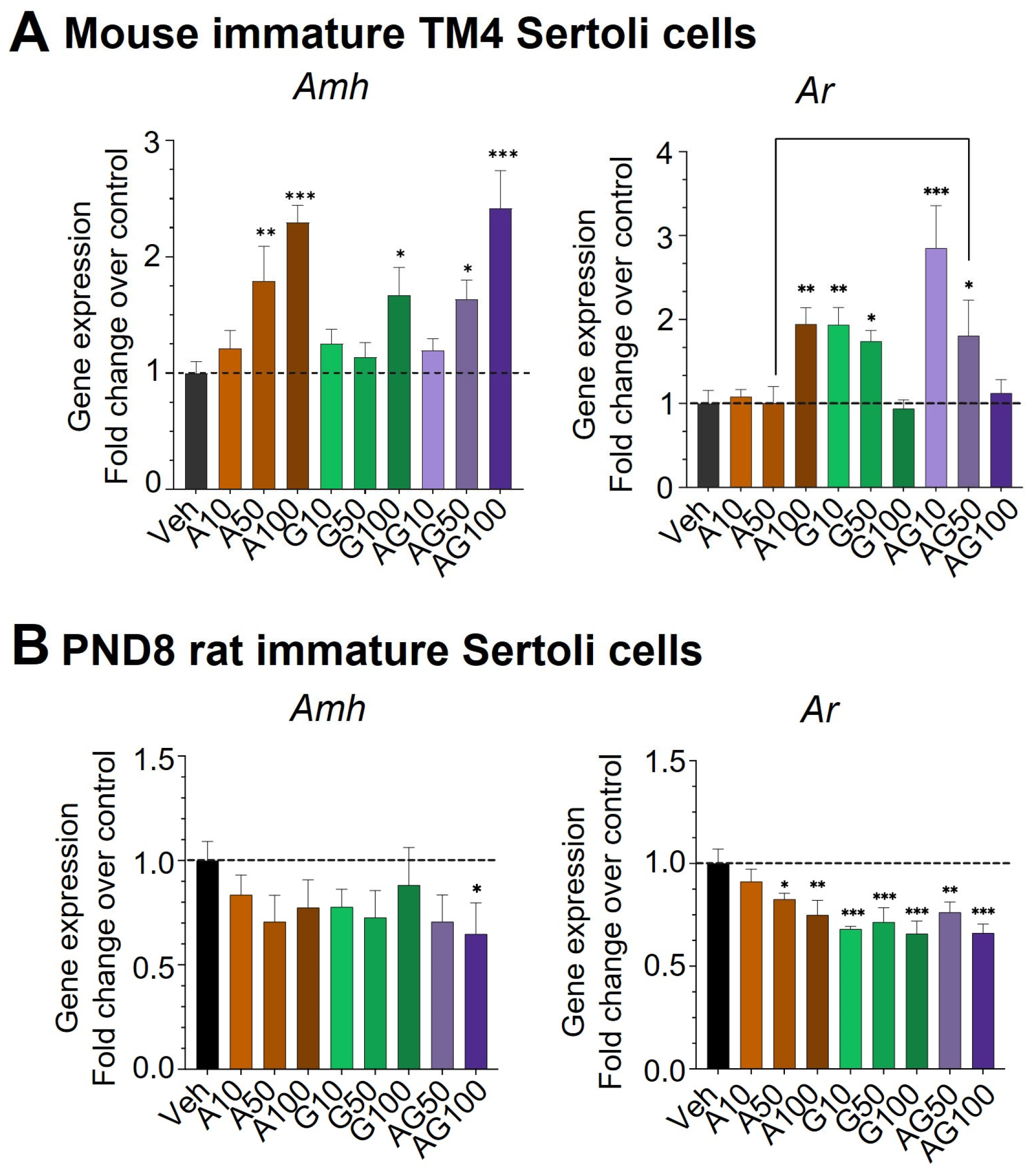
3.4. Dysregulation of Immature Sertoli Cell Differentiation by APAP and GEN
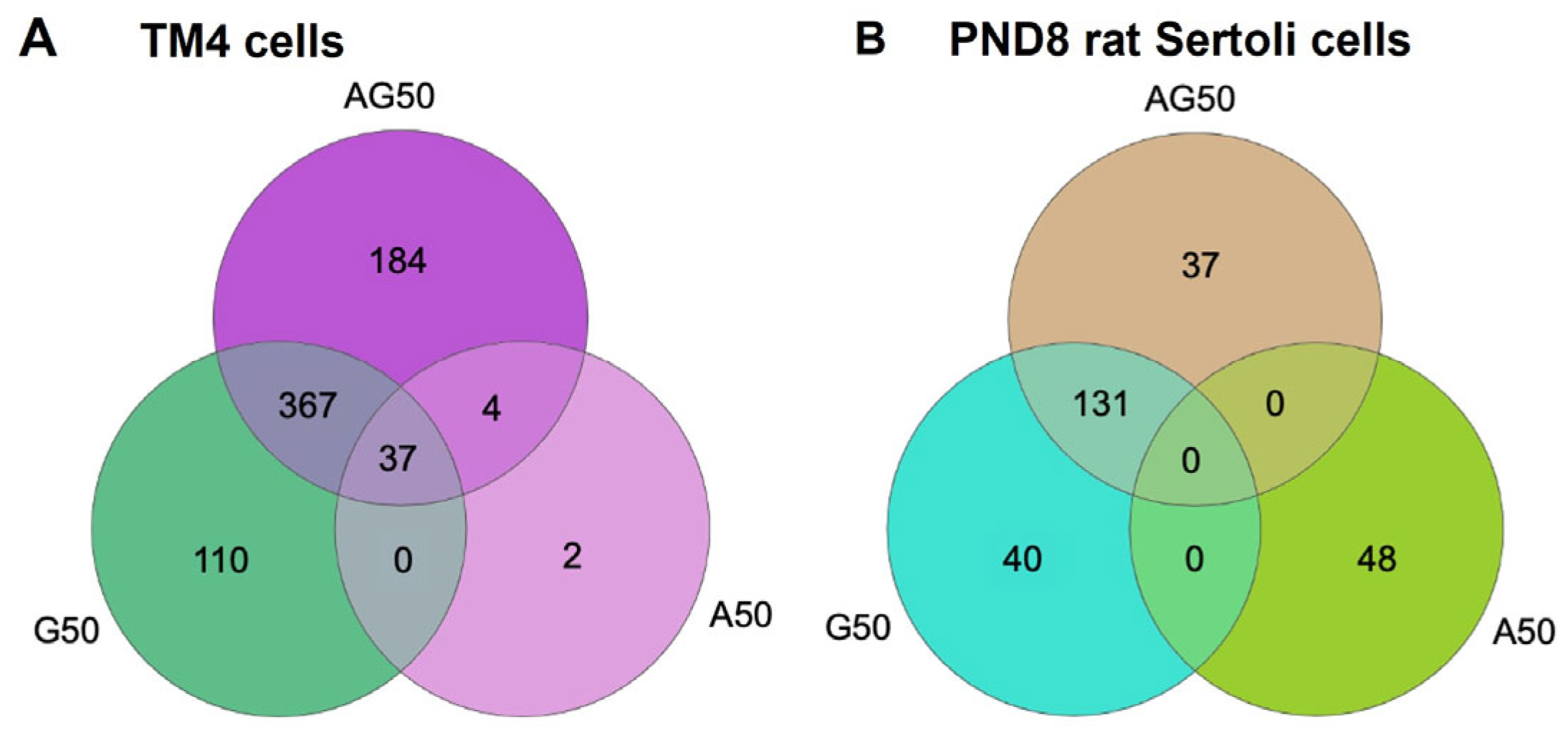
3.5. APAP and GEN Disrupt TM4 Cells and PND8 Sertoli Cell Transcriptomes
4. Discussion
4.1. Dysregulation of Sertoli Cell Development by APAP and GEN
4.2. Similarities and Differences between TM4 Cells and PND8 Immature Sertoli Cells
4.3. Is There a Link between APAP and GEN Effects on Sox9 and Eicosanoid Pathway?
4.4. Mechanisms Dysregulated in Immortalized and Primary Immature Sertoli Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Griswold, M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol. Reprod. 2018, 99, 87–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- França, L.R.; Hess, R.A.; Dufour, J.M.; Hofmann, M.C.; Griswold, M.D. The Sertoli cell: One hundred fifty years of beauty and plasticity. Andrology 2016, 4, 189–212. [Google Scholar] [CrossRef] [Green Version]
- Corpuz-Hilsabeck, M.; Culty, M. Impact of endocrine disrupting chemicals and pharmaceuticals on Sertoli cell development and functions. Front. Endocrinol. 2023, 14, 1095894. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Arguelles, D.; Campioli, E.; Culty, M.; Zirkin, B.; Papadopoulos, V. Fetal origin of endocrine dysfunction in the adult: The phthalate model. J. Steroid Biochem. Mol. Biol. 2013, 137, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Skakkebaek, N.E.; Rajpert-De Meyts, E.; Buck Louis, G.M.; Toppari, J.; Andersson, A.-M.; Eisenberg, M.L.; Jensen, T.K.; Jørgensen, N.; Swan, S.H.; Sapra, K.J. Male reproductive disorders and fertility trends: Influences of environment and genetic susceptibility. Physiol. Rev. 2016, 96, 55–97. [Google Scholar] [CrossRef] [Green Version]
- Walker, C.; Garza, S.; Papadopoulos, V.; Culty, M. Impact of endocrine-disrupting chemicals on steroidogenesis and consequences on testicular function. Mol. Cell. Endocrinol. 2021, 527, 111215. [Google Scholar] [CrossRef]
- Setchell, K.D.; Zimmer-Nechemias, L.; Cai, J.; Heubi, J.E. Exposure of infants to phyto-oestrogens from soy-based infant formula. Lancet 1997, 350, 23–27. [Google Scholar] [CrossRef]
- Suen, A.A.; Kenan, A.C.; Williams, C.J. Developmental exposure to phytoestrogens found in soy: New findings and clinical implications. Biochem. Pharmacol. 2022, 195, 114848. [Google Scholar] [CrossRef]
- Thuillier, R.; Manku, G.; Wang, Y.; Culty, M. Changes in MAPK pathway in neonatal and adult testis following fetal estrogen exposure and effects on rat testicular cells. Microsc. Res. Tech. 2009, 72, 773–786. [Google Scholar] [CrossRef]
- Walker, C.; Ghazisaeidi, S.; Collet, B.; Boisvert, A.; Culty, M. In utero exposure to low doses of genistein and di-(2-ethylhexyl) phthalate (DEHP) alters innate immune cells in neonatal and adult rat testes. Andrology 2020, 8, 943–964. [Google Scholar] [CrossRef]
- Jones, S.; Boisvert, A.; Duong, T.B.; Francois, S.; Thrane, P.; Culty, M. Disruption of rat testis development following combined in utero exposure to the phytoestrogen genistein and antiandrogenic plasticizer di-(2-ethylhexyl) phthalate. Biol. Reprod. 2014, 91, 64. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Boisvert, A.; Francois, S.; Zhang, L.; Culty, M. In utero exposure to di-(2-ethylhexyl) phthalate induces testicular effects in neonatal rats that are antagonized by genistein cotreatment. Biol. Reprod. 2015, 93, 92. [Google Scholar] [CrossRef] [PubMed]
- Tran-Guzman, A.; Moradian, R.; Cui, H.; Culty, M. In vitro impact of genistein and mono (2-ethylhexyl) phthalate (MEHP) on the eicosanoid pathway in spermatogonial stem cells. Reprod. Toxicol. 2022, 107, 150–165. [Google Scholar] [CrossRef]
- Tran-Guzman, A.; Culty, M. Eicosanoid Biosynthesis in Male Reproductive Development: Effects of Perinatal Exposure to NSAIDs and Analgesic Drugs. Front. Toxicol. 2022, 4, 842565. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, D.M.; Mazaud-Guittot, S.; Gaudriault, P.; Lesné, L.; Serrano, T.; Main, K.M.; Jégou, B. Analgesic use—Prevalence, biomonitoring and endocrine and reproductive effects. Nat. Rev. Endocrinol. 2016, 12, 381–393. [Google Scholar] [CrossRef]
- Zafeiri, A.; Mitchell, R.T.; Hay, D.C.; Fowler, P.A. Over-the-counter analgesics during pregnancy: A comprehensive review of global prevalence and offspring safety. Hum. Reprod. Update 2021, 27, 67–95. [Google Scholar]
- Ben Maamar, M.; Lesné, L.; Hennig, K.; Desdoits-Lethimonier, C.; Kilcoyne, K.R.; Coiffec, I.; Rolland, A.D.; Chevrier, C.; Kristensen, D.M.; Lavoué, V.; et al. Ibuprofen results in alterations of human fetal testis development. Sci. Rep. 2017, 7, 44184. [Google Scholar] [CrossRef] [Green Version]
- Rossitto, M.; Marchive, C.; Pruvost, A.; Sellem, E.; Ghettas, A.; Badiou, S.; Sutra, T.; Poulat, F.; Philibert, P.; Boizet-Bonhoure, B. Intergenerational effects on mouse sperm quality after in utero exposure to acetaminophen and ibuprofen. FASEB J. 2019, 33, 339–357. [Google Scholar] [CrossRef]
- Axelstad, M.; Hass, U.; Scholze, M.; Christiansen, S.; Kortenkamp, A.; Boberg, J. EDC IMPACT: Reduced sperm counts in rats exposed to human relevant mixtures of endocrine disrupters. Endocr. Connect. 2018, 7, 139–148. [Google Scholar] [CrossRef]
- Manku, G.; Mazer, M.; Culty, M. Neonatal testicular gonocytes isolation and processing for immunocytochemical analysis. In Germline Development; Springer: Berlin/Heidelberg, Germany, 2012; pp. 17–29. [Google Scholar]
- Manku, G.; Wing, S.S.; Culty, M. Expression of the ubiquitin proteasome system in neonatal rat gonocytes and spermatogonia: Role in gonocyte differentiation. Biol. Reprod. 2012, 87, 44. [Google Scholar] [CrossRef]
- Manku, G.; Culty, M. Dynamic changes in the expression of apoptosis-related genes in differentiating gonocytes and in seminomas. Asian J. Androl. 2015, 17, 403–414. [Google Scholar] [CrossRef]
- Brown, R.D.; Wilson, J.T.; Kearns, G.L.; Eichler, V.F.; Johnson, V.A.; Bertrand, K.M. Single-dose pharmacokinetics of ibuprofen and acetaminophen in febrile children. J. Clin. Pharmacol. 1992, 32, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Rozman, K.K.; Bhatia, J.; Calafat, A.M.; Chambers, C.; Culty, M.; Etzel, R.A.; Flaws, J.A.; Hansen, D.K.; Hoyer, P.B.; Jeffery, E.H.; et al. NTP-CERHR expert panel report on the reproductive and developmental toxicity of genistein. Birth Defects Res. B Dev. Reprod. Toxicol. 2006, 77, 485–638. [Google Scholar] [CrossRef] [PubMed]
- Manku, G.; Papadopoulos, P.; Boisvert, A.; Culty, M. Cyclooxygenase 2 (COX2) expression and prostaglandin synthesis in neonatal rat testicular germ cells: Effects of acetaminophen and ibuprofen. Andrology 2020, 8, 691–705. [Google Scholar] [CrossRef]
- Jefferson, W.N.; Williams, C.J. Circulating levels of genistein in the neonate, apart from dose and route, predict future adverse female reproductive outcomes. Reprod. Toxicol. 2011, 31, 272–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, O.; Desdoits-Lethimonier, C.; Lesne, L.; Legrand, A.; Guille, F.; Bensalah, K.; Dejucq-Rainsford, N.; Jegou, B. Paracetamol, aspirin and indomethacin display endocrine disrupting properties in the adult human testis in vitro. Hum. Reprod. 2013, 28, 1890–1898. [Google Scholar] [CrossRef] [Green Version]
- Swami, S.; Krishnan, A.V.; Moreno, J.; Bhattacharyya, R.S.; Gardner, C.; Brooks, J.D.; Peehl, D.M.; Feldman, D. Inhibition of prostaglandin synthesis and actions by genistein in human prostate cancer cells and by soy isoflavones in prostate cancer patients. Int. J. Cancer 2009, 124, 2050–2059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.R.; Hao, X.X.; Zhang, Y.; Deng, S.L.; Wang, Z.P.; Wang, Y.Q.; Wang, X.X.; Liu, Y.X. Androgen receptor in Sertoli cells regulates DNA double-strand break repair and chromosomal synapsis of spermatocytes partially through intercellular EGF-EGFR signaling. Oncotarget 2016, 7, 18722–18735. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.Q.; Wei, B.H.; Hao, S.L.; Yang, W.X. The PI3K/AKT signaling pathway: How does it regulate development of Sertoli cells and spermatogenic cells? Histol. Histopathol. 2022, 37, 621–636. [Google Scholar] [CrossRef]
- Ma, Y.; Zhou, Y.; Zou, S.S.; Sun, Y.; Chen, X.F. Exosomes released from Sertoli cells contribute to the survival of Leydig cells through CCL20 in rats. Mol. Hum. Reprod. 2022, 28, gaac002. [Google Scholar] [CrossRef]
- Shahidul Makki, M.; Cristy Ruteshouser, E.; Huff, V. Ubiquitin specific protease 18 (Usp18) is a WT1 transcriptional target. Exp. Cell Res. 2013, 319, 612–622. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.N.; Li, Z.S.; Ren, Y.; Jiang, T.; Wang, Y.Q.; Chen, M.; Zhang, J.; Hao, J.X.; Wang, Y.B.; Sha, R.N.; et al. The Wilms tumor gene, Wt1, is critical for mouse spermatogenesis via regulation of sertoli cell polarity and is associated with non-obstructive azoospermia in humans. PLoS Genet. 2013, 9, e1003645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honke, N.; Shaabani, N.; Zhang, D.E.; Hardt, C.; Lang, K.S. Multiple functions of USP18. Cell Death Dis. 2016, 7, e2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautam, M.; Bhattacharya, I.; Rai, U.; Majumdar, S.S. Hormone induced differential transcriptome analysis of Sertoli cells during postnatal maturation of rat testes. PLoS ONE 2018, 13, e0191201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Zhu, J.; Li, X.; Li, S.; Lan, Z.; Ko, J.; Lei, Z. Expression of genomic functional estrogen receptor 1 in mouse sertoli cells. Reprod. Sci. 2014, 21, 1411–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akpovi, C.D.; Murphy, B.D.; Erickson, R.P.; Pelletier, R.M. Dysregulation of testicular cholesterol metabolism following spontaneous mutation of the niemann-pick c1 gene in mice. Biol. Reprod. 2014, 91, 42. [Google Scholar] [CrossRef]
- Singh, N.P.; De Kumar, B.; Paulson, A.; Parrish, M.E.; Scott, C.; Zhang, Y.; Florens, L.; Krumlauf, R. Genome-Wide Binding Analyses of HOXB1 Revealed a Novel DNA Binding Motif Associated with Gene Repression. J. Dev. Biol. 2021, 9, 6. [Google Scholar] [CrossRef]
- Zhang, F.L.; Feng, Y.Q.; Wang, J.Y.; Zhu, K.X.; Wang, L.; Yan, J.M.; Li, X.X.; Wang, J.J.; Ge, W.; De Felici, M.; et al. Single cell epigenomic and transcriptomic analysis uncovers potential transcription factors regulating mitotic/meiotic switch. Cell Death Dis. 2023, 14, 134. [Google Scholar] [CrossRef]
- Altena, R.; Fehrmann, R.S.; Boer, H.; de Vries, E.G.; Meijer, C.; Gietema, J.A. Growth differentiation factor 15 (GDF-15) plasma levels increase during bleomycin- and cisplatin-based treatment of testicular cancer patients and relate to endothelial damage. PLoS ONE 2015, 10, e0115372. [Google Scholar] [CrossRef]
- Sridharan, S.; Simon, L.; Meling, D.D.; Cyr, D.G.; Gutstein, D.E.; Fishman, G.I.; Guillou, F.; Cooke, P.S. Proliferation of adult sertoli cells following conditional knockout of the Gap junctional protein GJA1 (connexin 43) in mice. Biol. Reprod. 2007, 76, 804–812. [Google Scholar] [CrossRef] [Green Version]
- Bogdan, M.; Meca, A.D.; Turcu-Stiolica, A.; Oancea, C.N.; Kostici, R.; Surlin, M.V.; Florescu, C. Insights into the Relationship between Pentraxin-3 and Cancer. Int. J. Mol. Sci. 2022, 23, 5302. [Google Scholar] [CrossRef] [PubMed]
- Doni, A.; Paffoni, A.; Nebuloni, M.; Ragni, G.; Pasqualini, F.; Valentino, S.; Bonetti, S.; Mantovani, A.; Somigliana, E.; Garlanda, C. The long pentraxin 3 is a soluble and cell-associated component of the human semen. Int. J. Androl. 2009, 32, 255–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, D.M.; Hass, U.; Lesné, L.; Lottrup, G.; Jacobsen, P.R.; Desdoits-Lethimonier, C.; Boberg, J.; Petersen, J.H.; Toppari, J.; Jensen, T.K. Intrauterine exposure to mild analgesics is a risk factor for development of male reproductive disorders in human and rat. Hum. Reprod. 2011, 26, 235–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valeri, C.; Lovaisa, M.M.; Racine, C.; Edelsztein, N.Y.; Riggio, M.; Giulianelli, S.; Venara, M.; Bedecarrás, P.; Ballerini, M.G.; di Clemente, N.; et al. Molecular mechanisms underlying AMH elevation in hyperoestrogenic states in males. Sci. Rep. 2020, 10, 15062. [Google Scholar] [CrossRef]
- Sharpe, R.M.; McKinnell, C.; Kivlin, C.; Fisher, J.S. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003, 125, 769–784. [Google Scholar] [CrossRef]
- Behringer, R.R.; Finegold, M.J.; Cate, R.L. Müllerian-inhibiting substance function during mammalian sexual development. Cell 1994, 79, 415–425. [Google Scholar] [CrossRef]
- Knebelmann, B.; Boussin, L.; Guerrier, D.; Legeai, L.; Kahn, A.; Josso, N.; Picard, J. Anti-Müllerian hormone Bruxelles: A nonsense mutation associated with the persistent Müllerian duct syndrome. Proc. Natl. Acad. Sci. USA 1991, 88, 3767–3771. [Google Scholar] [CrossRef]
- Rey, R.A.; Musse, M.; Venara, M.; Chemes, H.E. Ontogeny of the androgen receptor expression in the fetal and postnatal testis: Its relevance on Sertoli cell maturation and the onset of adult spermatogenesis. Microsc. Res. Tech. 2009, 72, 787–795. [Google Scholar] [CrossRef]
- Rotgers, E.; Jørgensen, A.; Yao, H.H.-C. At the crossroads of fate—Somatic cell lineage specification in the fetal gonad. Endocr. Rev. 2018, 39, 739–759. [Google Scholar] [CrossRef]
- Fröjdman, K.; Harley, V.R.; Pelliniemi, L.J. Sox9 protein in rat sertoli cells is age and stage dependent. Histochem. Cell Biol. 2000, 113, 31–36. [Google Scholar] [CrossRef]
- Mather, J.P. Establishment and characterization of two distinct mouse testicular epithelial cell lines. Biol. Reprod. 1980, 23, 243–252. [Google Scholar] [PubMed]
- Wang, Y.-M.; Sullivan, P.M.; Petrusz, P.; Yarbrough, W.; Joseph, D.R. The androgen-binding protein gene is expressed in CD1 mouse testis. Mol. Cell. Endocrinol. 1989, 63, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Lucas, T.F.G.; Lazari, M.F.M.; Porto, C.S. Differential role of the estrogen receptors ESR1 and ESR2 on the regulation of proteins involved with proliferation and differentiation of Sertoli cells from 15-day-old rats. Mol. Cell. Endocrinol. 2014, 382, 84–96. [Google Scholar] [CrossRef]
- Savard, C.; Gawhary, S.; Boyer, A.; Chorfi, Y. Assessment of Zearalenone-Induced Cell Survival and of Global Gene Regulation in Mouse TM4 Sertoli Cells. Toxins 2022, 14, 98. [Google Scholar] [CrossRef]
- Boizet-Bonhoure, B.; Déjardin, S.; Rossitto, M.; Poulat, F.; Philibert, P. Using experimental models to decipher the effects of acetaminophen and NSAIDs on reproductive development and health. Front. Toxicol. 2022, 4, 167. [Google Scholar] [CrossRef]
- Moniot, B.; Declosmenil, F.; Barrionuevo, F.; Scherer, G.; Aritake, K.; Malki, S.; Marzi, L.; Cohen-Solal, A.; Georg, I.; Klattig, J. The PGD2 pathway, independently of FGF9, amplifies SOX9 activity in Sertoli cells during male sexual differentiation. Development 2009, 136, 1813–1821. [Google Scholar] [CrossRef] [Green Version]
- Kugathas, S.; Audouze, K.; Ermler, S.; Orton, F.; Rosivatz, E.; Scholze, M.; Kortenkamp, A. Effects of common pesticides on prostaglandin D2 (PGD2) inhibition in SC5 mouse Sertoli cells, evidence of binding at the COX-2 active site, and implications for endocrine disruption. Environ. Health Perspect. 2016, 124, 452–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, D.; Hiramatsu, R.; Mizusaki, H.; Widjaja, L.; Combes, A.N.; Kanai, Y.; Koopman, P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J. Biol. Chem. 2007, 282, 10553–10560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, D.; Martinson, F.; Bradford, S.; Wilson, M.J.; Combes, A.N.; Beverdam, A.; Bowles, J.; Mizusaki, H.; Koopman, P. Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev. Biol. 2005, 287, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Rossitto, M.; Ujjan, S.; Poulat, F.; Boizet-Bonhoure, B. Multiple roles of the prostaglandin D2 signaling pathway in reproduction. Reproduction 2015, 149, R49–R58. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, D.M.; Skalkam, M.L.; Audouze, K.; Lesné, L.; Desdoits-Lethimonier, C.; Frederiksen, H.; Brunak, S.; Skakkebæk, N.E.; Jégou, B.; Hansen, J.B. Many putative endocrine disruptors inhibit prostaglandin synthesis. Environ. Health Perspect. 2011, 119, 534–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, S.P.; Windschüttl, S.; Matzkin, M.E.; Rey-Ares, V.; Terradas, C.; Ponzio, R.; Puigdomenech, E.; Levalle, O.; Calandra, R.S.; Mayerhofer, A. Reactive oxygen species (ROS) production triggered by prostaglandin D2 (PGD2) regulates lactate dehydrogenase (LDH) expression/activity in TM4 Sertoli cells. Mol. Cell. Endocrinol. 2016, 434, 154–165. [Google Scholar] [CrossRef] [PubMed]
- van den Driesche, S.; Macdonald, J.; Anderson, R.A.; Johnston, Z.C.; Chetty, T.; Smith, L.B.; McKinnell, C.; Dean, A.; Homer, N.Z.; Jorgensen, A. Prolonged exposure to acetaminophen reduces testosterone production by the human fetal testis in a xenograft model. Sci. Transl. Med. 2015, 7, ra280–ra288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zirkin, B.R.; Papadopoulos, V. Leydig cells: Formation, function, and regulation. Biol. Reprod. 2018, 99, 101–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zirkin, B.R.; Papadopoulos, V. Leydig cell androgen synthesis. In Encyclopedia of Reproduction; Elsevier: Baltimore, MD, USA, 2018; pp. 215–221. [Google Scholar]
- Meroni, S.B.; Galardo, M.N.; Rindone, G.; Gorga, A.; Riera, M.F.; Cigorraga, S.B. Molecular mechanisms and signaling pathways involved in sertoli cell proliferation. Front. Endocrinol. 2019, 10, 224. [Google Scholar] [CrossRef] [Green Version]
- Ni, F.-D.; Hao, S.-L.; Yang, W.-X. Multiple signaling pathways in Sertoli cells: Recent findings in spermatogenesis. Cell Death Dis. 2019, 10, 541. [Google Scholar] [CrossRef] [Green Version]
- Lehraiki, A.; Chamaillard, C.; Krust, A.; Habert, R.; Levacher, C. Genistein impairs early testosterone production in fetal mouse testis via estrogen receptor alpha. Toxicol. In Vitr. 2011, 25, 1542–1547. [Google Scholar] [CrossRef]
- Lucas, T.F.G.; Royer, C.; Siu, E.R.; Lazari, M.F.M.; Porto, C.S. Expression and Signaling of G Protein-Coupled Estrogen Receptor 1 (GPER) in Rat Sertoli Cells1. Biol. Reprod. 2010, 83, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Chimento, A.; De Luca, A.; Nocito, M.C.; Avena, P.; La Padula, D.; Zavaglia, L.; Pezzi, V. Role of GPER-Mediated Signaling in Testicular Functions and Tumorigenesis. Cells 2020, 9, 2115. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Karin, M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010, 21, 11–19. [Google Scholar] [CrossRef] [Green Version]







| Gene | Forward Primer | Reverse Primer | Product Size (bp) |
|---|---|---|---|
| Rat | |||
| Gapdh | CCATTCTTCCACCTTTGATGCT | TGTTGCTGTAGCCATATTCATTGT | 98 |
| Sox9 | TGCTGAACGAGAGCGAGAAG | ATGTGAGTCTGTTCGGTGGC | 160 |
| Amh | GTGGGTGGCAGCAGCACTAGG | CGGGCTGTTTGGCTCTGATTCCCG | 69 |
| Ar | CGGTCGAGTTGACATTAGTGAAGGACC | ATTCCTGGATGGGACTGATGGT | 66 |
| Cox1 | AGGTGTACCCACCTTCCGTA | GGTTTCCCCTATAAGGATGAGGC | 242 |
| Cox2 | ACGTGTTGACGTCCAGATCA | CTTGGGGATCCGGGATGAAC | 234 |
| Ptgds | ATTCAAGCTGGTTCCGGGAG | CAGGAACGCGTACTCATCGT | 242 |
| Esr1 | GCCACTCGATCATTCGAGCA | CCTGCTGGTTCAAAAGCGTC | 107 |
| Ppar-α | TGCGGACTACCAGTACTTAGGG | GCTGGAGAGAGGGTGTCTGT | 72 |
| Mouse | |||
| Rps29 | TGAAGGCAAGATGGGTCAC | GCACATGTTCAGCCCGTATT | 127 |
| Cox1 | CCTCTTTCCAGGAGCTCACA | TCGATGTCACCGTACAGCTC | 70 |
| Cox2 | CAGGACTCTGCTCACGAAGG | ATCCAGTCCGGGTACAGTCA | 231 |
| Ptgds | GGCTCCTGGACACTACACCT | CTGGGTTCTGCTGTAGAGGGT | 160 |
| Ptges | GGCTCCTCCAAAGACGGAAA | TGGCACAGCATGGGTCTTAG | 226 |
| Ptgdr2 | CACGTGTCGGTGCTGTTG | GATGAGTCCGTTTTCCACCA | 63 |
| Amh | GGGGAGACTGGAGAACAGC | AGAGCTCGGGCTCCCATA | 67 |
| Ar | ACCAGATGGCGGTCATTCAG | TGTGCATGCGGTACTCATTG | 135 |
| Sox9 | TCGGACACGGAGAACACC | GCACACGGGGAACTTATCTT | 96 |
| Esr1 | TCTCCTCAAACACATCCCGTG | GGCGAGTTACAGACTGGCTC | 96 |
| Gper | CCTGGACGAGCAGTATTACGATATC | TGCTGTACATGTTGATCTG | 77 |
| Err-α | CGCTGTCAGCTGGAGGAA | ACCTTGGGCTCCGGCA | 199 |
| Err-β | TCTTCCCAGCTCCCACAGTA | CCCCATGCAAGCTTCGTAGT | 106 |
| TM4 APAP (50 µM) | |||||
|---|---|---|---|---|---|
| Downregulated | Upregulated | ||||
| Gene | p-value | Fold change | Gene | p-value | Fold change |
| Ereg | 2.36 × 10−11 | −4.90 | Dag1 | 1.13 × 10−4 | 2.09 |
| Hbegf | 8.14 × 10−10 | −3.37 | Rpa1 | 1.29 × 10−4 | 2.11 |
| Lif | 1.39 × 10−7 | −2.96 | Ubc | 2.08 × 10−7 | 2.20 |
| Cth | 1.14 × 10−11 | −2.84 | Mki67 | 1.08 × 10−14 | 2.29 |
| H1f1 | 4.41 × 10−18 | −2.55 | Kif20a | 4.78 × 10−6 | 2.32 |
| H1f0 | 3.34 × 10−21 | −2.54 | Ckap2l | 2.18 × 10−6 | 2.43 |
| Il1rl1 | 5.63 × 10−8 | −2.51 | Acat2 | 1.76 × 10−4 | 3.28 |
| Mtmr10 | 4.93 × 10−5 | −2.51 | Mettl26 | 1.93 × 10−5 | 8.46 |
| H4f16 | 4.61 × 10−10 | −2.39 | |||
| Prkg2 | 2.33 × 10−10 | −2.35 | |||
| PND8 rat Sertoli APAP (50 µM) | |||||
| Downregulated | Upregulated | ||||
| Gene | p-value | Fold change | Gene | p-value | Fold change |
| Hoxb1 | 4.11 × 10−2 | −13.14 | Ppp1r3d | 3.85 × 10−2 | 12.17 |
| Senp18 | 3.59 × 10−2 | −11.41 | |||
| Hjv | 3.63 × 10−2 | −10.06 | |||
| Sypl2 | 4.26 × 10−2 | −7.85 | |||
| Hs3st2 | 3.37 × 10−2 | −7.70 | |||
| Mal | 4.63 × 10−2 | −7.57 | |||
| Nmur2 | 4.35 × 10−2 | −7.48 | |||
| Esrrg | 4.85 × 10−2 | −5.88 | |||
| Synb | 4.50 × 10−2 | −5.66 | |||
| Akain1 | 4.52 × 10−2 | −5.23 | |||
| TM4 GEN (50 µM) | |||||
| Downregulated | Upregulated | ||||
| Gene | p-value | Fold change | Gene | p-value | Fold change |
| Fosl1 | 5.86 × 10−28 | −15.38 | Akr1c14 | 1.29 × 10−18 | 3.57 |
| Ereg | 6.71 × 10−35 | −14.81 | Mboat2 | 8.25 × 10−8 | 3.85 |
| Tgtp1 | 1.06 × 10−17 | −13.57 | Gja1 | 3.97 × 10−38 | 3.89 |
| Gbp3 | 8.73 × 10−60 | −13.20 | Dhcr24 | 4.89 × 10−5 | 4.01 |
| Tgtp2 | 1.37 × 10−19 | −13.16 | Nbl1 | 1.07 × 10−5 | 4.17 |
| Hbegf | 1.64 × 10−42 | −12.08 | Scd2 | 1.10 × 10−72 | 4.88 |
| Ccl20 | 3.98 × 10−5 | −10.50 | Lars2 | 9.28 × 10−5 | 5.29 |
| Ifit3 | 6.09 × 10−85 | −9.99 | Acat2 | 3.36 × 10−9 | 5.35 |
| Iigp1 | 2.80 × 10−36 | −8.57 | Mettl26 | 3.78 × 10−6 | 7.98 |
| Usp18 | 1.67 × 10−54 | −8.45 | Mettl7a1 | 2.43 × 10−11 | 8.48 |
| PND8 rat Sertoli GEN (50 µM) | |||||
| Downregulated | Upregulated | ||||
| Gene | p-value | Fold change | Gene | p-value | Fold change |
| Depdc1b | 7.54 × 10−4 | −10.00 | Ptx3 | 1.17 × 10−43 | 2.62 |
| H2ac10 | 1.39 × 10−37 | −7.87 | Mdm2 | 5.71 × 10−67 | 2.93 |
| E2f8 | 4.22 × 10−8 | −5.69 | Bbc3 | 2.14 × 10−21 | 3.02 |
| Sgo2 | 2.73 × 10−6 | −4.99 | Csf2rb | 6.13 × 10−5 | 3.17 |
| Cenpq | 1.56 × 10−3 | −4.96 | Cd80 | 4.64 × 10−9 | 3.36 |
| Pbk | 1.09 × 10−4 | −4.94 | Abcb1b | 2.99 × 10−101 | 3.66 |
| Cdca3 | 8.23 × 10−8 | −4.71 | Star | 2.53 × 10−188 | 4.36 |
| Depdc1 | 1.17 × 10−4 | −4.71 | Cdkn1a | 1.35 × 10−155 | 4.86 |
| Hist1h2bc | 7.92 × 10−5 | −4.67 | Il6 | 3.29 × 10−8 | 5.18 |
| Uhrf1 | 1.10 × 10−21 | −4.44 | Gdf15 | 2.02 × 10−22 | 9.17 |
| TM4 APAP + GEN (50 µM) | |||||
| Downregulated | Upregulated | ||||
| Gene | p-value | Fold change | Gene | p-value | Fold change |
| Fosl1 | 1.19 × 10−27 | −22.36 | Greb1 | 4.85 × 10−12 | 3.54 |
| Ereg | 2.19 × 10−31 | −17.81 | Thra | 3.82 × 10−6 | 3.60 |
| Mx1 | 3.19 × 10−8 | −15.73 | Nbl1 | 3.52 × 10−4 | 3.63 |
| Il1rl1 | 1.39 × 10−53 | −15.63 | Ulk1 | 1.74 × 10−6 | 3.75 |
| Tgtp2 | 8.87 × 10−17 | −14.41 | Mreg | 3.26 × 10−6 | 3.76 |
| Gbp3 | 3.49 × 10−49 | −13.68 | Scd2 | 1.59 × 10−43 | 3.88 |
| Ccl20 | 7.76 × 10−5 | −12.90 | Dzip1 | 1.14 × 10−6 | 4.05 |
| Dusp5 | 1.04 × 10−7 | −12.49 | Acacb | 6.33 × 10−4 | 4.18 |
| Hbegf | 3.01 × 10−34 | −12.06 | B4gat1 | 1.30 × 10−3 | 4.64 |
| Ifit3 | 6.59 × 10−75 | −11.33 | Acat2 | 1.39 × 10−7 | 5.23 |
| PND8 rat Sertoli APAP + GEN (50 µM) | |||||
| Downregulated | Upregulated | ||||
| Gene | p-value | Fold change | Gene | p-value | Fold change |
| H2ac10 | 3.82 × 10−39 | −8.28 | Pde4d | 2.74 × 10−12 | 2.49 |
| Pimreg | 9.09 × 10−4 | −6.10 | Nqo1 | 9.70 × 10−23 | 2.54 |
| E2f8 | 9.53 × 10−8 | −5.40 | Csf2rb | 1.12 × 10−3 | 2.56 |
| Uhrf1 | 6.39 × 10−26 | −5.24 | Il6 | 8.36 × 10−4 | 2.73 |
| Cdca3 | 1.40 × 10−8 | −5.23 | Eda2r | 5.03 × 10−26 | 2.82 |
| Pbk | 2.67 × 10−4 | −4.45 | Cd80 | 3.24 × 10−9 | 3.40 |
| Hist1h2bg | 5.98 × 10−7 | −4.40 | Cdkn1a | 8.58 × 10−94 | 3.40 |
| Ckap2l | 5.00 × 10−10 | −4.20 | Abcb1b | 3.69 × 10−113 | 3.94 |
| Hist1h2bc | 2.33 × 10−4 | −4.17 | Star | 3.91 × 10−179 | 4.21 |
| Cdca5 | 5.13 × 10−6 | −4.14 | Gdf15 | 8.82 × 10−15 | 5.88 |
| TM4 Cells | PND8 Rat Sertoli Cells | |||||
|---|---|---|---|---|---|---|
| Pathway Description | Enrichment Score | p-Value | Genes | Enrichment Score | p-Value | Genes |
| Necroptosis | 18.7 | 7.39 × 10−9 | 22 | 3.4 | 3.50 × 10−2 | 4 |
| Viral carcinogenesis | 10.1 | 4.01 × 10−5 | 20 | 9.9 | 5.17 × 10−5 | 9 |
| Transcriptional misregulation in cancer | 13.5 | 1.36 × 10−6 | 21 | 7.0 | 9.35 × 10−4 | 7 |
| MicroRNAs in cancer | 6.8 | 1.10 × 10−3 | 14 | 4.0 | 1.80 × 10−2 | 5 |
| p53 signaling pathway | 5.9 | 2.61 × 10−3 | 8 | 12.5 | 3.73 × 10−6 | 7 |
| Cellular senescence | 6.0 | 2.47 × 10−3 | 14 | 10.8 | 1.95 × 10−5 | 9 |
| TNF signaling pathway | 4.5 | 1.16 × 10−2 | 9 | 3.9 | 2.10 × 10−2 | 4 |
| Viral protein interaction with cytokine and cytokine receptor | 5.2 | 5.72 × 10−3 | 8 | 3.1 | 4.32 × 10−2 | 3 |
| TM4 Cells | |||
|---|---|---|---|
| Pathway Description | Enrichment Score | p-Value | Genes |
| Ribosome biogenesis in eukaryotes | 15.8 | 1.40 × 10−7 | 15 |
| NOD-like receptor signaling pathway | 8.2 | 0.000270 | 16 |
| AMPK signaling pathway | 4.6 | 0.0100 | 10 |
| NF-kappa B signaling pathway | 4.0 | 0.0185 | 8 |
| IL-17 signaling pathway | 3.6 | 0.0280 | 7 |
| Steroid biosynthesis | 3.5 | 0.0291 | 3 |
| TGF-beta signaling pathway | 3.4 | 0.0343 | 7 |
| Cytosolic DNA-sensing pathway | 3.0 | 0.0500 | 5 |
| PND8 Rat Sertoli Cells | |||
|---|---|---|---|
| Pathway Description | Enrichment Score | p-Value | Genes |
| Foxo signaling pathway | 8.6 | 0.000180 | 7 |
| Melanoma | 3.7 | 0.0252 | 3 |
| Glioma | 3.6 | 0.0283 | 3 |
| PI3K-Akt signaling pathway | 3.5 | 0.0298 | 7 |
| Pancreatic cancer | 3.5 | 0.0316 | 3 |
| Rheumatoid arthritis | 3.5 | 0.0316 | 3 |
| Hematopoietic cell lineage | 3.4 | 0.0351 | 3 |
| JAK-STAT signaling pathway | 3.3 | 0.0368 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corpuz-Hilsabeck, M.; Mohajer, N.; Culty, M. Dysregulation of Immature Sertoli Cell Functions by Exposure to Acetaminophen and Genistein in Rodent Cell Models. Cells 2023, 12, 1804. https://doi.org/10.3390/cells12131804
Corpuz-Hilsabeck M, Mohajer N, Culty M. Dysregulation of Immature Sertoli Cell Functions by Exposure to Acetaminophen and Genistein in Rodent Cell Models. Cells. 2023; 12(13):1804. https://doi.org/10.3390/cells12131804
Chicago/Turabian StyleCorpuz-Hilsabeck, Maia, Nicole Mohajer, and Martine Culty. 2023. "Dysregulation of Immature Sertoli Cell Functions by Exposure to Acetaminophen and Genistein in Rodent Cell Models" Cells 12, no. 13: 1804. https://doi.org/10.3390/cells12131804





